Abstract
The species Xanthomonas oryzae is comprised of two designated pathovars, both of which cause economically significant diseases of rice in Asia and Africa. Although X. oryzae is not considered endemic in the United States, an X. oryzae-like bacterium was isolated from U.S. rice and southern cutgrass in the late 1980s. The U.S. strains were weakly pathogenic and genetically distinct from characterized X. oryzae pathovars. In the current study, a draft genome sequence from two U.S. Xanthomonas strains revealed that the U.S. strains form a novel clade within the X. oryzae species, distinct from all strains known to cause significant yield loss. Comparative genome analysis revealed several putative gene clusters specific to the U.S. strains and supported previous reports that the U.S. strains lack transcriptional activator-like (TAL) effectors. In addition to phylogenetic and comparative analyses, the genome sequence was used for designing robust U.S. strain-specific primers, demonstrating the usefulness of a draft genome sequence in the rapid development of diagnostic tools.
INTRODUCTION
The species Xanthomonas oryzae is comprised of pathovars oryzae and oryzicola, the causative agents of bacterial leaf blight (BLB) and bacterial leaf streak (BLS) on rice, respectively (27). Although the two pathovars are closely related, BLB is a vascular disease characterized by marginal leaf lesions, while BLS affects parenchyma cells and results in leaf streaking. Both pathovars can cause substantial losses to rice production (27). X. oryzae has been designated a USDA select agent in the United States, and movement is restricted by several international quarantines (25, 29).
The finished genomes of three Asian X. oryzae pv. oryzae strains and one X. oryzae pv. oryzicola strain are available, facilitating in-depth comparative genomic analyses (22, 28, 35) (GenBank accession no. AAQN01000001). Whole-genome alignment revealed that the sequenced X. oryzae pv. oryzae strains, MAFF 311018, KACC 10331, and PXO99A, are very closely related (24, 35). X. oryzae pv. oryzicola clusters with the X. oryzae group, forming a branch distinct from X. oryzae pv. oryzae strains (24, 35). All the X. oryzae genomes are characterized by large numbers of insertion sequence (IS) elements, the major contributors to sequence diversity within the species (28, 35), and by various numbers of secreted transcriptional activator-like (TAL) effectors required for full virulence (35, 42). The African X. oryzae pv. oryzae strains are different from Asian strains and more closely related to Asian X. oryzae pv. oryzicola. A specific and intriguing feature of African X. oryzae pv. oryzae strains is that the genome contains a smaller number of TAL effector and IS elements than the Asian strains (8). Bacterial blight has also been observed in South America (12, 23); strains isolated in Colombia are closely related to Asian strains (15, 17).
Although X. oryzae is not historically considered indigenous to the United States (27), strains of a yellow bacterium causing mild BLB-like symptoms were collected from rice fields in Texas and Louisiana in 1987 (17). The bacterium was classified as X. oryzae based on serological tests and fatty acid profiling (17). However, the U.S. strains were weakly virulent on rice (17), and restriction enzyme length polymorphism (RFLP) fingerprint profiles suggested that they were genetically distinct from X. oryzae pv. oryzae and oryzicola (34). Most of the U.S. strains were shown to carry one or more plasmids (41). These findings led the authors to speculate that U.S. Xanthomonas strains were highly divergent from those found in Asia and had not entered the United States in the recent past. Although the X. oryzae-like strains were collected in subsequent years in U.S. fields of rice and the native southern cutgrass (Leersia hexandra) (9), there were no reports of any significant yield loss in the field, and there have been no subsequent reports of X. oryzae-related problems in the United States. However, due to the numerous global regulatory measures aimed at preventing the spread of bacterial pathogens, improved characterization of any U.S. strains of X. oryzae will be critical for development of diagnostic tools and crop protection.
Recent technological advances have substantially increased the speed and cost-efficiency of genome sequencing. However, the final steps of completion, such as gap closure and analysis of repetitive elements, remain time-consuming and expensive. As more bacterial species are sequenced, a draft sequence of genomes closely related to a finished reference genome will serve as a valuable resource for comparative analysis. Draft genome sequences have been used for rapid development of diagnostic tools, comparison of gene repertoires, and identification of putative virulence factors (2, 4, 11).
In this work, we characterized the phylogeny and gene content of two U.S. Xanthomonas strains by analyzing high-quality draft genome sequence data. Comparative genomics and multilocus sequence typing revealed that the U.S. Xanthomonas strains, while part of the X. oryzae species, form a group substantially divergent from a clade formed by X. oryzae pv. oryzae, X. oryzae pv. oryzicola, and Xanthomonas campestris pv. leersiae. Genome-based diagnostic primers were developed and tested to identify this novel clade.
MATERIALS AND METHODS
Bacterial strains, genome sequencing, and genome assembly methods.
Table 1 lists bacterial strains used for whole-genome and single-gene sequencing and for primer testing. DNA was extracted using the genomic DNA extraction kit (Qiagen, Valencia, CA). The DNA was sheared, a library was constructed (Illumina, San Diego, CA), and the DNA was end sequenced (76-bp paired-end reads) using an Illumina Genome Analyzer II (Illumina, San Diego, CA) at the Michigan State University Research Technology Support Facility. The paired-end reads were trimmed to 40 bp to remove low-quality regions at the 3′ end of the reads. A custom PerlScript was used to clean and remove reads with low-complexity and low-quality regions. Low-quality regions were defined as an average quality score of ≤20 over a 10-bp window along the read or ≥2 N bases in the read. The low-complexity threshold was defined as >85% of the base composition composed of a single nucleotide. In cases where one read passed the cleaning process and the paired end failed, the good read was retained in a single read file for use in the assembly.
Table 1.
Strains used in this study
| Species | Strain(s)a | Country |
|---|---|---|
| X. oryzae, unknown pathovar | X8-1A, X11-5A, Ru87-7, X4-2C, X7-2D, X212-3-1, X37-2, X1-8, X44-D, X211-2, X45-A1, X13-5C, X54-A1, X7-5A, X211-1 (C. Gonzalez), X207-A1 (C. Gonzalez) | United States |
| X. oryzae pv. oryzae | PXO99A, PXO86 | Philippines |
| Xoo475-304 | Japan | |
| Xoo94 | China | |
| Xoo228 | Korea | |
| CFBP 1947, CFBP 1948 | Cameroon | |
| MAI1, MAI2 | Mali | |
| NAI1, NAI8 | Niger | |
| BAI3, BAI4 | Burkina Faso | |
| CIAT 1185 | Colombia | |
| XOO4 | Thailand | |
| IXO51 | India | |
| MXO49 | Malaysia | |
| X. oryzae pv. oryzicola | BLS256, BLS177, BLS333 | Philippines |
| MAI8, MAI10 | Mali | |
| X. campestris pv. leersiae | NCPPB 4346 (N. Parkinson) | China |
| Pseudomonas syringae pv. syringae | ICPB PS296 (L. E. Claflin) | |
| Ralstonia solanacearum | K60 (B. A. Hetrick) | |
| X. campestris pv. campestris | Xcc X1g10 (L. E. Claflin) | |
| X. axonopodis pv. vesicatoria | 85-10 (C. Gonzalez) | United States |
| Xanthomonas translucens pv. hordei | 2181 (L. E. Claflin) | United States |
| X. vasicola |
Strains are from the collections of J. E. Leach and V. Verdier unless otherwise noted.
The Velvet short read assembler (version 0.7.53; PubMed identifier 18349386) (43) was used to assemble the genomes in conjunction with the VelvetOptimiser.pl PerlScript provided with Velvet. VelvetOptimiser.pl automates the searching of hash length, expected coverage, and coverage cutoff parameter space to generate an optimal assembly. The program was provided the cleaned paired-end reads, single reads from the cleaning process described above, an estimated insert size of 300 bp, and a minimum contig length of 200 bp. The Velvet parameters used in the final assembly for X11-5A were a hash length of 31, expected coverage of 19.58-fold, and a k-mer coverage cutoff of 9.79-fold. The hash length for X8-1A was 31, the expected coverage was 18.02-fold, and the k-mer coverage cutoff was 9.01-fold. Sanger sequencing of individual X. oryzae genes was performed at the Proteomics and Metabolomics Facility at Colorado State University.
Contigs from the Velvet assembly were assembled into a pseudomolecule, or draft chromosome sequence, using the genome of PXO99A as a reference sequence; contigs in each strain not mapping to PXO99A were concatenated into a second pseudomolecule named “assembly 2.” Open reading frames (ORFs) were predicted and annotated using the Institute for Genome Sciences annotation engine (http://ae.igs.umaryland.edu/cgi/index.cgi).
Pathogenicity assays.
Seedlings of the rice Oryza sativa subsp. japonica cv. Lemont and the O. sativa subsp. indica cv. IR64 were grown for 21 days under controlled conditions (28°C, 80% humidity) in a growth chamber. Strains PXO99A, X11-5A, and X8-1A were grown for 48 h on plates of nutrient agar (BD, Franklin Lakes, NJ) at 28°C and suspended to 109 CFU/ml in water. Plants were inoculated using the clip inoculation method (18). Lesion length and length of leaf curling were measured at several time points over 10 to 16 days postinoculation (dpi). PXO99A caused rapid leaf curling from clipped ends of leaves well in advance of lesion development, whereas X11-5A and X8-1A caused distinct lesions and very little leaf curling. Therefore, the extent of leaf curling caused by PX099A was compared to lesion length caused by the two U.S. strains. Additionally four accessions of Oryza glaberrima (TOG5497, TOG5672, TOG6202, TOG6308) and four cultivars of O. sativa (O. sativa subsp. japonica cv. Azucena, Nipponbare, and Curinga and O. sativa subsp. indica cv. TN1) were grown for 45 days in a greenhouse and clip-inoculated with strains X8-1A and X11-5A and X. oryzae pv. oryzae strain BAI3 (Burkina Faso) as a control. Lesion length (mm) was measured 16 and 21 dpi on two leaves on each of five replicate plants.
Multilocus sequence typing (MLST) and phylogenetic analyses.
The ORFs of the genes gyrB, fusA, dnaK, glnA, groEL, atpD, gapA, recA, and efp were identified in genomic sequences of X11-5A, X8-1A, MAI-1, NAI-8, and other Xanthomonas strains (GenBank accession numbers: X. oryzae pv. oryzicola, AAQN01000001; X. oryzae pv. oryzae, CP000967, AE013598, and AP008229; Xanthomonas vasicola, ACHT00000000; and X. campestris pv. campestris, CP000050). For MLST of unsequenced strains, fusA, gyrB, and gapA were amplified from selected X. oryzae strains using published primer sequences (1), and PCR products were sequenced. Other Xanthomonas fusA, gyrB, and gapA gene sequences were downloaded at the Plant Associated and Environmental Microbes Database (1). Sequences were aligned by ClustalW (21) with the Mega4 program (39). Phylogenetic analyses were performed using the MrBayes program for Bayesian analysis (16), using the general time-reversible model with inverse-gamma rates of evolution for 100,000 generations. Phylogenetic trees were drawn and formatted in Mega4.
Comparative genome analyses.
Average nucleotide identity (ANI) values were calculated using the SpeciesJ program with the default settings for BLAST-based analysis (33). Briefly, the program divides a query genome into 1,020-nucleotide fragments and then calculates the average percentage of identical nucleotides shared with the corresponding fragments in a reference genome. Unique fragments with no homology to the reference genome are not included in the analysis. Strains with ANI values of 95 to 96 or greater are typically considered to be within the same species (10, 33). PXO99A and BLS256 genomes were used as the query against the target X. oryzae genomes of strains KACC, MAFF, PXO99A, and BLS256 and the rough assemblies of U.S. strains X11-5A and X8-1A.
The number of common ORFs shared between species was determined using BLASTN (3) searches of predicted ORFs in the genomes of X. oryzae strains KACC, MAFF, PXO99A, and BLS256 and X. campestris strain 8004 against the draft assemblies of U.S. strains X11-5A and X8-1A. The criterion of 70% identity over 150 or more nucleotides was used to classify ORFs as present or absent from strains. Vmatch (www.vmatch.de) was used to search unassembled short reads of U.S. and African strains for evidence of TAL effector sequences.
Primer design and testing.
The program ePrimer3 from the EMBOSS package (32) was used to design primers within U.S. strain-specific ORFs or at the junction of strain-specific regions with conserved regions. The PrimerSearch program (32) was used to confirm the specificity of the primers (cutoff of 20% mismatch) against the sequenced X. oryzae genomes named above, as well as the genome sequences of Xanthomonas albilineans, Xanthomonas axonopodis pv. citri, X. axonopodis pv. vesicatoria, X. campestris pv. campestris, X. vasicola, and Xylella fastidiosa 9a5c (GenBank accession numbers FP565176, AE008923, AM039952, CP000050, ACHT00000000, and AE003849, respectively). Fifteen primer pairs with different predicted locations and product sizes were chosen for testing. Each 25-μl PCR mixture contained 1.5 mM MgCl2, 0.2 μM forward and reverse primers, 1× PCR buffer (Invitrogen, Carlsbad, CA), 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.5 units Taq polymerase (Invitrogen, Carlsbad, CA), and 10 ng template DNA. After an initial denaturing step, PCR was conducted for 28 cycles of 30 s at 94°C, 30 s at 55°C, and 70 s at 72°C. Primers were tested against DNA from all strains listed in Table 2.
Table 2.
General characteristics of draft genomes of U.S. X. oryzae strains and completed X. oryzae genomes
| Strain | Pathovar | Country of origin | Total sequence length (bp) | No. of predicted genes | % GC | % ANI with: |
Source or reference | |
|---|---|---|---|---|---|---|---|---|
| PXO99A | BLS256 | |||||||
| X11-5A | Undesignated | United States (Texas) | 4,641,765a | 4,655 | 64 | 97 | 97.11 | This study |
| X8-1A | Undesignated | United States (Louisiana) | 4,679,331a | 4,886 | 64 | 97.04 | 97.06 | This study |
| PXO99A | X. oryzae pv. oryzae | Philippines | 5,240,075b | 5,083 | 63.6 | 100 | 97.8 | 35 |
| KACC | X. oryzae pv. oryzae | Korea | 4,941,439b | 4,637 | 63.7 | 99.49 | 97.8 | 22 |
| BLS256 | X. oryzae pv. oryzicola | Philippines | 4,831,739b | 4,614 | 64 | 97.76 | 100 | GenBank accession no. AAQN00000000 |
Metrics for genome reported in Table S1 in the supplemental material.
Single contig representing the circular chromosome.
Nucleotide sequence accession numbers.
This whole genome shotgun project was deposited at GenBank under accession numbers AFHK00000000 (X11-5A) and AFHL00000000 (X8-1A). Assemblies and annotation data can be downloaded from the Comprehensive Phytopathogen Genomics Resource at http://cpgr.plantbiology.msu.edu/us_xo_anno_download.shtml. Illumina reads were deposited in the NCBI Sequence Read Archive under accession number SRP00669. Novel housekeeping gene sequences used in this study have been deposited in GenBank under accession numbers JF830787 to JF830795.
RESULTS
Pathogenicity testing and selection of U.S. X. oryzae strains for sequence characterization.
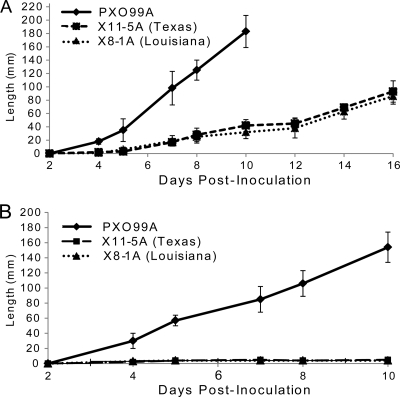
U.S. strains of Xanthomonas were previously identified as X. oryzae and divided into groups based on biocin production and plasmid profiling (41). To determine if disease development varied between individual U.S. X. oryzae strains and to identify virulent strains for sequencing, 16 U.S. strains from various groups were tested for virulence on Oryza sativa subsp. japonica cv. Lemont. Lesion length was monitored over 16 days. Inoculation with U.S. X. oryzae strains resulted in mean lesion lengths of between 2 and 10 cm for each individual strain (data not shown). The two strains causing the largest lesions in the initial screen were X11-5A and X8-1A, isolated from fields in Texas and in Louisiana, respectively (17). Based on this observation, X11-5A and X8-1A were selected for genome sequencing. Additional clip experiments confirmed that X11-5A and X8-1A caused lesions on O. sativa subsp. japonica cv. Lemont, although the lesions were smaller and slower to form than those caused by the highly virulent Asian X. oryzae pv. oryzae strain PXO99A (Fig. 1A). O. sativa subsp. indica cv. IR64 is a widely grown cultivar known to be susceptible to most Asian strains of X. oryzae. No lesions or other symptoms were observed on IR64 inoculated with U.S. X. oryzae strains (Fig. 1B). In contrast, X. oryzae pv. oryzae PXO99A caused long lesions on IR64.
Fig. 1.
Symptom development on leaves of the rice cultivars Oryza sativa subsp. japonica cv. Lemont (A) and O. sativa subsp. indica IR64 (B) following inoculation with X. oryzae strains PXO99A, X11-5A, and X8-1A. The length of leaf curling caused by PXO99A was compared to lesion lengths in X115-A and X8-1A. Curling caused by PXO99A extended the length of the leaf after 10 days on O. sativa subsp. japonica cv. Lemont. Error bars represent the means of six replicate plants ± standard deviations (SD). The experiment was repeated once with similar results.
Strains X8-1A and X11-5A did not cause lesions on four additional accessions of O. sativa or four accessions of O. glaberrima tested. Strain BAI3 caused lesions averaging 205 ± 14 mm in length on the O. glaberrima and O. sativa accessions, except on accession TOG6202 (142 ± 3 mm).
Genome sequence of U.S. Xanthomonas strains.
The U.S. Xanthomonas strains were sequenced to approximately 70-fold sequence coverage and assembled (see Table S1 in the supplemental material). Over 90% of the assembled sequence of each genome was covered by contigs greater than 10 kbp in length. Table 2 contains a summary of the characteristics of the X11-5A and X8-1A draft genomes compared with genome sequences of X. oryzae pv. oryzae and X. oryzae pv. oryzicola. ANI values among the U.S. strain assemblies and X. oryzae genomes were greater than 95% (Table 2), the minimum level of identity shared among members of the same species (10, 19). This result places the U.S. strains in the species X. oryzae according to widely adopted standards in prokaryotic taxonomy (33). Gene content was predicted and annotated as described in Materials and Methods.
Phylogenetic analysis of X. oryzae housekeeping genes.
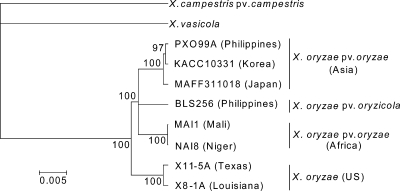
Concatenated housekeeping nucleotide sequences from X11-5A and X8-1A were aligned with corresponding sequences obtained from all published X. oryzae genome sequences (36) and sequences obtained from two draft genomes of X. oryzae strains NAI-8 and MAI-1 from West Africa. Bayesian phylogenetic analysis of the MLST alignments revealed that, although clustering with X. oryzae, the U.S. strains form a branch separate from that of X. oryzae pv. oryzae and oryzicola (Fig. 2). Phylogenetic analysis of sequences of the predicted hrp, gum, and rpf virulence clusters supported this finding (data not shown).
Fig. 2.
Phylogenetic relationships between U.S., Asian, and African Xanthomonas oryzae strains. Bayesian analysis was performed for 100,000 generations using a GTR +1 model of evolution based on a concatenated data set of nine complete housekeeping genes totaling 13,425 bp. Bayesian probabilities are shown next to each branch. X. vasicola was among the closest relatives to the X. oryzae group in previous studies.
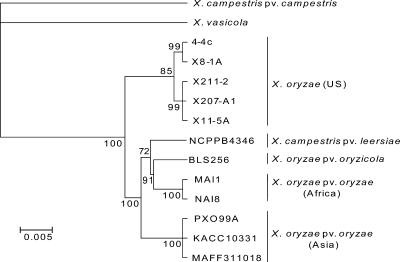
To determine whether the sequenced U.S. strains were closely related to other U.S. strains of Xanthomonas on rice, including those isolated in different years, we conducted a smaller-scale MLST experiment using additional field isolates. Fragments of the housekeeping genes fusA, gapA, and gyrB were amplified from three additional U.S. strains: 4-4C, isolated from Lemont rice in Bazoria County, TX, in 1987 (17), and X207A1 and X211-2, isolated in 1990 from Lemont rice and southern cutgrass, respectively, in the same county (C. Gonzalez, personal communication). X. campestris pv. leersiae strain NCPPB4346, a pathogen of southern cutgrass, was previously shown to group distinctly from X. oryzae (30); this strain was also included in the analysis to investigate possible relatedness to the U.S. strains. Analysis of the alignment showed that the U.S. strains 4-4C, X207A1, and X211-2 are very closely related to X8-1A and X11-5A (Fig. 3). X. campestris pv. leersiae was grouped in the cluster of Asian and African strains. The African strains also grouped with X. oryzicola in this reduced data set, confirming previous findings (8). Although South American strains were not included in the phylogenetic analysis, we sequenced a fragment of gyrB from the Colombian X. oryzae strain CIAT1185 to confirm previous findings that Colombian strains group with the Asian X. oryzae pv. oryzae strains (data not shown) (15, 17). Together, these results demonstrate that the U.S. strains of X. oryzae form a closely related group that is distinct from the economically important X. oryzae pv. oryzae and oryzicola of Asia, Africa, and South America.
Fig. 3.
Phylogenetic tree based on a reduced sequence set, including additional Xanthomonas strains from rice and southern cutgrass. Bayesian analysis was performed based on a concatenated data set of three partial housekeeping genes totaling 1,440 bp.
Predicted gene content of U.S. Xanthomonas strains.
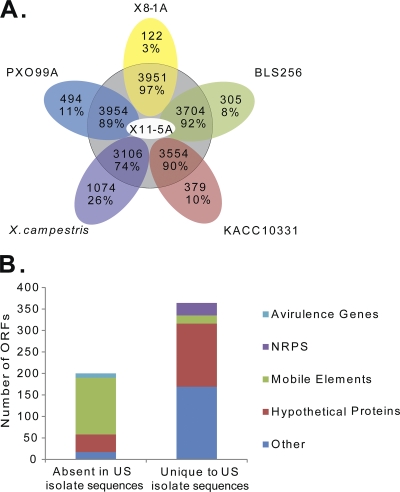
Predicted ORFs from U.S. X. oryzae strains were compared with those of sequenced Xanthomonas genomes using reciprocal BLASTN searches. Over 97% of predicted ORFs in the genome of U.S. strain X11-5A have predicted homologs in the genome of U.S. strain X8-1A (Fig. 4A) and vice versa. In contrast, 92% of predicted ORFs from X. oryzae pv. oryzicola strain BLS256, and 89 to 90% of those predicted in X. oryzae pv. oryzae strains KACC and PXO99A, have predicted homologs in the U.S. X. oryzae genomes (Fig. 4A). These results support the observation that the U.S. X. oryzae strains are more closely related to one another than to X. oryzae strains from Asia and suggest that the U.S. strains are slightly more similar to X. oryzae pv. oryzicola than to X. oryzae pv. oryzae.
Fig. 4.
Comparison of predicted ORFs in the draft sequence of U.S. X. oryzae isolates with those in other previously sequenced Xanthomonas genomes. (A) Numbers and percentages of ORFs in the genomic sequences of X. oryzae strains X8-1A, KACC10331, PXO99A, and BLS256 and X. campestris strain 8004, with and without BLAST hits in the draft genome assembly of strain X11-5A. (B) Functional composition of predicted genes with BLAST hits among all X. oryzae genomes except the U.S. draft sequences (left column) and predicted genes shared among the U.S. draft sequences with no BLAST hits in other X. oryzae strains (right column).
Genes present in Asian X. oryzae strains and absent in U.S. strains.
BLASTN analysis identified 200 ORFs in the genome of KACC with homologs in the genome of X. oryzae pv. oryzae PXO99A and X. oryzae pv. oryzicola BLS256 but not in the draft genomic sequence of the U.S. strains. Most genes absent in the U.S. X. oryzae sequences are copies of insertion sequence (IS) elements or other transposable elements or are hypothetical genes (Fig. 4B). The lack of these transposable elements may partially result from the incomplete nature of the genomic sequence; comparison of annotated genes in the U.S. genomes with those in previously sequenced genomes suggest that transposable elements are likely underrepresented in the U.S. sequenced genomes (data not shown). Both genomes are predicted to encode intact copies of the major virulence clusters, including hrp, gum, rax, and rpf operons.
The U.S. strain genome sequences lack any evidence of genes associated with clustered regularly interspersed palindromic repeats (CRISPRs). CRISPR-associated genes are involved in a virus resistance mechanism (37); these genes are present in many bacterial species, including all three sequenced genomes of X. oryzae pv. oryzae (13, 35), but a search of the publicly available genome sequence of strain BLS256 revealed that they are lacking in X. oryzae pv. oryzicola. X. oryzae pv. oryzae and oryzicola are characterized by the presence of 8 to 26 copies of TAL effector genes, which encode secreted proteins required for full virulence (42). TAL effectors are characterized by extensive central repeat regions that bind to promoter elements on host DNA, resulting in modification of host gene expression (6). No TAL effectors were detected in the genome assemblies of the two U.S. strains. Analysis of the unassembled short reads failed to find any evidence of TAL effectors in either U.S. strain genome, while the same analysis performed on sequences from non-U.S. X. oryzae strains demonstrated high coverage of TAL sequence (data not shown). In addition, two previous reports demonstrated that TAL effector probes do not hybridize to DNA blots from U.S. strains (7, 34). A BLASTN search revealed that the other predicted type III-secreted effectors reported in X. oryzae genomes (36, 40) were nearly all represented in both U.S. X. oryzae sequences, with the exception of those in the XopU and XopO families (see Table S2 in the supplemental material).
U.S. strain-specific genes.
BLAST analysis identified 364 predicted ORFs in the genomic sequence of X11-5A, with close matches in the other U.S. strain sequenced, X8-1A, but no matches with previously sequenced X. oryzae genomes. While most of the genes missing from the U.S. strains had no predicted function, over half of the U.S. strain-specific genes were assigned a predicted function (Fig. 4B). The fragmented assemblies used here cannot be used to determine genome-scale gene arrangement; however, analysis of gene arrangement within individual contigs identified 175 unique predicted genes arranged in 47 clusters of two or more in both U.S. strains. The predicted composition of 9 U.S. strain-specific clusters, containing 70 predicted genes, is described in Table S3 in the supplemental material. Cluster 2 encodes a predicted homolog of the bla gene for ampicillin resistance. Accordingly, we observed that strains X11-5A and X8-1A both grow robustly on nutrient agar amended with 100 μg/ml ampicillin, while growth of Asian strains PXO99 and PXO86 is completely suppressed.
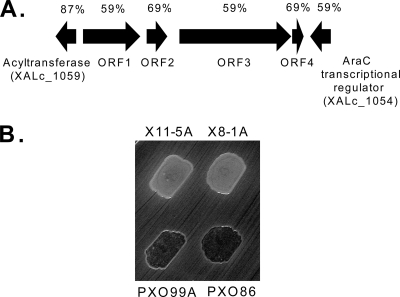
Over 20 predicted ORFs in both U.S. genomes shared significant identity to predicted nonribosomal peptide synthase genes in the genome of X. albilineans. The genome of X. albilineans encodes multiple clusters of nonribosomal peptide synthases (NRPS) thought to function in antibiosis against bacterial competitors and chlorosis in plants (5, 31). The four predicted ORFs comprising one such X. albilineans cluster share 59 to 69% nucleotide identity with predicted NRPS genes in the U.S. X. oryzae strains (Fig. 5A). With the exception of the putative NRPS, the rest of the genome shares no more similarity to X. albilineans than with the X. oryzae pv. oryzae or X. oryzae pv. oryzicola genomes (30).
Fig. 5.
U.S. strains of Xanthomonas harbor sequences similar to X. albilineans NRPS genes and thrive in coculture with Escherichia coli. (A) Map of X. albilineans genes from locus XAL_1054 to XAL_1059, a partially predicted NRPS cluster. Percent nucleotide identity shared with a predicted U.S. X. oryzae ORF is shown below each arrow. (B) Growth of U.S. X. oryzae strains (top) and common Asian strains (bottom) on a lawn of E. coli DH5α.
Due to the fragmented nature of the genome assembly, further study will be needed to determine the structure and genomic location of the NRPS genes. However, high coverage and nucleotide polymorphisms in some of the contigs strongly suggest the presence of multiple copies of NRPS clusters in the genome. Given that NRPS genes are often involved in the production of antibiotic peptides (14, 38), we hypothesized that the U.S. X. oryzae strains may have increased capacity for interspecies competition compared with that of X. oryzae pv. oryzae. When cultures of U.S. and Asian X. oryzae strains were incubated on a lawn of Escherichia coli DH5α, the U.S. strains grew well, but two strains from the Philippines did not (Fig. 4B). There was no difference in the appearance of the strains when incubated on nutrient agar without E. coli (data not shown). These results suggest that U.S. strains of X. oryzae have an increased ability to compete with bacteria in the environment, possibly as a result of antibiosis by NRPS genes present in the genome.
Genomic differences between the U.S. X. oryzae strains.
Comparative analysis between the two X. oryzae assemblies revealed a large contig present in the X8-1A genome which is absent in the genome of X11-5A. The contig was identified as a 31-kb plasmid similar to X. axonopodis pv. vesicatoria plasmid pXAV38, sharing 97% identity with 74% of the pXAV38 genome. Electrophoresis of the X8-1A genomic DNA confirmed the presence of this plasmid. Despite previous reports of a plasmid in strain X11-5A (41), we found no evidence of a plasmid in the genomic sequence of strain X11-5A; it is possible that a plasmid may have been lost during lab culture.
There were 12 additional gene clusters predicted in X8-1A but not X11-5A, and six gene clusters were found in X11-5A but not X8-1A. These clusters, five of which are described in Table S4 in the supplemental material, include ORFs with predicted involvement in adhesion, modulation of active oxygen species, and O-antigen modification. Because we found no difference in virulence between the two strains, it is unlikely that any of these clusters has a significant role in virulence. The genome of strain X11-5A contains a fragment corresponding to residues 130 to 257 of the X. oryzae pv. oryzicola effector protein AvrRxo1, although the putative gene product would lack the probable N-terminal signal necessary for secretion.
Design of primers amplifying U.S. X. oryzae strains.
Previously, we identified primers specific to Asian pathovars of X. oryzae that did not amplify products from U.S. strains (20). Here, 15 primer sets were designed based on ORFs specific to U.S. strain genomes or based on the junction sites of predicted U.S.-specific genomic islands. Thirteen of the primer sets successfully differentiated X8-1A and X11-5A from the previously sequenced X. oryzae strains; seven primers with robust results in preliminary assays were tested against a panel of 16 U.S. X. oryzae strains, 25 strains of X. oryzae pv. oryzae and oryzicola, and six other Xanthomonas, Pseudomonas, and Ralstonia strains listed in Table 2. Five of these primer pairs, reported as USX1 to USX5 in Table 3, consistently amplified a product specific to all U.S. strains.
Table 3.
U.S. Xanthomonas-specific PCR primers
| Primer | Forward sequence | Reverse sequence | Product length (bp) | Start positiona |
|---|---|---|---|---|
| USX1 | GCGCCTGCACAACAATATC | GTACTGCACCACCGTCTGC | 309 | 4127617 |
| USX2 | TCCTCAAAGTTTCCCAGTGC | GGCGTTGGTAAGACGAAGTC | 302 | 1625928 |
| USX3 | ATGCAACACCTGCATTTACG | CGACACAGAAAACAGGCTCA | 306 | 11089 |
| USX4 | TGGTGGCGAGCTTCTACTATG | GTAGGTCGTCCCAGTTCAGC | 327 | 2395182 |
| USX5 | AGTCGCGCTGTTCTCTCAGT | AAGCAACAGCAGACCACCAT | 310 | 1221500 |
Position of forward primer in assembly 1 of the sequence of strain X11-5A.
DISCUSSION
With the technological and fiscal advances in next-generation sequencing technologies, research can readily generate whole-genome sequences for an array of isolates of interest. These draft genome sequences are highly informative for comparative analyses of gene content, phylogenetic analyses, and development of diagnostic tests. Because sequencing and automated assembly can be performed in a short period of time, this strategy could be used to identify novel virulence islands and diagnostic markers of an outbreak pathogen well in advance of the following growing season. In this study, we generated a genome sequence by using next-generation sequencing technologies to characterize two U.S. strains of X. oryzae. Analysis of the draft sequences enabled placement of the U.S. strains within a novel subgroup of X. oryzae strains, identification of many genes missing or uniquely present in the U.S. strains, and development of robust diagnostic primers for identification of the U.S. strains.
Phylogenetic analyses placed the U.S. strains within a group of X. oryzae strains distinct from all known Asian and African strains, demonstrating that the diversity of the X. oryzae species extends far beyond the previously sequenced model organisms. To a lesser extent, X. campestris pv. leersiae strain NCPPB4346 and African strains of X. oryzae pv. oryzae were genetically distinct from Asian strains of X. oryzae pv. oryzae, supporting previous RFLP studies (7) (unpublished data). Further studies are being conducted to characterize the relationship between the African and Asian strains of X. oryzae. Little is known about the L. hexandra pathogen X. campestris pv. leersiae, but it is likely a member of the species X. oryzae. Our phylogenetic analysis did not include strains isolated from South America (23), although gyrB sequencing and diagnostic primer amplification of one Colombian strain supports previous phenotypic and phylogenetic analyses grouping South American strains within the X. oryzae pv. oryzae (8, 12).
The phylogenetic separation of U.S. strains from other groups supports the hypothesis that X. oryzae has persisted in the United States for many years (17). Although bacterial blight is one of the oldest known diseases of rice (26), the existence of distantly related U.S. strains suggests that the divergence of X. oryzae pv. oryzae and oryzicola, and perhaps the entry of TAL effectors into X. oryzae, may have happened in the 300 years since the first importation of rice into the United States (assuming that the rice trade is a historically feasible means of X. oryzae spread). Given the extent of trade and travel between the United States, Asia, and Africa at that time, it is also possible that an X. oryzae progenitor could have entered the United States by way of Africa or traveled by way of other grassy hosts, such as Leersia spp. Several of the diagnostic primers tested in this study, initially assumed to be U.S. strain specific, amplified a product from African X. oryzae strains, suggesting that there may be some genomic similarities between the African and U.S. X. oryzae strains. Although we are currently limited to speculation in this area, the discovery of strains in Asia or Africa sharing U.S. strain-specific features could provide valuable clues regarding the origin and evolution of the X. oryzae species and TAL effectors.
Despite the weak pathogenicity and severely limited range of host cultivars of the U.S. strains compared to those of Asian and African X. oryzae pv. oryzae, we did not identify many ORFs common to the Asian pathovars that are lacking among the U.S. strains; TAL effectors are the only such genes with a known or suspected virulence function. This study, along with two previous reports that the U.S. strains lack TAL effectors (34), is compatible with speculation that the TAL effectors could have entered the X. oryzae genome in a single event (40). We hypothesize that an ancestor of X. oryzae, possibly a weak pathogen or commensal organism of rice, acquired increased survival on rice with the transfer of TAL effectors from another Xanthomonas species. Radiation and diversification of the TAL effectors may have conferred a selective virulence advantage to the descendant X. oryzae pv. oryzae and oryzicola. The U.S. strains of X. oryzae may provide a valuable platform for study of the individual or collective contributions of TAL effectors to virulence.
U.S. X. oryzae genomes are predicted to share several gene clusters highly similar to those in other environmental Xanthomonas spp., including X. campestris, X. axonopodis pv. vesicatoria, and X. albilineans. These findings underscore the extensive genetic variation and genomic plasticity among X. oryzae species and suggest that U.S. X. oryzae has shared a niche with other Xanthomonas spp. common in the United States. The U.S. strain-specific genes have suspected involvement in environmental survival and interspecies competition, including predicted genes for oxidative stress resistance, antibiotic resistance, and antibiosis. These gene clusters could equip these strains for survival in the U.S. environment, which is drier and cooler than that encountered in the rice fields of Southeast Asia or West Africa.
This work demonstrates that U.S. strains of X. oryzae form a closely related group genetically and phenotypically distinct from both defined X. oryzae pathovars, as well as from X. campestris pv. leersiae. Given that the U.S. X. oryzae strains were isolated from both rice and asymptomatic native grasses in several seasons, it is very plausible that this group of X. oryzae still colonizes rice or Leersia fields in the southern United States. The clear distinctions between U.S. strains of X. oryzae and X. oryzae pv. oryzae should be taken into consideration in regulatory policy discussions related to bacterial leaf blight of rice. The U.S. strain-specific diagnostic primers developed here will serve as a valuable resource for regulatory programs to distinguish indigenous X. oryzae in U.S. rice fields from closely related X. oryzae of pathogenic importance. We are currently investigating whether these differences warrant the designation of a novel taxon within the X. oryzae species.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by grants USDA-CSREES-2006-55605-16645 and -2006-55605-04558 to C. R. Buell, N. A. Tisserat, and J. E. Leach and the CSU Infectious Diseases Supercluster. V. Verdier (IRD, CSU) is currently supported by a Marie Curie IOF Fellowship (EU grant PIOF-GA-2009-235457).
We thank C. Gonzales, R. Koebnik, and A. Bogdanove for thoughtful discussions and materials.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Almeida N. F., et al. 2010. PAMDB, a multilocus sequence typing and analysis database and website for plant-associated microbes. Phytopathology 100:208–215 [DOI] [PubMed] [Google Scholar]
- 2. Almeida N. F., et al. 2009. A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 22:52–62 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhattacharyya A., et al. 2002. Draft sequencing and comparative genomics of Xylella fastidiosa strains reveal novel biological insights. Genome Res. 12:1556–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birch R. G. 2001. Xanthomonas albilineans and the antipathogenesis approach to disease control. Mol. Plant Pathol. 2:1–11 [DOI] [PubMed] [Google Scholar]
- 6. Boch J., et al. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512 [DOI] [PubMed] [Google Scholar]
- 7. Choi S. H. 1993. Restriction-modification systems and genetic variability of Xanthomonas oryzae pv. oryzae. Ph.D. dissertation. Kansas State University, Manhattan, KS [Google Scholar]
- 8. Gonzalez C., et al. 2007. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol. Plant Microbe Interact. 20:534–546 [DOI] [PubMed] [Google Scholar]
- 9. Gonzalez C. F., Xu G. W., Li H. L., Cosper J. W. 1991. Leersia hexandra, an alternative host for Xanthomonas campestris pv. oryzae in Texas. Plant Dis. 75:159–162 [Google Scholar]
- 10. Goris J., et al. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57:81–91 [DOI] [PubMed] [Google Scholar]
- 11. Greub G., et al. 2009. High throughput sequencing and proteomics to identify immunogenic proteins of a new pathogen: the dirty genome approach. PLoS One 4:e8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guevara Y., Maselli A. 1999. El tizón bacteriano del arroz en Venezuela. Agronomía Trop. 49:505–516 [Google Scholar]
- 13. Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashimi S. M., Wall M. K., Smith A. B., Maxwell A., Birch R. G. 2007. The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob. Agents Chemother. 51:181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu J., Zhang Y., Qian W., He C. 2007. Avirulence gene and insertion element-based RFLP as well as RAPD markers reveal high levels of genomic polymorphism in the rice pathogen Xanthomonas oryzae pv. oryzae. Syst. Appl. Microbiol. 30:587–600 [DOI] [PubMed] [Google Scholar]
- 16. Huelsenbeck J. P., Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 17. Jones R. K., et al. 1989. Identification of low-virulence strains of Xanthomonas campestris pv. oryzae from rice in the United States. Phytopathology 79:984–990 [Google Scholar]
- 18. Kauffman H. E., Reddy A. P. K., Hsieh S. P. Y., Merca S. D. 1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57:537–541 [Google Scholar]
- 19. Konstantinidis K. T., Ramette A., Tiedje J. M. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lang J. M., et al. 2010. Genomics based diagnostic marker development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis. 94:311–319 [DOI] [PubMed] [Google Scholar]
- 21. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 22. Lee B.-M., et al. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lozano J. C. 1977. Identification of bacterial leaf blight in rice, caused by Xanthomonas oryzae, in America [Central America, South America]. Plant Dis. Rep. 61:644–648 [Google Scholar]
- 24. Lu H., et al. 2008. Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS One 3:e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mew T. W., Bridge J., Hibino H., Bonman J. M., Merca S. D. 1988. Rice pathogens of quarantine importance, p. 101–105 In Rice seed health. The International Rice Research Institute, Manila, Philippines [Google Scholar]
- 26. Mizukami T., Wakimoto S. 1969. Epidemiology and control of bacterial leaf blight of rice. Annu. Rev. Phytopathol. 7:51–72 [Google Scholar]
- 27. Nino-Liu D. O., Ronald P. C., Bogdanove A. J. 2006. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7:303. [DOI] [PubMed] [Google Scholar]
- 28. Ochiai H., Inoue Y., Takeya M., Sasaki A., Kaku H. 2005. Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. 39:275–287 [Google Scholar]
- 29. OEPP/EPPO 1980. Data sheets on quarantine organisms no. 2, Xanthomonas campestris pv. oryzae. OEPP/EPPO bulletin 10. European and Mediterranean Plant Protection Organization, Paris, France [Google Scholar]
- 30. Parkinson N., Cowie C., Heeney J., Stead D. 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int. J. Syst. Evol. Microbiol. 59:264–274 [DOI] [PubMed] [Google Scholar]
- 31. Pieretti I., et al. 2009. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rice P., Longden I., Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 33. Richter M., Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryba-White M. 1995. Comparison of Xanthomonas oryzae pv. oryzae strains from Africa, North America, and Asia by restriction fragment length polymorphism analysis. Int. Rice Res. Notes 20:25–26 [Google Scholar]
- 35. Salzberg S., et al. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song C., Yang B. 2010. Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 23:893–902 [DOI] [PubMed] [Google Scholar]
- 37. Sorek R., Kunin V., Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 38. Strieker M., Tanovic A., Marahiel M. A. 2010. Nonribosomal peptide synthetases: structures and dynamics. Curr. Opin. Struct. Biol. 20:234–240 [DOI] [PubMed] [Google Scholar]
- 39. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 40. White F. F., Potnis N., Jones J. B., Koebnik R. 2009. The type III effectors of Xanthomonas. Mol. Plant Pathol. 10:749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu G. W., Gonzalez C. F. 1991. Plasmid, genomic, and bacteriocin diversity in U.S. strains of Xanthomonas campestris pv. oryzae. Phytopathology 81:628–631 [Google Scholar]
- 42. Yang B., White F. F. 2004. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant Microbe Interact. 17:1192–1200 [DOI] [PubMed] [Google Scholar]
- 43. Zerbino D. R., Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.