Abstract
Arctic soils are increasingly susceptible to petroleum hydrocarbon contamination, as exploration and exploitation of the Arctic increase. Bioremediation in these soils is challenging due to logistical constraints and because soil temperatures only rise above 0°C for ∼2 months each year. Nitrogen is often added to contaminated soil in situ to stimulate the existing microbial community, but little is known about how the added nutrients are used by these microorganisms. Microbes vary widely in their ability to metabolize petroleum hydrocarbons, so the question becomes: which hydrocarbon-degrading microorganisms most effectively use this added nitrogen for growth? Using [15N]DNA-based stable isotope probing, we determined which taxonomic groups most readily incorporated nitrogen from the monoammonium phosphate added to contaminated and uncontaminated soil in Canadian Forces Station-Alert, Nunavut, Canada. Fractions from each sample were amplified with bacterial 16S rRNA and alkane monooxygenase B (alkB) gene-specific primers and then sequenced using lage-scale parallel-pyrosequencing. Sequence data was combined with 16S rRNA and alkB gene C quantitative PCR data to measure the presence of various phylogenetic groups in fractions at different buoyant densities. Several families of Proteobacteria and Actinobacteria that are directly involved in petroleum degradation incorporated the added nitrogen in contaminated soils, but it was the DNA of Sphingomonadaceae that was most enriched in 15N. Bacterial growth in uncontaminated soils was not stimulated by nutrient amendment. Our results suggest that nitrogen uptake efficiency differs between bacterial groups in contaminated soils. A better understanding of how groups of hydrocarbon-degraders contribute to the catabolism of petroleum will facilitate the design of more targeted bioremediation treatments.
INTRODUCTION
Soils in the Arctic are increasingly vulnerable to contamination by hydrocarbons as exploration and exploitation of Arctic resources becomes more technically feasible. While hydrocarbon spills are commonly degraded year-round by naturally occurring soil bacteria at lower latitudes, the remediation of High Arctic soils is limited to about 2 months of summer where soil temperatures rise above 0°C. Certain psychrophilic bacteria remain at least somewhat active over parts of the winter, since microbial respiration has been recorded at temperatures as low as −15°C (39), and yet most significant hydrocarbon degradation is known to occur above 0°C (40). As a result, bioremediation treatments that make optimum use of the short summer are highly desirable. In addition, logistical constraints generally limit the treatment of contaminated Arctic soils to what can be easily transported or found on site. An in situ treatment that is commonly applied to hydrocarbon-contaminated polar soils is biostimulation (1), the addition of nutrients to soil in an attempt to promote the growth and activity of nutrient-limited bacteria. Although this generally increases the rates of hydrocarbon breakdown, the degree to which this occurs varies between sites (13, 34, 46). An understanding of how these nutrients are used within the soil microbial community will help in developing treatments that optimize hydrocarbon bioremediation over the Arctic summer.
The nitrogen content of Arctic soils is generally low (28, 37), and nitrogen has been shown to be a major limiting factor in Arctic bioremediation (30), but biostimulation studies have not specifically examined which organisms are active in incorporating added nutrients. Nitrogen-based fertilizers are applied as blanket treatments to contaminated soil, but not all microorganisms have the same capacity for nutrient assimilation and subsequent growth. This may be due to differences in nitrogen uptake mechanisms or to competition from co-occurring species (17, 22). Bacteria also vary in the rate and efficiency with which they degrade alkane substrates (33, 38, 43), so an understanding of which species are most effectively incorporating added nutrients in contaminated Arctic soils will help in determining whether these treatments can be further optimized for more rapid mineralization of hydrocarbons.
Stable isotope probing (SIP) is a popular approach for characterizing microbial activity in situ. While large-scale sequencing has provided an unprecedented view of microbial diversity, other approaches are still required to determine which microorganisms are active under various environmental conditions. Many studies have used DNA-SIP to examine uptake of carbon sources in environmental samples (e.g., 2, 28, 45), and a few have used it to look at nitrogen uptake (6, 7, 41). A recent review (9) recommends the use of large-scale sequencing of SIP fractions to determine the activity of less-abundant species, since traditional SIP approaches using denaturing gradient gel electrophoresis and clone libraries tend to only identify the major substrate consumers (2, 45). While [15N]DNA-based SIP (15N-SIP) is often complicated by a small degree of DNA separation between labeled and unlabeled DNA molecules (0.016 g ml−1 for 100% 15N-labeled DNA compared to 0.036 g ml−1 for 100% 13C-labeled DNA [5]), mass sequencing provides large amounts of sequence data that make enrichment visible even in the presence of high levels of background DNA.
In the present study, we added 14N- and 15N-labeled mono-ammonium phosphate (MAP) to hydrocarbon-contaminated and uncontaminated Arctic soils, separated DNA by weight in CsCl gradients, and amplified 16S rRNA and alkB genes from DNA isolated from several different buoyant density ranges. These amplicons were sequenced, and the 14N- and 15N-amended samples were compared among CsCl fractions with equal densities to determine the degree of 15N enrichment for various phylogenetic groups. While cross-feeding is considered a drawback in studies that look only to identify a primary consumer (32), we were interested in identifying all bacteria that used added nitrogen to facilitate growth during the incubation. We found that a wide diversity of bacteria incorporated nitrogen in the contaminated soils, but the degree of 15N enrichment varied greatly between taxonomic groups, suggesting that some bacteria are benefiting more from the addition of nitrogen than others.
MATERIALS AND METHODS
Site description.
Samples were treated and incubated on site at Canadian Forces Station (CFS)-Alert, on the northern tip of Ellesmere Island, Nunavut, in the Canadian High Arctic. CFS-Alert (82°31′N, 62°17′W) is the northernmost permanent human settlement in the world and is located at an elevation of 30.5 m. The annual precipitation averages 153.8 mm, while the average daily temperature is −18.0°C. The average annual daily maximum is −14.7°C, and the average annual daily minimum is −21.3°C (http://www.climate.weatheroffice.ec.gc.ca/index.html). The contaminated soil that was used in the present study experienced a large JP-8 fuel spill in 1999 from a ruptured pipeline. JP-8 fuel has a maximum allowance of 25% aromatic compounds by volume, while the remainder consists of aliphatic hydrocarbons (https://assist.daps.dla.mil/quicksearch/basic_profile.cfm?ident_number=33505). The soil in the area is naturally unvegetated, contains many colluvial blocks and rubble, and is dominated mainly by shale. Uncontaminated soil was taken from an adjacent area outside of the spill site that was similarly unvegetated.
Collection of samples, in situ incubation, and soil analyses.
A composite soil sample was collected from multiple locations within the delineated fuel spill, and a second sample was collected from the adjacent uncontaminated soil. Each sample was separated into three: one portion was left untreated, and the other two were amended with either 14N-labeled (Tri-County Agromart, Trenton, Ontario, Canada) or 15N-labeled (Icon Services, Inc., Mt. Marion, NY) MAP at a concentration of 250 mg/kg of soil. Our lab previously found that MAP was the most effective nitrogen-based fertilizer for promoting bioremediation at our site (16). Each treatment was separated into three 50-g subsamples, which were placed inside 50 ml Falcon (BD, Franklin Lakes, NJ) tubes. These were incubated in situ with open tops between 14 July and 17 August 2009. The soil temperature was measured throughout this time period by using an iButton temperature probe (Maxim, Sunnyvale, CA). After the incubation, the Falcon tubes were sealed and returned to the lab on ice. The soil water content was determined by drying soil overnight at 100°C, while the total soil carbon was determined by mass loss after combustion at 600°C. Samples incubated in parallel were sent to Maxxam Analytics (Montreal, Quebec, Canada), where soil was analyzed for pH, total N, and F1-F4 hydrocarbons according to the protocol set forth by The Canadian Council of Ministers of the Environment (http://www.maxxam.ca/solutions/sol_env_CCME_Petr_Hydroc_0805.pdf).
DNA extraction and ultracentrifugation in CsCl gradients.
Prior to extraction, soil was prewashed based on the protocol of Fortin et al. (15). Total soil DNA was then extracted from 10-g subsamples from each replicate by using MoBio DNA PowerSoil isolation kits (MoBio Laboratories, Carlsbad, CA). CsCl gradient fractionation, DNA precipitation, and DNA quantitation were carried out as described previously (5, 6). In brief, equal amounts of DNA from each replicate were combined within each treatment (a total of 4 μg for each contaminated soil treatment and 2 μg for each uncontaminated soil treatment), and the volume was adjusted to 0.45 ml with TE buffer (50 mM Tris-HCl, 15 mM EDTA [pH 8.0]) and added to 13-by-51-mm polyallomer Quick-Seal centrifuge tubes (Beckman, Fullerton, CA), along with 4.3 ml of 1.762 g of CsCl ml−1 in gradient buffer (15 mM Tris-HCl, 15 mM KCl, 15 mM EDTA [pH 8.0]). The tubes were spun for 65 h at 46,000 rpm (168,500 × g maximum) in a Vti80 rotor (Beckman), and the resulting gradient was collected in 100-μl fractions from the bottom of each tube. The buoyant density of each fraction was determined by measuring 15-μl volumes from each sample on a Reichert AR200 refractometer (Depew, NY). Buckley et al. (5) used an electrical tape mask to measure 5-μl volumes on this model, and our trials showed that even without a mask, volumes as small as 5 μl produced refractive index values that were equivalent to those produced by 100-μl volumes when samples were placed centrally on the refractometer prism. Each fraction was ethanol precipitated, and DNA-density profiles were calculated for each treatment by using the PicoGreen dsDNA quantitation assay (Invitrogen, Carlsbad, CA).
PCR of selected density ranges and amplicon pyrosequencing.
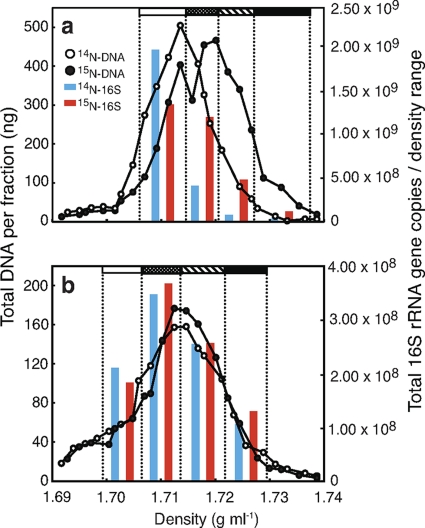
Based on the PicoGreen DNA profiles (Fig. 1), equal density ranges of 0.006 to 0.010 g ml−1 were selected from 14N- and 15N-incubated samples (i.e., samples that had been incubated with 14N and 15N, respectively). As mentioned above, 100% 15N-labeled DNA would have a buoyant density 0.016 g ml−1 more than identical but unlabeled DNA. Each density range was amplified separately with both 16S rRNA and alkB gene primers containing unique multiplex identifier (MID) tags. We used MID-1 through MID-34 from the extended MID set recommended by Roche Diagnostics (35). Each forward primer began at the 5′ end with the Primer A-key (5′-CGTATCGCCTCCCTCGCGCCA-3′), followed by the library key sequence (5′-TCAG-3′), the appropriate MID sequence, and finally the template-specific sequence. Reverse primers were similarly designed, except that the Primer-A key was replaced with the Primer B-key (5′-CTATGCGCCTTGCCAGCCCGC-3′), and no MID sequences were added. The template-specific sequences used in 16S rRNA gene amplification were the forward primer Univ-9F (5′-GAGTTTGATYMTGGCTC-3′) and the reverse primer BR534/18 (5′-ATTACCGCGGCTGCTGGC-3′) (44), while the alkB genes were amplified by using the degenerate primer set of alkB-1f (5′-AAYACNGCNCAYGARCTNGGNCAYAA-3′) and alkB-1r (5′-GCRTGRTGRTCNGARTGNCGYTG-3′) developed by Kloos et al. (20). Reactions were carried out in 50-μl volumes containing 1 μl of template DNA, either 40 pmol of each 16S rRNA gene or 80 pmol of each alkB primer, 8 μl of 1.25 mM deoxynucleoside triphosphates, 1 mM MgCl2, and 2.5 U of Taq DNA polymerase in 5 μl of the 10× Taq DNA polymerase buffer provided (GE Healthcare, Baie d'Urfé, Canada). Cycling conditions for 16S rRNA gene amplifications involved an initial 5 min denaturing step at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, and a final elongation step of 15 min at 72°C. Cycling conditions for the alkB primers were identical, except that the annealing temperature began at 65°C, and was lowered by 1°C in each of the first 10 cycles. PCR products were pooled in an equimolar ratio, except that 10 times less product was added from each alkB gene amplification than was added from each 16S rRNA gene amplification. The pooled sample was then analyzed on a Roche/454 GS FLX sequencer using Titanium chemistry at the DNA Sequencing Facility at the University of Pennsylvania. Only 1/8 of a plate was used, since this was sufficient for the desired sequencing depth.
Fig. 1.
Quantitative profiles of DNA distribution in CsCl SIP gradients, as assessed by PicoGreen, in both hydrocarbon-contaminated (a) and uncontaminated (b) Arctic soils. Density ranges that were amplified for pyrosequencing occur between the dotted vertical lines and are indicated by shaded bars at the top of each graph. 16S rRNA gene copies as determined by qPCR are also shown for each density range.
qPCR.
For each density range, quantitative real-time PCR (qPCR) was performed to determine relative 16S rRNA and alkB gene copy numbers. These values were combined with sequencing results to yield relative numbers for each taxonomic group. All qPCRs were performed in 20-μl volumes using the SYBR green QuantiTect PCR mix (Qiagen, Mississauga, Ontario, Canada) in a Rotor-Gene 3000 apparatus (Corbett Life Science, Sydney, New South Wales, Australia). Bacterial 16S rRNA gene copy numbers were assessed with the Eub338/Eub518 primer set described by Fierer et al. (14), and conditions were as described in Yergeau et al. (47). Real-time qPCR of alkB genes was performed with the primers described above but did not contain the additional adaptor, library, or MID sequences, and cycling conditions were as described above, with the addition of 15 cycles. Standards were made from 10-fold dilutions of linearized plasmids containing the gene fragment of interest that was cloned from amplified pure culture DNA (16S rRNA gene, R2 = 0.992; alkB, R2 = 0.994).
Analysis.
Differences in mean soil carbon and extracted DNA were statistically analyzed by one-way analysis of variance and Tukey's post hoc test. Sequence data were analyzed mainly by using the RDP pyrosequencing pipeline (http://pyro.cme.msu.edu/). The sequences were then deconvoluted and binned according to their MID tags, and the MID and forward primer were trimmed by using the Pipeline Initial Process tool. This resulted in 32 distinct datasets (16 16S rRNA gene datasets and 16 alkB datasets, each with 4 density ranges × 2 isotope labels [14N and 15N] × 2 soil types [contaminated and uncontaminated]). Datasets containing MID sequences associated with the 16S rRNA gene amplifications were individually classified using the RDP Classifier tool with an 80% bootstrap cutoff. Datasets derived from alkB gene amplifications were compared to a BLAST database made from all of the alkB sequences contained in GenBank (613 sequences as of November 2010) using blastall. The best hit from a blastx search was then used to determine class-level phylogeny.
To calculate overall diversity in each of the two soil types, the relative number of copies from each taxonomic group was calculated for each fraction, and the copy numbers were combined from all 14N and 15N fractions within each soil type. To calculate the relative enrichment of each 16S rRNA gene defined taxonomic group within the 15N-treated soils, the proportion of 16S rRNA gene sequences for each group was determined within the defined CsCl density ranges. The proportion of sequences representing a taxonomic group within a given density range was then multiplied by the number of 16S rRNA gene copies within that same range (as defined by 16S rRNA gene qPCR) to give the relative number of copies for each taxonomic group. Relative copy numbers within the 15N-incubated soils were then corrected for slight differences in total DNA and overall phylogenetic composition between the 14N- and 15N-incubated soils. Enrichment of a specific taxonomic group was determined by dividing its relative copy numbers from a 15N density range by its relative copy numbers from the corresponding 14N density range. Since the calculation of enrichment involves the combination of sequence and qPCR data, enrichment can occur even when the percentages of taxonomic groups remain similar (i.e., 20% of 109 sequences is more than 20% of 108 sequences). Each group is compared to itself within a given density range rather than to the other members of the community, so deficits are expected to occur for each enriched group at a lighter density range, rather than a given group within each range. The same calculation was performed with the alkB gene sequences but was instead based on the number of alkB gene copies as determined by qPCR.
Sequence accession numbers.
The sequence data generated in the present study were deposited in GenBank and are accessible through the accession numbers JF358018 to JF409700.
RESULTS
Soil characteristics.
Over the course of the incubation, the mean soil temperature was 7.4°C ± 3.0°C (as measured by the iButton temperature probe), while the mean air temperature was 4.5°C ± 3.6°C, and the total precipitation was 1.1 mm (http://www.climate.weatheroffice.ec.gc.ca/index.html). The addition of MAP was an effective biostimulation treatment at our site and led to an ∼4-fold decrease in the F2 hydrocarbon fraction (C10 to C16) by the end of the incubation (Table 1) . A summary of other soil characteristics can be found in Table 1.
Table 1.
Characteristics from petroleum hydrocarbon-contaminated and uncontaminated CFS-Alert soils collected for in situ incubations
| Treatment | pH | Water content |
Total N (mg/kg) | Mean ± SD |
Total petroleum hydrocarbon content (ppm) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| % Start | % End | % Total Ca | DNA (μg/g of soil)b | F1 (C6 to C10) | F2 (C10 to C16) | F3 (C16 to C34) | F4 (C34 to C50) | |||
| Contaminated | ||||||||||
| No MAP | 7.36 | 18.23 | 4.6 | 5.0 | 5.85 ± 0.53A | 1.12 ± 0.42c | ND | 4,300 | 15 | <10 |
| MAP at 250 mg/kg | 6.85 | 18.23 | 4.7 | 40.6 | 5.56 ± 0.13A | 6.24 ± 0.99A | 27 | 1,000 | 31 | <10 |
| Uncontaminated | ||||||||||
| No MAP | 7.63 | 19.95 | 3.5 | 0.1 | 5.55 ± 0.47A | 4.59 ± 0.55AB | <10 | <10 | <10 | <10 |
| MAP at 250 mg/kg | 7.52 | 19.95 | 4.7 | 38.4 | 5.59 ± 0.79A | 2.85 ± 0.39BC | <10 | <10 | <10 | <10 |
No significant differences were detected according to one-way ANOVA (P = 0.695).
Different superscript capital letters indicate significantly (P < 0.05) different averages according to Tukey's HSD test. One-way analysis of variance was significant (P = 0.0024). The amount of extractable DNA from 10 g (dry equivalent) of soil is indicated.
ND, not determined.
Ultracentrifugation and DNA quantification.
Total DNA recovered in the gradients was 3,407 and 3,861 ng from the 14N- and 15N-incubated contaminated samples, respectively, while 1,615 and 1,493 ng were recovered from the 14N- and 15N-incubated uncontaminated samples, respectively. Quantification of DNA within fractions following centrifugation showed a strong shift in the DNA profile from the 15N-incubated contaminated soil toward the heavier densities within the gradient (Fig. 1a), while there was no obvious shift in the DNA profile in the 15N-incubated uncontaminated soil (Fig. 1b). This is not entirely surprising, since MAP addition led to a significant increase in extractable DNA in the contaminated soil, likely a reflection of newly formed DNA in these samples, but a slight decrease in extractable DNA in the uncontaminated soil (Table 1).
General community composition within MAP-treated soils.
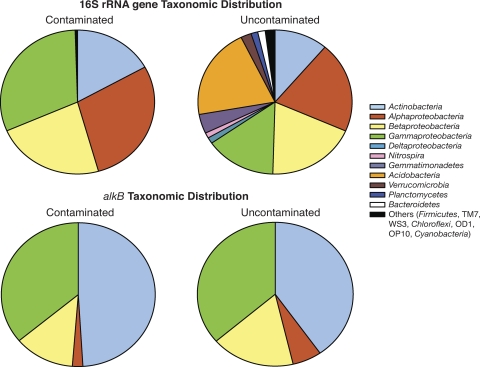
We received a total of 45,984 classifiable 16S rRNA gene sequences and 5,698 classifiable alkB sequences, with an average of 3,577 16S rRNA gene and 394 alkB sequences per contaminated density range, and 2,171 16S rRNA gene and 318 alkB sequences per uncontaminated density range. The average read length was 271 bases. When all density ranges of the 14N- and 15N-treated contaminated soil were combined and adjusted for relative 16S rRNA gene copy numbers, the majority of the sequences retrieved belonged to the Actinobacteria and Proteobacteria, while nine different phyla each made up at least 1% of the sequences from the combined density ranges of the uncontaminated soil (Fig. 2). The alkB sequences were dominated by the Actinobacteria, as well as the Gammaproteobacteria, while the Betaproteobacteria and Alphaproteobacteria dropped in relative abundance compared to the 16S rRNA gene sequence data (Fig. 2). There were 10 bacterial families that each made up at least 1% of the community in the contaminated soil, compared to 9 in the uncontaminated soil. The Sphingomonadaceae dominated the contaminated soil and were also the second-most represented family in the uncontaminated soil, although they represented only ca. 5% of the total bacterial community in the uncontaminated soil as opposed to just >20% in the contaminated soil (Table 2) . The Xanthomonadaceae were the most represented family in the uncontaminated soil and represented a similar percentage of the total community in the contaminated soil (Table 2).
Fig. 2.
Overall phylogenetic diversity of bacteria from the combination of all fractions within the contaminated and uncontaminated soils amended with MAP. The top panel depicts 16S rRNA gene diversity, while the bottom panel depicts the diversity of alkB sequences across all fractions.
Table 2.
Bacterial families representing approximately 1% or more of the overall community in their respective treatments, along with the most commonly identified genera from within these families
| Family | Affiliation | Main genera | % Populationa |
|---|---|---|---|
| Contaminated | |||
| Sphingomonadaceae | Alphaproteobacteria | 20.33 | |
| Sphingobium | 8.81 | ||
| Sphingomonas | 0.28 | ||
| Pseudomonadaceae | Gammaproteobacteria | 17.69 | |
| Pseudomonas | 5.85 | ||
| Azomonas | 2.68 | ||
| Comamonadaceae | Betaproteobacteria | 13.08 | |
| Polaromonas | 3.79 | ||
| Variovorax | 3.05 | ||
| Hydrogenophaga | 0.71 | ||
| Alcaligenaceae | Betaproteobacteria | 7.57 | |
| Pusillimonas | <0.01 | ||
| Achromobacter | <0.01 | ||
| Pigmentiphaga | <0.01 | ||
| Xanthomonadaceae | Gammaproteobacteria | 5.11 | |
| Pseudoxanthomonas | 0.63 | ||
| Thermomonas | 0.39 | ||
| Lysobacter | 0.12 | ||
| Nocardiaceae | Actinobacteria | 4.13 | |
| Rhodococcus | 3.36 | ||
| Caulobacteraceae | Alphaproteobacteria | 3.04 | |
| Brevundimonas | 2.34 | ||
| Phenylobacterium | 0.34 | ||
| Caulobacter | 0.16 | ||
| Microbacteriaceae | Actinobacteria | 2.82 | |
| Salinibacterium | 0.13 | ||
| Agrococcus | 0.03 | ||
| Nocardioidaceae | Actinobacteria | 1.72 | |
| Pimelobacter | 0.78 | ||
| Aeromicrobium | 0.17 | ||
| Nocardioides | 0.12 | ||
| Sinobacteraceae | Gammaproteobacteria | 1.32 | |
| Nevskia | 1.30 | ||
| Steroidobacter | 0.02 | ||
| Uncontaminated | |||
| Xanthomonadaceae | Gammaproteobacteria | 5.46 | |
| Lysobacter | 0.81 | ||
| Thermomonas | 0.52 | ||
| Sphingomonadaceae | Alphaproteobacteria | 5.39 | |
| Sphingomonas | 0.19 | ||
| Sphingobium | 0.18 | ||
| Sinobacteraceae | Gammaproteobacteria | 3.53 | |
| Nevskia | 3.38 | ||
| Steroidobacter | 0.15 | ||
| Gemmatimonadaceae | Gemmatimonadetes | 3.42 | |
| Gemmatimonas | 3.42 | ||
| Comamonadaceae | Betaproteobacteria | 2.45 | |
| Polaromonas | 0.57 | ||
| Variovorax | 0.26 | ||
| Ramlibacter | 0.25 | ||
| Burkholderiales | Betaproteobacteria | 1.87 | |
| Methylibium | 0.66 | ||
| Oxalobacteraceae | Betaproteobacteria | 1.79 | |
| Duganella | 0.42 | ||
| Janthinobacterium | 0.18 | ||
| Massilia | 0.11 | ||
| Planctomycetaceae | Planctomycetes | 1.13 | |
| Pirellula | 0.23 | ||
| Zavarzinella | 0.11 | ||
| Caulobacteraceae | Alphaproteobacteria | 1.04 | |
| Brevundimonas | 0.68 | ||
| Caulobacter | 0.25 | ||
| Phenylobacterium | 0.07 | ||
| Nitrospiraceae | Nitrospira | 0.95 | |
| Nitrospira | 0.95 |
Family percent values are indicated in bold.
Enriched groups within 15N fractions.
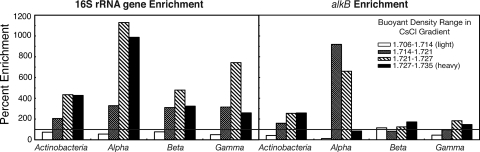
It was expected that organisms that incorporated the 15N label would be enriched in the 15N sample at the higher densities but at a deficit in the lighter density range. The 16S rRNA gene sequence data suggested that most phylogenetic groups were enriched to some degree in the 15N-incubated contaminated sample; however, the degree to which they were enriched varied widely, with the Alphaproteobacteria demonstrating the highest level of 15N enrichment (Fig. 3). Overall, the Alphaproteobacteria made up a much smaller proportion of the alkB gene sequences retrieved (Fig. 2), but they were still identified as the most highly enriched phylogenetic group. They were at a slight 15N deficit in the heaviest density range, but this may simply be due to a small number of sequences migrating to lower densities than were included in our analysis, while the bulk of enrichment occurred in DNA that had a higher natural buoyant density. The alkB gene sequence data also showed the Actinobacteria to be enriched in the heavy 15N fractions, while the Betaproteobacteria and Gammaproteobacteria were not enriched to any great degree (Fig. 3). No clear enrichment was seen in the 15N-treated uncontaminated soil (see the supplemental material).
Fig. 3.
Percent enrichment of major bacterial groups in the 15N-amended contaminated soil compared to the 14N-amended control. The black horizontal line occurs at 100% which indicates no enrichment, while everything above the line is enriched in the 15N treatment, and everything below is at a deficit compared to the 14N control.
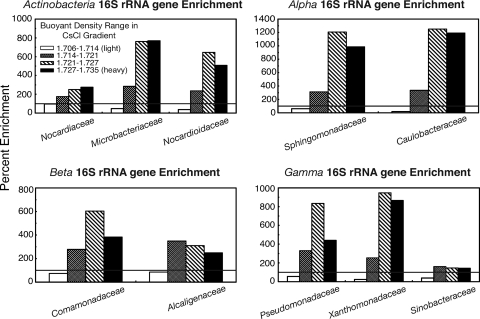
Each of the major phylogenetic groups that were identified in the contaminated soil were dominated by a few major families, and the degree of enrichment for each family representing at least 1% of the total bacterial community is shown in Fig. 4. In addition to being the most highly represented family, the Sphingomonadaceae, along with the Caulobacteraceae, were the most highly 15N-enriched groups in the heavier fractions according to the 16S rRNA gene data (Fig. 4). The Sinobacteraceae were 15N enriched to a much lower extent than the other two major Gammaproteobacteria families (Xanthomonadaceae and Pseudomonadaceae), while the Nocardiaceae were much less efficient at 15N incorporation than the other major families of Actinobacteria (Nocardioidaceae and Microbacteriaceae) (Fig. 4).
Fig. 4.
Percent enrichment of the bacterial families that made up at least 1% of the community in the 15N-amended contaminated soil compared to the 14N-amended control. The black horizontal line occurs at 100% which indicates no enrichment, while everything above the line is enriched in the 15N treatment, and everything below is at a deficit compared to the 14N control.
DISCUSSION
Phylogenetic distribution varied widely between the MAP-treated contaminated and uncontaminated soils, as has been observed previously in other Arctic and alpine environments (3, 21). The vast majority of 16S rRNA gene sequences retrieved from the contaminated soil identified most closely with groups of Proteobacteria and Actinobacteria that are known to be involved in the degradation of hydrocarbons. In addition, the 4-fold reduction of hydrocarbons in the sample was similar to that observed by Yergeau et al. (46) and suggests that the biostimulation treatment was quite effective despite the limited time of the incubation. A much more diverse bacterial community was seen in the MAP-treated uncontaminated soils, where the Acidobacteria appeared as the dominant phylum and Xanthomonadaceae appeared as the dominant family. Both groups have represented large portions of the microbial community in other Arctic soils treated with nitrogen-based fertilizer (8, 31). The proportion of Gemmatimonadetes, Alphaproteobacteria, Actinobacteria, and Verrucomicrobia in the uncontaminated soil was similar to that observed in other Arctic and lower-latitude soils, while Betaproteobacteria and Gammaproteobacteria were more highly represented in our soils (11).
Sequence data from the 14N- and 15N-incubated soils were compared within each density range and expressed as a percentage to demonstrate the relative amount of 15N enrichment between taxonomic groups. Because of the labor-intensive nature of SIP experiments, we chose to combine replicates at the CsCl gradient stage and were therefore unable to perform statistical analyses on the resulting enrichment data. Although extrapolation of the results must be done with caution, the effects that we observed were surprisingly strong. The 16S rRNA gene enrichment data showed that the Actinobacteria, along with the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria, were overrepresented in the heavier fractions of the 15N-treated contaminated soil. Although all of these major groups were enriched in the heaviest 15N density ranges, it was the Alphaproteobacteria that stood out as being enriched 10- to 11-fold relative to controls. Alphaproteobacteria were also confirmed as being the most 15N-enriched group according to the alkB gene data. A higher percentage of enrichment relative to other taxonomic groups indicates increased cell replication due to the addition of fertilizer. The prevalence of Alphaproteobacteria and Gammaproteobacteria was greater in other nitrogen-treated Arctic soils compared to controls (8).
No bacterial groups were clearly enriched in the uncontaminated soils. It may be that bacteria are not capable of assimilating the added fertilizer in the absence of sufficient amounts of carbon to permit growth. A previous study showed that the addition of a carbon source increased extractable DNA from 1.8 to 12.9 μg of DNA g−1 soil, while also largely increasing the incorporation of added 15N, as demonstrated by DNA quantification profiles (7). It may also be that a longer incubation period is required to detect 15N enrichment in the uncontaminated soils, since another 15N-SIP study allowed soils to incubate with 15N substrates for 2 months prior to sampling (7).
The Sphingomonadaceae represented roughly 20% of the bacterial community in the MAP-treated hydrocarbon-contaminated soils and were also among the most heavily 15N-enriched bacterial groups. Members of this family have previously been detected in contaminated polar soils and are generally associated with a high capacity for polyaromatic hydrocarbon (PAH) degradation (1). Sequences from Comamonadaceae represented just >13% of the contaminated soil community, and they have also been identified as degraders of aromatics (24, 29). The dominance of these groups was somewhat surprising, since previous work on hydrocarbon-contaminated CFS-Alert soils showed that untreated soils contained a high ratio of alkane to PAH-degrading genes (46). The limited label incorporation in the alkB gene sequences of both the Betaproteobacteria and Gammaproteobacteria also suggests that alkane degradation was not the dominant form of metabolism in these soils at the time of sampling. One possibility is that the aliphatic hydrocarbons were degraded very quickly during the incubation and that nitrogen was acquired secondarily by PAH-degrading bacteria during community succession, since microbial communities and their associated nutrients have the potential to turnover on the scale of days to months (36). The potential of cross-feeding to cause such high specific SIP labeling has not previously been shown; however, strong secondary label acquisition has been observed after only a week as a result of what was presumed to be micropredation (26). Future 15N-SIP studies using a time course design (as in reference 26) will yield more detailed information on the order of nitrogen incorporation and species succession within biostimulated soils.
It has been suggested that PAH-degrading bacteria in soils do not exhibit much in situ growth (19), but the extremely high level of 15N incorporation by the Sphingomonadaceae suggests otherwise. Strains of Sphingomonas have been shown to effectively degrade PAHs in the presence of both high and low concentrations of nitrogen (23), indicating that they are a diversely adapted group. Sphingmonadaceae also seem to be well adapted to this environment in the absence of contamination, since they were the second-most represented family in the uncontaminated soil. In studies of other contaminated soils at CFS-Alert, it was the pseudomonads that dominated the soil microbial communities for the first year following aeration and the addition of MAP, at which point the sphingomonads replaced them as the most common microorganisms (E. Yergeau et al., unpublished data). The density of pseudomonads is suspected to be a major factor in determining the rate of bioremediation in contaminated Arctic soils (Yergeau et al., unpublished). The Pseudomonadaceae, the second-most represented family in our contaminated soil sequences, were likely responsible for the dominance of Gammaproteobacteria within our alkB sequences and may also have contributed to PAH degradation, sincce they have been shown to possess both aliphatic and aromatic degrading genes in these High Arctic soils (42). The successful growth of Caulobacteraceae in this and other contaminated CFS-Alert soils (Yergeau et al., unpublished) is unexpected, since it has not previously been identified as a hydrocarbon degrader in the Arctic, being more commonly linked with heavy metal resistance (4, 18). However, concentrations of arsenic and nickel are naturally high in the soils surrounding CFS-Alert (unpublished results), perhaps leading the Caulobacteraceae to be uniquely adapted to this region. More detailed studies of these organisms may help to clarify whether they actually do play an important role in Arctic bioremediation.
One of the major advantages of applying SIP to bioremediation studies is that it is relatively easy to maintain in situ conditions. Our samples were left to incubate under on-site temperatures and precipitation and were treated similarly to other biostimulated soils at CFS-Alert. Some major concerns in previous SIP studies were the addition of unrealistic amounts of substrate, cross-feeding, and disturbance of the soil in order to apply the substrate (27, 32). Because we were simulating an in situ remediation treatment, soil disturbance and nutrient addition were considered to be part of the “natural” environment. We were also not concerned with cross-feeding, since we were interested in identifying all active bacteria that accessed and incorporated added nitrogen through any route during the incubation and to then determine which bacteria had most successfully incorporated the nitrogen amendment into their DNA. All previous SIP studies of hydrocarbon-contaminated soils have used 13C-labeled substrates (10, 27), but it was unknown whether the organisms degrading these substrates were the same ones that most successfully incorporated other macronutrients in the soil.
To our knowledge, this is the first study to combine DNA-SIP and large-scale sequencing. It is often difficult to determine the heavy fraction within a CsCl gradient (12), and recent SIP experiments have selected specific small density ranges that the authors define as “heavy” and “light” from which to construct clone libraries (2, 6, 7, 45). Previous 15N studies have used a secondary gradient that allows wider separation of labeled and unlabeled DNA (5), but this can only be easily used to target a few specific fractions of the initial gradient at a time. The benefit of large-scale sequencing is that enrichment can be observed even in the presence of a high background of unlabeled DNA, without prior knowledge of which fractions will contain the labeled organisms. High-throughput analysis of environmental samples is a major hurdle in SIP studies (9), and an unbiased, rapid method of fraction screening could allow automation of future SIP experiments. Although this method certainly benefits 15N-SIP studies due to the lower proportion of nitrogen than carbon in DNA, it could also benefit 13C-SIP studies in which label incorporation is incomplete.
In conclusion, 15N-DNA SIP combined with large-scale sequencing determined the relative incorporation of nitrogen between bacterial groups in a contaminated Arctic soil. Alphaproteobacteria showed the highest degree of nitrogen incorporation, were dominant members of the fertilized contaminated soil, and were therefore the greatest beneficiaries of this biostimulation treatment. It is important that research into new bioremediation treatments proceed with a better understanding of how these treatments are specifically affecting microbial ecology and interactions, a sentiment that has been expressed in recent reviews of advances in environmental biotechnology (25, 27). Further studies on the nitrogen-utilizing and hydrocarbon-mineralizing capacity of specific groups of Arctic bacteria will help in determining whether biostimulation is in fact producing the most efficient bioremediating community.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the NRCan PERD Program.
We thank the staff at CFS-Alert, Drew Craig, Don Kovanen, Diane Labbé, Sylvie Sanschagrin, Claude Masson, Josée Sirois, and Julie Champagne for their technical advice and support with this project.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Aislabie J., Saul D. J., Foght J. M. 2006. Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10:171–179 [DOI] [PubMed] [Google Scholar]
- 2. Antony C. P., et al. 2010. Active methylotrophs in the sediments of Lonar Lake, a saline and alkaline ecosystem formed by meteor impact. ISME J. 4:1470–1480 [DOI] [PubMed] [Google Scholar]
- 3. Atlas R. M., Schofield E. A., Morelli F. A., Cameron R. E. 1976. Effects of petroleum pollutants on Arctic microbial populations. Environ. Pollut. 10:35–43 [Google Scholar]
- 4. Braz V. S., Marques M. V. 2005. Genes involved in cadmium resistance in Caulobacter crescentus. FEMS Microbiol. Lett. 251:289–295 [DOI] [PubMed] [Google Scholar]
- 5. Buckley D. H., Huangyutitham V., Hsu S. F., Nelson T. A. 2007. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl. Environ. Microbiol. 73:3189–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buckley D. H., Huangyutitham V., Hsu S. F., Nelson T. A. 2007. Stable isotope probing with 15N2 reveals novel noncultivated diazotrophs in soil. Appl. Environ. Microbiol. 73:3196–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buckley D. H., Huangyutitham V., Hsu S. F., Nelson T. S. 2008. 15N2-DNA-stable isotope probing of diazotrophic methanotrophs in soil. Soil Biol. Biochem. 40:1272–1283 [Google Scholar]
- 8. Campbell B. J., Polson S. W., Hanson T. E., Mack M. C., Schuur E. A. G. 2010. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ. Microbiol. 12:1842–1854 [DOI] [PubMed] [Google Scholar]
- 9. Chen Y., Murrell J. C. 2010. When metagenomics meets stable isotope probing: progress and perspectives. Trends Microbiol. 18:157–163 [DOI] [PubMed] [Google Scholar]
- 10. Chen Y., Vohra J., Murrell J. C. 2010. Applications of DNA-stable isotope probing in bioremediation studies. Methods Mol. Biol. 599:129–139 [DOI] [PubMed] [Google Scholar]
- 11. Chu H., et al. 2010. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12:2998–3006 [DOI] [PubMed] [Google Scholar]
- 12. Cupples A. M., Shaffer E. A., Chee-Sanford J. C., Sims G. K. 2007. DNA buoyant density shifts during 15N-DNA stable isotope probing. Microbiol. Res. 162:328–334 [DOI] [PubMed] [Google Scholar]
- 13. Delille D., Coulon F. 2008. Comparative mesocosm study of biostimulation efficiency in two different oil-amended sub-Antarctic soils. Microb. Ecol. 56:243–252 [DOI] [PubMed] [Google Scholar]
- 14. Fierer N., Jackson J. A., Vilgalys R., Jackson R. B. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fortin N., Beaumier D., Lee K., Greer C. W. 2004. Soil washing improves the recovery of total community DNA from polluted and high organic content sediments. J. Microbiol. Methods 56:181–191 [DOI] [PubMed] [Google Scholar]
- 16. Greer C. W. 2009. Bioremediation of contaminated sites in the Canadian Arctic: monitoring performance and the effects of biostimulation using molecular methods, p. 321–340 In Bej A. K., Aislabie J., Atlas R. M. (ed.), Polar microbiology: the ecology, diversity, and bioremediation potential of microorganisms in extremely cold environments. CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 17. Hibbing M. E., Fuqua C., Parsek M. R., Peterson S. B. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu P., Brodie E. L., Suzuki Y., McAdams H. H., Andersen G. L. 2005. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 187:8437–8449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnsen A. R., Wick L. Y., Harms H. 2005. Principles of microbial PAH-degradation in soil. Environ. Pollut. 133:71–84 [DOI] [PubMed] [Google Scholar]
- 20. Kloos K., Munch J. C., Schloter M. 2006. A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization. J. Microbiol. Methods 66:486–496 [DOI] [PubMed] [Google Scholar]
- 21. Labbé D., Margesin R., Schinner F., Whyte L. G., Greer C. W. 2007. Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated alpine soils. FEMS Microbiol. Ecol. 59:466–475 [DOI] [PubMed] [Google Scholar]
- 22. Leigh J. A., Dodsworth J. A. 2007. Nitrogen regulation in Bacteria and Archaea. Annu. Rev. Microbiol. 61:349–377 [DOI] [PubMed] [Google Scholar]
- 23. Leys N. M., Bastiaens L., Verstraete W., Springael D. 2005. Influence of the carbon/nitrogen/phosphorus ratio on polycyclic aromatic hydrocarbon degradation by Mycobacterium. Appl. Microbiol. Biotechnol. 66:726–736 [DOI] [PubMed] [Google Scholar]
- 24. Liang Y., et al. 2011. Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. ISME J. 5:403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lovley D. R. 2011. Powering microbes with electricity: direct electron transfer from electrodes to microbes. Environ. Microbiol. Rep. 3:27–35 [DOI] [PubMed] [Google Scholar]
- 26. Lueders T., Kindler R., Miltner A., Friedrich M. W., Kaestner M. 2006. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl. Environ. Microbiol. 72:5342–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madsen E. L. 2006. The use of stable isotope probing techniques in bioreactor and field studies on bioremediation. Curr. Opin. Microbiol. 17:92–97 [DOI] [PubMed] [Google Scholar]
- 28. Martineau C., Whyte L. G., Greer C. W. 2010. Stable isotope probing analysis of the diversity and activity of methanotrophic bacteria in soils from the Canadian High Arctic. Appl. Environ. Microbiol. 76:5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Master E. R., Mohn W. W. 1998. Psychrotolerant bacteria isolated from Arctic soil that degrade polychlorinated biphenyls at low temperatures. Appl. Environ. Microbiol. 64:4823–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohn W. W., Stewart G. R. 2000. Limiting factors for hydrocarbon biodegradation at low temperature in Arctic soils. Soil. Biol. Biochem. 32:1161–1172 [Google Scholar]
- 31. Neufeld J. D., Mohn W. W. 2005. Unexpectedly high bacterial diversity in Arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl. Environ. Microbiol. 71:5710–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neufeld J. D., Wagner M., Murrell J. C. 2007. Who eats what, where and when? Isotope-labeling experiments are coming of age. ISME J. 1:103–110 [DOI] [PubMed] [Google Scholar]
- 33. Obuekwe C. O., Al-Jadi Z. K., Al-Saleh E. S. 2009. Hydrocarbon degradation in relation to cell-surface hydrophobicity among bacterial hydrocarbon degraders from petroleum-contaminated Kuwait desert environment. Int. Biodeter. Biodegr. 63:273–279 [Google Scholar]
- 34. Powell S. M., Ferguson S. H., Snape I., Siciliano S. D. 2006. Fertilization stimulates anaerobic fuel degradation of Antarctic soils by denitrifying microorganisms. Environ. Sci. Technol. 40:2011–2017 [DOI] [PubMed] [Google Scholar]
- 35. Roche Diagnostics 2009. Using multiplex identifier (MID) adaptors for the GS FLX titanium chemistry–extended MID set. Technical Bulletin: Genome Sequencer FLX System. TCB no. 005-2009. Roche, Branchburg, NJ [Google Scholar]
- 36. Schmidt S. K., et al. 2007. Biogeochemical consequences of rapid microbial turnover and seasonal succession in soil. Ecology 88:1379–1385 [DOI] [PubMed] [Google Scholar]
- 37. Shaver G. R., Chapin F. S. 1980. Response to fertilization by various plant-growth forms in an Alaskan tundra: nutrient accumulation and growth. Ecology 61:662–675 [Google Scholar]
- 38. Sorkhoh N. A., Ghannoum M. A., Ibrahim A. S., Stretton R. J., Radwan S. S. 1990. Crude oil and hydrocarbon-degrading strains of Rhodococcus rhodochrous isolated from soil and marine environments in Kuwait. Environ. Pollut. 65:1–17 [DOI] [PubMed] [Google Scholar]
- 39. Steven B., Niederberger T. D., Bottos E. M., Dyen M. R., Whyte L. G. 2007. Development of a sensitive radiorespiration method for detecting microbial activity at subzero temperatures. J. Microbiol. Methods 71:275–280 [DOI] [PubMed] [Google Scholar]
- 40. Walworth J., Braddock J., Woolard C. 2001. Nutrient and temperature interactions in bioremediation of cryic soils. Cold Regions Sci. Technol. 32:85–91 [Google Scholar]
- 41. Wawrik B., Callaghan A. V., Bronk D. A. 2009. Use of inorganic and organic nitrogen by Synechococcus spp. and diatoms on the West Florida Shelf as measured using stable isotope probing. Appl. Environ. Microbiol. 75:6662–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whyte L. G., Bourbonnière L., Greer C. 1997. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl. Environ. Microbiol. 63:3719–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whyte L. G., Bourbonnière L., Greer C. W. 1998. Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl. Environ. Microbiol. 64:2578–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilmotte A., Van der Auwera G., De Wachter R. 1993. Structure of the 16S rRNA of the thermophilic cynobacterium Chlorogloeopsis HTF (′Mastigocladus laminosus HTF') strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96–100 [DOI] [PubMed] [Google Scholar]
- 45. Winderl C., Penning H., von Netzer F., Meckenstock R. U., Lueders T. 2010. DNA-SIP identifies sulfate-reducing Clostridia as important toluene degraders in tar oil-contaminated aquifer sediment. ISME J. 4:1314–1325 [DOI] [PubMed] [Google Scholar]
- 46. Yergeau E., et al. 2009. Microarray and real-time PCR analyses of the responses of High Arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl. Environ. Microbiol. 75:6258–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yergeau E., Hogues H., Whyte L. G., Greer C. W. 2010. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR, and microarray analyses. ISME J. 4:1206–1214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.