Abstract
Resistance to HIV infection in a cohort of commercial sex workers living in Nairobi, Kenya, is linked to mucosal and antiinflammatory factors that may be influenced by the vaginal microbiota. Since bacterial vaginosis (BV), a polymicrobial dysbiosis characterized by low levels of protective Lactobacillus organisms, is an established risk factor for HIV infection, we investigated whether vaginal microbiology was associated with HIV-exposed seronegative (HESN) or HIV-seropositive (HIV+) status in this cohort. A subset of 44 individuals was selected for deep-sequencing analysis based on the chaperonin 60 (cpn60) universal target (UT), including HESN individuals (n = 16), other HIV-seronegative controls (HIV-N, n = 16), and HIV+ individuals (n = 12). Our findings indicate exceptionally high phylogenetic resolution of the cpn60 UT using reads as short as 200 bp, with 54 species in 29 genera detected in this group. Contrary to our initial hypothesis, few differences between HESN and HIV-N women were observed. Several HIV+ women had distinct profiles dominated by Escherichia coli. The deep-sequencing phylogenetic profile of the vaginal microbiota corresponds closely to BV+ and BV− diagnoses by microscopy, elucidating BV at the molecular level. A cluster of samples with intermediate abundance of Lactobacillus and dominant Gardnerella was identified, defining a distinct BV phenotype that may represent a transitional stage between BV+ and BV−. Several alpha- and betaproteobacteria, including the recently described species Variovorax paradoxus, were found to correlate positively with increased Lactobacillus levels that define the BV− (“normal”) phenotype. We conclude that cpn60 UT is ideally suited to next-generation sequencing technologies for further investigation of microbial community dynamics and mucosal immunity underlying HIV resistance in this cohort.
INTRODUCTION
It has been 50 years since the earliest known case of HIV type 1 infection (59), 30 since the first reported case of AIDS (22), and 15 since the first formal report of relative HIV resistance in a cohort of commercial sex workers (CSW) from Nairobi, Kenya, many of whom are never infected with HIV despite years of exposure (19). Although several immunogenetic factors have been identified in HIV-exposed seronegative (HESN) women in this cohort (6, 38), no determinant has been observed in all HESN women and some have seroconverted (31). Therefore, HESN status is currently described as a relative rather than an absolute characteristic, defined by active sex work while remaining HIV seronegative after 3 to 7 years of follow-up.
The mucosal immune environment is likely to play an important role in HIV resistance. Mucosal surfaces of the female genital tract function as robust physical and immunological barriers in the context of episodic sexual intercourse, cyclical hormonal changes, and the complex ecology of resident microbiota. Genital infections induce mucosal inflammation and are strongly associated with HIV acquisition (18), but few studies have examined the role of commensal bacteria in mediating vaginal inflammation and HIV susceptibility (1).
Bacterial vaginosis (BV) is a continuum of physical symptoms related to a polymicrobial shift of the vaginal microbiota from Lactobacillus dominated to mixed anaerobes, including Gardnerella, Prevotella, Atopobium, and other organisms (35, 57). BV is strongly associated with a number of reproductive health problems, such as premature birth (21), pelvic inflammatory disease, and increased vulnerability to most sexually transmitted infections (32), including HIV (5).
Despite the lack of a classic inflammatory response in BV (3), possibly abrogated by antibody- and cytokine-cleaving proteases of BV organisms (13), BV is strongly associated with increased levels of proinflammatory cytokines such as interleukin-1β (IL-1β), IL-6, and IL-8 (40) and decreased mucosal defense factors such as elafin (52). In contrast, vaginal lactobacilli metabolize epithelial glycogen and produce lactic acid and reactive oxygen species such as hydrogen peroxide (H2O2), widely believed to protect the reproductive tract from exogenous pathogens (7, 9, 28). The possibility of antiinflammatory effects of lactobacilli in the vagina has not been explored, although several studies of the gut epithelium indicate induction of epithelial defense molecules, antiinflammatory signaling, and increased T-regulatory cells by specific strains (10, 33, 55).
Increased mucosal factors with possible antimicrobial or antiinflammatory effects, including elafin, have been observed in a subset of HESN women (11, 30), and recent data indicate a reduced baseline immune activation level, or “immune quiescence,” in some HESN women (12). These factors may explain why HIV is not transmitted in this group of highly exposed women, since a critical determinant of infection is increased concentration and activation status of target immune cells infiltrating mucosal surfaces during inflammatory processes (23). Therefore, HESN women may be protected from HIV infection by an unknown combination of innate and mucosal immune responses causing a viral reproductive rate of <1 and extinction of founder viral populations (23).
Although the role of commensal bacteria in suppressing HIV replication in macrophages has recently been investigated (1), no previous studies have addressed whether characteristics of the vaginal microbiota predict HESN status. Using laboratory diagnosis of BV based on microscopic analysis of vaginal swab smears and Nugent scoring (41), we recently conducted a retrospective study of nearly 1,000 CSW in this cohort and determined that HESN women were just as likely as HIV-N women to have BV, while HIV+ individuals were significantly more likely to have BV than HESN and HIV-N women combined (48). Since microscopy can only define bacterial morphotypes and not species or strains, we used culture-based and molecular techniques to address the hypothesis that HESN women have increased levels of specific Lactobacillus organisms and/or decreased levels of specific BV-related organisms compared to those found in HIV-N and HIV+ women.
Phylogenetic analyses were based on the chaperonin 60 (cpn60) universal target (UT), a region of the gene encoding the 60-kDa chaperonin (CPN60, HSP60, or GroEL) in almost all organisms (20). An information-rich, protein-encoding region of 549 to 567 bp, this target has many advantages over the more commonly used 16S rRNA gene for phylogenetic characterization of microbial communities, as we have recently shown (49). Uniform distribution of sequence differences throughout the cpn60 UT allows accurate taxonomic assignment with reads as short as 150 bp (49), and the presence of typically a single copy per genome enhances quantitative analysis (16, 17). A curated online database of more than 12,000 cpn60 sequences is available for sequence comparisons (http://www.cpndb.ca/cpnDB/home.php) (27).
MATERIALS AND METHODS
Participant recruitment and sample collection.
The Majengo CSW cohort has been active since the early 1980s in a market area of central Nairobi. In the context of a long-standing collaboration between the University of Manitoba and the University of Nairobi, clinical staff provide year-round basic medical services to approximately 700 CSW while recruiting individuals for biyearly research visits (“resurveys”) where biological samples, including physician-collected midvaginal swabs, are acquired. All study procedures were reviewed and approved by the Ethics Review boards of the University of Manitoba and the University of Nairobi.
Vaginal swab samples from 243 CSW attending research visits during July 2007 were collected and processed as previously described (49), including samples from HIV-exposed seronegative (HESN; active CSW remaining HIV negative for longer than 3 years of follow-up), HIV-negative controls (HIV-N; active CSW with less than 3 years of follow-up), and HIV-seropositive (HIV+) CSW. BV diagnosis by microscopy of Gram-stained swab smears using Nugent's criteria (41) was conducted by two independent evaluators assigning slides to three categories: BV negative (BV−), BV intermediate (BVI), and BV positive (BV+). Slides with discrepant diagnoses were reanalyzed by both evaluators until a consensus diagnosis was reached. A total of 127/242 women (52%) had BVI or BV+ diagnoses by Nugent score, with no significant differences between HESN, HIV-N, and HIV+ individuals (by chi-square test; data not shown).
A subset of 96 individuals, including 32 each HESN, HIV-N, and HIV+ women, were selected for culture-based analysis and strain isolation. A total of 179 isolates were identified by cpn60 UT-based sequencing. In-depth clone libraries were generated for a subset of 10 individuals (4 HIV-N and 6 HESN women) as previously described (49), resulting in 7,180 clones for which a full-length cpn60 UT was generated. A subset of 44 individuals including 16 HESN, 16 HIV-N, and 12 HIV+ women was selected for deep sequencing.
Generation of cpn60 amplicons.
Isolated strains were recovered from Lactobacillus-selective Rogosa or modified Brucella medium (45). The cpn60 UT was amplified with an annealing temperature of 42°C as previously described (26), using degenerate forward primer H729 (5′-CGC CAG GGT TTT CCC AGT CAC GAC GAI III GCI GGI GAY GGI ACI ACI AC-3′) and reverse primer H730 (5′-AGC GGA TAA CAA TTT CAC ACA GGA YKI YKI TCI CCR AAI CCI GGI GCY TT-3′ [M13 sequencing primer landing sites are in bold]). Amplicons were purified using an automated MagnaPure system and sequenced in both directions using an ABI 3730xl genetic analyzer (Genomics Core, National Microbiology Laboratory). Resulting sequences were assembled and trimmed using Lasergene 8 software (DNAStar, Madison, WI). Full-length UT sequences (∼552 bp) were compared to those in the cpnDB to determine the nearest neighbor using FASTA (42).
For clone libraries and deep sequencing, DNA was isolated from previously frozen samples and a cpn60 UT amplicon was generated as previously described (49). For clone libraries, cpn60 sequences in whole vaginal specimens were amplified using a 1:3 ratio of two degenerate cpn60 UT primer sets (set 1, forward, H279, 5′-GAI III GCI GGI GAY GGI ACI ACI AC-3′; reverse, H280, 5′-YKI YKI TCI CCR AAI CCI GGI GCY TT-3′; set 2, forward, H1612, 5′-GAI III GCI GGY GAC GGY ACS ACS AC-3′; reverse, H1613, 5′-CGR CGR TCR CCG AAG CCS GGI GCC TT-3′) at four different annealing temperatures (42, 50, 55, and 60°C) prior to pooling, cloning, and ligation as previously described (49). Resulting clones were assembled, manually inspected, and trimmed to remove vector sequences in Gap4 (8). For deep sequencing, all primers were modified 5′ with GS-FLX sequencing primers while forward primers were modified with a 6-bp bar code unique to each sample (see Table S7 in the supplemental material). Water-only controls were negative for each amplification. Bar codes were used to pool 16 samples in a single region of the GS-FLX picotiter plate, with samples from HESN, HIV-N, and HIV+ individuals run in separate regions. The pyrosequencing setup was optimized to ensure the best sequence yield, and reactions were executed with GS-FLX kits and protocols (454 Life Sciences) in the Genomics Core facility of the National Microbiology Laboratory.
Bioinformatic pipeline.
Just under 1,000,000 reads were generated for 64 samples in two GS-FLX runs, including the 44 samples discussed in detail in this paper. Raw deep sequence SFF files were parsed and binned according to specimen bar code as previously described (49). All deep-sequencing reads were assembled using the Next-Generation Sequencing assembly algorithm newbler (454 Life Sciences, Branford, CT). Assembly was performed by using the cDNA mode for newbler, an overlapMinMatchIdentity of 91%, and an overlapMinMatchLength of 137 (or at least 25% of the cpn60 UT) and by using the enumerated cpn60 UT primer sequences for screening. The use of a DNA sequence assembly algorithm means that each consensus sequence is a multiple alignment of experimental DNA sequences. The consensus sequence for each assembly then represents a proxy for each of the reads in terms of taxonomic mapping. In other words, the consensus sequence is applied as the representative sequence for the operational taxonomic unit (OTU) containing the set of sequences within the multiple alignment. Manual inspection of each assembly was conducted through the use of Tablet software to inspect the corresponding ace file (39).
In order to keep the number of OTUs manageable in downstream analyses, singletons (14.6% of the total reads) were excluded from further analysis. For the 44 samples described here, a total of 838 assemblies encompassing 648,508 reads were defined by newbler. The majority of these assemblies (513 or 61%) were non-cpn60 UT sequences, encompassing 44,947 or 6.9% of the total reads, and were excluded from further analysis. Manual checking for chimeric sequences was undertaken in order to ensure that 100-bp fragments from the beginning and end of OTU sequences matched identical sequences in the cpnDB at similar identity levels. Based on this analysis, five OTUs representing 19 clones (0.3% of the total) were removed from the analysis. No deep-sequencing OTUs were removed from the data set based on this analysis. The relative abundance of each OTU in the total data set and per individual was calculated as a percentage of the total number of reads or normalized to the median number of reads per sample.
Sequence alignment and construction of phylogeny.
For tree-based analyses, sequences generated from isolates, from clones, and by deep sequencing were trimmed 5′ to remove primers and 3′ at 200 bp so as to be equal in length for alignment and tree building. Deep-sequencing OTUs <200 bp long or containing internal stop codons were excluded (21 OTUs representing 831 reads or <0.1% of the total reads). Trimmed sequences 100% identical to each other were identified using blastclust (2) and collapsed into a single OTU, including the sum of isolates, clones, and/or reads for all sequences (Fig. 1; see Table S1 in the supplemental material).
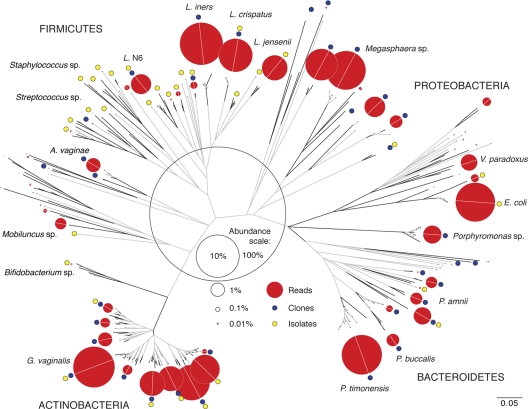
Fig. 1.
Phylogeny of 420 unique cpn60 UT sequences generated from vaginal specimens of African CSW by deep sequencing, clone library construction, and selective culture. Branches are collapsed into clusters for visual clarity (see Table S1 in the supplemental material). The relative abundance of deep-sequencing reads for each cluster is proportional to the area of the red circles, as indicated in the abundance scale. Blue and yellow circles indicate the presence of clones and culture-based isolates, respectively. Trimmed cpn60 UT sequences (200 bp, no internal stop codons) were aligned using Clustal in MEGA 4.0 and bootstrap values based on 500 replicates conducted using the neighbor-joining method. Branch segments proceeding from unsupported nodes (bootstrap value, <50%) are shown in gray. The scale bar indicates phylogenetic distance in base pair substitutions per site. Detailed abundance information and the species and strain designations for each branch and cluster are shown in Table S1 in the supplemental material.
Sequences were aligned using the Clustal function and default parameters in MEGA 4.0 (53). Phylogeny was inferred using the neighbor-joining method (47), and evolutionary distances were computed using the maximum-likelihood method (54), with statistical support for nodes calculated by bootstrap using 500 replicates.
Principal-component analysis (PCA) using FastUniFrac.
The resulting phylogenetic tree (Newick format), excluding clone and isolate sequences (a total of 250 nonredundant deep-sequencing OTUs), was imported into the FastUniFrac environment (http://128.138.212.43/fastunifrac//index.psp) (36) along with the number of reads per OTU for each specimen and metadata associated with each sample, including serostatus (HESN, HIV-N, HIV+) and BV diagnosis by Nugent score (BV−, BVI, BV+). Principal components were defined based on the UniFrac metric (36), resulting in the identification of a distinct category of samples called molecular BVI or mBVI.
Analysis of α diversity measures using mothur.
Files containing data used for UniFrac analysis were adapted for and imported into the mothur environment for OTU-based α diversity analysis of individual samples (http://www.mothur.org/) (50). Richness and diversity were assessed by calculating Chao and Simpson values using the summary.shared command.
Data visualization and hierarchical clustering using Genespring.
Initially, a heat map based on tiers of abundance was constructed manually for each sample and organized by higher-order phylotype (see Fig. 3). After evaluating the visualization capabilities of the gene expression data analysis tool Genespring (Agilent, Ontario, Canada), we adapted read abundance data and imported them into Genespring to capitalize on its unique capabilities. OTU abundance data were scaled to the median abundance level and to the baseline of samples. Hierarchical clustering was performed by clustering on both entities (OTUs) and conditions (study subgroups: HESN/BV−, HESN/mBVI, HESN/BV+, HIV-N/BV−, HIV-N/mBVI, HIV-N/BV+, HIV+/BV−, HIV+/mBVI, HIV+/BV+, HIV+/outlier) using a Euclidian similarity measure with complete linkage. Only OTUs present in at least 11/44 samples, or 25% of the samples, were selected for these analyses, based on the uncollapsed OTU set (n = 90).
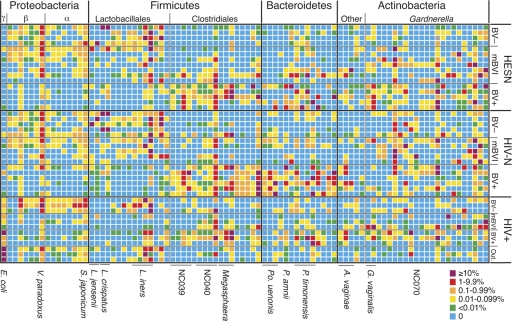
Fig. 3.
Heat map of deep-sequencing OTUs detected in at least 25% of the total study group members, organized in rows by HIV serostatus and BV status. OTUs are organized into columns by higher-order phylotype. The scale indicates the percentage of the total reads for each individual. Selected OTUs referred to in text are indicated at the bottom. A full listing of the OTUs, along with abundance information and statistical comparisons by HESN or HIV+ status and BV status are provided in Tables S4 and S5 in the supplemental material.
Statistical tests.
Distribution of major phylogenetic groups in HESN/HIV-N/HIV+ and BV−/mBVI/BV+ was evaluated by collapsing OTUs into six higher-order phylotypes (Gardnerella, other Actinobacteria, Lactobacillales, Clostridiales, Bacteroidetes, and Proteobacteria). Intergroup comparisons of overall density and diversity indices, as well as phylotype density and normalized abundance for each individual OTU and phylogenetic subgroup, were executed using boxplot and Mann-Whitney test functions in R (v2.11.0). A P value greater than 0.05 after Bonferroni correction for multiple comparisons was considered significant. Tests within Genespring were performed via Mann-Whitney unpaired test using a Benjamini Hochberg False Discovery Correction.
RESULTS
Deep-sequencing read assembly, OTU definition, and tiers of abundance.
Using newbler (454 Life Sciences), raw reads with an average length of 250 bp were assembled into 325 consensus sequences matching cpnDB sequences, representing 603,561 reads from vaginal specimens of 44 individuals (16 HESN, 16 HIV-N, 12 HIV+). The median number of reads per individual was 12,468 (range, 2,580 to 48,629).
The range of the proportion of the total number of reads attributed to each OTU was very large, at >7% to <0.0002%. Therefore, the concept of “tiers of abundance” was developed, based on relative abundances of >1% (tier 1), 0.1 to 0.99% (tier 2), 0.01 to 0.099% (tier 3), and <0.01% (tier 4) (see Fig. S1A in the supplemental material). Most reads were observed in tier 1, and most of the OTUs representing a very small number of reads were observed in tier 4 (Fig. S1B).
Abundance and phylogeny of deep-sequencing reads, clones, and isolates.
The phylogeny of deep-sequencing OTUs is shown in the context of the cpn60 UT generated from 179 isolates and 7,190 clones from the same study group (Fig. 1; see Table S1 in the supplemental material). In order to directly compare sequences and generate the phylogeny, all OTUs were trimmed to the first 200 bp of the cpn60 UT. OTUs were collapsed into a single branch (in Fig. 1) and row (in Table S1) if 100% identical over the full sequence. This resulted in 420 OTUs, including 73 for isolates, 138 for clones, and 248 for reads.
Only 5 OTUs were detected by all 3 methods, representing 22% of the clones and 8% of the reads. Of 248 OTUs defined for pyrosequencing data, 21 were identical to OTUs detected in clone libraries (representing 41% of the clones and 38% of the reads) and 15 were identical to isolate OTUs (representing 22% of the reads). In contrast, we detected 114 OTUs in clone libraries only and 56 among isolates only, as well as 2 OTUs in both isolates and clones but not in deep-sequencing data.
Sequence heterogeneity.
Extensive sequence heterogeneity was observed at branch tips for the most abundant phylotypes (e.g., Gardnerella vaginalis, Lactobacillus iners, L. crispatus, Prevotella timonensis, Porphyromonas sp.) (Fig. 1). Although branch tips were clearly linked by a shared valid node, relationships between branch tips were generally not reproducible by bootstrap.
Most dramatically, a large number of OTUs sharing a valid node with and ranging from 89 to 100% identical to G. vaginalis ATCC 14018 was observed (Fig. 1), with a median of 13 and a maximum of 51 G. vaginalis OTUs observed per individual. This heterogeneity was also observed in clone libraries and isolates, with representatives very close (but not always identical) to deep-sequencing OTUs (see Table S1 in the supplemental material).
Species and strain level resolution of deep-sequencing OTUs.
Nearly all of the OTUs belonged to four phyla well known to predominate in human microbiota (Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes) (14). Over half of the OTUs from deep sequencing (140 or 56%), representing 65% of the total reads, matched species or strain level designations in the cpnDB at ≥95% identity. A total of 54 species and strains in 29 genera were detected by deep sequencing in this study.
A further 29 species in 10 genera (including 8 Streptococcus, 6 Staphylococcus, and 5 Lactobacillus species) were detected only by culture, indicating that culture-based techniques may be more sensitive than deep sequencing for detecting these organisms in vaginal specimens (15).
Defining core microbiota.
Most tier 1 organisms were observed in the majority of the study group members (see Fig. S1C in the supplemental material); however, no single OTU was observed in all of the women due to sequence heterogeneity, as described above. Three groups of OTUs 96 to 100% identical to G. vaginalis, L. iners, and P. timonensis were each observed in 43 out of 44 women, indicating that these three species constitute a core microbiota in this cohort.
Although the most abundant OTU was identical to E. coli, it was observed in less than half of the study group. In fact, virtually all E. coli reads were found in HIV+ individuals only and samples from three HIV+ individuals were mostly E. coli reads, indicating a highly skewed distribution for this OTU (see Table S2 in the supplemental material).
Several low-abundance OTUs in terms of the total number of reads were observed in a high proportion of the study group members (see Table S3 in the supplemental material), indicating that minor constituents of the microbiota were frequently detected. Interestingly, the median proportion of the total number of reads per individual was rarely greater than 1% for even the most abundant OTU, with a range across several orders of magnitude (see Fig. S1D in the supplemental material). This observation indicates that even the most frequently detected OTUs were observed at very low levels in most individuals, a feature likely to be missed in lower-resolution data sets.
Phylogeny of deep-sequencing reads.
Besides L. iners, other Lactobacillus OTUs included L. crispatus (6% of the total reads), L. jensenii (3.5%), and N6 (2.2%), a cultured isolate 90% identical to both L. vaginalis and L. reuteri. Surprisingly, L. gasseri was not detected by deep sequencing, although it was well represented among cultured isolates.
Most of the Clostridiales OTUs were >85% identical to database sequences, indicating the identification of several novel organisms in this order. One group of OTUs, representing 8.4% of the total reads and observed in 89% of the study group members, was virtually identical to a Megasphaera sp. reference sequence from the Human Microbiome Project (56).
Although P. timonensis was the most abundant Bacteroidetes species (>8% of the total reads), others included Prevotella melaninogenica (1.4%), Prevotella amnii (0.8%), Prevotella buccalis (0.25%), and Prevotella zoogleoformans (0.25%). Several OTUs similar to the newly described species Porphyromonas uenonis made up nearly 2% of the total reads.
All alpha- and betaproteobacterial OTUs were detected only by deep sequencing, mostly in very small numbers (0.001 to 0.06% of the total reads). The betaproteobacterium Variovorax paradoxus was detected in 89% of the study group members. This organism has not previously been reported in the vaginal microbiota, although it has been isolated from the human mouth (4). A group of OTUs ∼90% identical to Sphingobium and Sphingomonas was also observed in most individuals. These genera have recently been reported in 16S rRNA gene-based deep-sequencing studies of the vaginal microbiota of American women (46).
Comparison of samples from HESN and HIV-N women.
No differences in BV diagnosis by Nugent score between HESN and HIV-N women were observed. Since preliminary analysis of isolates and clone libraries confirmed fundamental differences in the profile of the vaginal microbiota according to BV diagnosis and selection of a truly random sample of individuals was not feasible, we decided to select for a balanced representation of BV− and BV+ samples in order to ensure that any observed differences could be attributed to HESN or HIV+ status rather than to differences in BV diagnosis.
Contrary to the hypothesis that motivated this study, no clear differences between the deep-sequencing profiles of HESN and HIV-N women were observed. Analysis of OTUs collapsed to the phylum and order levels across individuals, sorted from left to right based on the descending proportion of Lactobacillus reads, shows that the two groups are virtually identical (Fig. 2A), while PCA of abundance-weighted phylogenetic trees for each individual, based on UniFrac (37), showed no clustering of HESN individuals (Fig. 2B). Analysis of the richness, diversity, and abundance of higher-order phylotypes also showed no differences between HESN and HIV-N individuals (see Fig. S2 in the supplemental material).
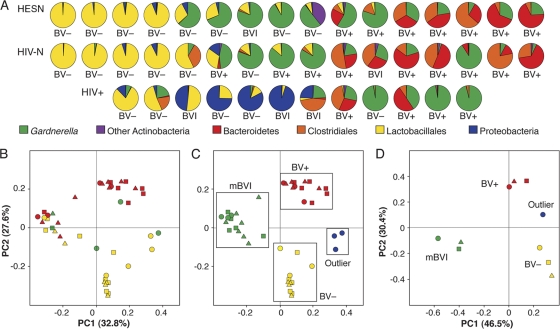
Fig. 2.
Distribution of higher-order phylotypes in each of 44 individuals reveals patterns associated with BV diagnosis by Nugent score and HIV serostatus (A). Each pie chart represents a single sample, sorted from left to right by decreasing frequency of the total reads for all Lactobacillales OTUs. Abundance-weighted UniFrac PCA of 44 samples for BV diagnosis by Nugent score is shown (B to D). Groups: HESN (triangles), HIV-N (squares), HIV+ (circles); BV− (yellow), BVI (green), and BV+ (red). Reclassification of samples based on molecular pattern is also shown (C). Groups: BV− (yellow), mBVI (green), BV+ (red), and outliers (blue). PCA of group level phylogenies demonstrates that samples do not cluster by HESN or HIV+ status (D).
Visualization of the distribution of OTU detected in at least 25% of the study group revealed the idiosyncrasy of individual profiles at the species and strain levels (Fig. 3), reinforcing the concept that the profile of the microbiota at high phylogenetic resolution is as unique as a fingerprint (14). A single Gardnerella-like OTU (NC070) was observed in 11 of 16 HIV-N women, compared to only 1 of 16 HESN women. Although statistically significant after correction for multiple comparisons (see Table S4 in the supplemental material), this OTU was also not observed in any HIV+ women; therefore, its absence seems an unlikely biomarker of HESN status.
Comparison of samples from HIV+ with those from HESN and HIV-N women.
Phylum and order level analyses showed that HIV+ women were visually distinct from HESN and HIV-N women (Fig. 2A). While 8/32 (25%) HESN and HIV-N women had samples with >95% Lactobacillus OTUs, the maximum proportion of Lactobacillus in HIV+ women did not exceed 80%. With the exception of three women with very high levels of Proteobacteria, HIV+ women did not cluster together by PCA (Fig. 2B), and no differences in richness or diversity between HIV+ and HESN or HIV-N women were observed (see Fig. S2 in the supplemental material).
An OTU identical to E. coli was significantly more abundant in HIV+ than in HESN and HIV-N individuals (Fig. 3; see Table S4 in the supplemental material). Although the difference is not statistically significant after correction for multiple comparisons, L. crispatus was detected in only 1 of 12 HIV+ women (8%), compared to 19 of 32 (59%) HESN and HIV-N women (Fig. 3; see Table S4 in the supplemental material). These observations may reflect an altered microbiota in HIV+ individuals consistent with the well-known association between BV and HIV+ serostatus (51).
A molecular definition of BV.
Several molecular patterns largely consistent with the BV diagnosis by Nugent score were revealed in phylum and order level analyses of deep-sequencing reads by individual (Fig. 2A). Those with the highest abundance of Lactobacillus were BV negative (BV−) by Nugent score, while those with the lowest abundance of Lactobacillus were all diagnosed as BV positive (BV+). Interestingly, samples with intermediate levels of Lactobacillus were dominated by Gardnerella OTUs and had a BV−, BVI, or BV+ diagnosis by Nugent score. These observations demonstrate a close correspondence between the traditional BV diagnosis and the total Lactobacillus level as defined by deep sequencing but also suggest a distinct molecular pattern associated with intermediate levels of Lactobacillus.
PCA demonstrated that samples with the highest proportion of Lactobacillus (BV−) and the lowest proportion of Lactobacillus (BV+) cluster together unambiguously (Fig. 2B). A third cluster corresponded to samples dominated by Gardnerella, while 3 samples dominated by E. coli formed a fourth cluster (Fig. 2B). Since traditional BV diagnoses as defined by Nugent scoring included samples with clearly different phylogenetic patterns as defined by deep sequencing, we redefined samples in molecular terms for downstream analyses (BV+, mBVI, BV−, and outlier) (Fig. 2C). The characteristics of mBVI samples (with all possible Nugent diagnoses) were compared to those of BV+ and BV− samples, based on the hypothesis that these samples are truly BV intermediate but are not defined this way by Nugent diagnosis due to the lower resolution of this technique.
As observed in other studies (29, 34), BV+ samples had higher median richness (Chao index) and higher median diversity (Simpson index) than BV− samples (see Fig. S3A and B in the supplemental material). As expected based on previous observations, BV+ samples had the highest abundance of Bacteroidetes and Clostridiales reads, while mBVI samples had the highest abundance of Gardnerella-like reads. Except for Gardnerella, mBVI samples had intermediate abundances of other phylotypes compared to BV+ and BV− samples. As well as having the highest abundance of Lactobacillales, BV− samples had the highest abundance of Proteobacteria reads (Fig. S3C).
In order to control for individual idiosyncrasies, samples were collapsed into 10 groups based on serostatus (HESN/HIV-N/HIV+) and mBV status (BV+/mBVI/BV−/outlier). As expected, PCA (Fig. 2D) and hierarchical clustering by an independent OTU-based approach conducted using Genespring (see Fig. S4 in the supplemental material) confirmed that HESN or HIV+ status does not define these samples phylogenetically.
Defining biomarkers of BV.
In order to determine OTUs significantly associated with BV, nonparametric pairwise statistical comparisons by BV diagnosis were carried out using two methods and software packages (R and Genespring) (see Table S5 in the supplemental material), for a subset of 90 OTUs present in at least 25% of the study group members. OTUs significantly different between groups after Bonferroni correction for multiple comparisons and/or significantly different by both methods were considered to be potential biomarkers of BV.
As expected, several OTUs in the phylum Bacteroidetes and the order Clostridiales were found to be significantly more abundant in BV+ than in BV− and/or mBVI samples, including those nearest P. amnii and the clostridial clones NC030 and NC040 (see Table S5 in the supplemental material). OTUs identical to L. iners were significantly less abundant in BV+ samples than in BV− samples. Consistent with previous observations, the proteobacterium V. paradoxus was found to be significantly more abundant in BV− samples.
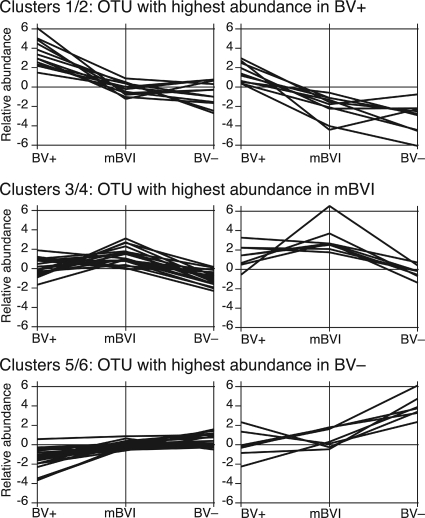
By k-means clustering of OTUs using Genespring (k = 6), three major patterns relative to BV status were identified, i.e., those with the highest abundance in BV+ (clusters 1 and 2), mBVI (clusters 3 and 4), and BV− (clusters 5 and 6) samples (Fig. 4; see Table S6 in the supplemental material). Consistent with UniFrac PCA results, OTUs closest to Clostridiales and Bacteroidetes dominate clusters 1 and 2, Gardnerella OTUs dominate clusters 3 and 4, and Lactobacillus OTUs dominate clusters 5 and 6. Several alpha- and betaproteobacterial OTUs were found alongside Lactobacillus and V. paradoxus in clusters 5 and 6, indicating that these may also be associated with BV− status (see Table S6 in the supplemental material).
Fig. 4.
k-means clustering of OTUs using Genespring software reveals patterns associated with BV+ (clusters 1 and 2), mBVI (clusters 3 and 4), and BV− (clusters 5 and 6) status. A list of the OTUs associated with each cluster is shown in Table S6 in the supplemental material.
DISCUSSION
Using the GS-FLX platform (454 Life Sciences), we recently described the first cpn60-based phylogenetic analysis by deep sequencing (49), comparing cpn60-based to 16S rRNA gene variable region 2 and 3-based phylogenies in matched samples. While the overall structure of the microbiota was similar, resolution (i.e., the number of distinct phylotypes detected per sample) was greatly increased by using cpn60 UT (49). Our findings indicate that cpn60 UT is ideally suited to deep sequencing using next-generation platforms.
The main advantage of defining consensus sequences using an assembly method, as opposed to selecting a representative sequence after clustering, is that the consensus sequence is the OTU summarizing all of the reads in each assembly for phylogenetic inference. This approach allows visual inspection of the aligned sequences used to create the consensus sequence. The impact of sequencing error in OTU assignment may also be mitigated by using a consensus-based approach, although this possibility was not formally evaluated.
Extensive sequence heterogeneity may, in part, represent technical error arising from sequencing or sequence assembly. However, heterogeneity was more extensive in some phylotypes than in others. Extensive Gardnerella heterogeneity was first observed in clone libraries and isolates and confirmed by deep sequencing. A similar observation in three independent data sets indicates that deep-sequencing error is unlikely to be the sole explanation for these differences.
Distinct G. vaginalis biotypes have been described in early culture-based studies, as well as more recently (24, 43), and recent genome level analyses indicate extensive genomic rearrangement in this organism (58). A recent study of HIV+ African women detected four distinct 16S rRNA gene v6-based G. vaginalis phylotypes (29), and we have recently observed similar patterns based on cpn60 UT sequences from published G. vaginalis genomes. The possible biological significance of the heterogeneity observed in this study is unknown.
Since no unambiguous rule for collapsing OTUs was established, we decided that the OTU as defined by newbler assembly was the most direct and reproducible basis for OTU definition. Whether this set of OTUs over- or underestimates the true sequence diversity was not established in this study, however, potential errors in OTU calling are restricted to the highest resolution (i.e., at branch tips) and are therefore unlikely to affect the study's main conclusions.
As in our previous study, we observed differences in the overall proportion of specific OTUs in clone libraries versus deep-sequencing reads. Although we used the same primers to amplify the cpn60 UT for either method, different molecular and bioinformatic biases associated with the two methods are likely. Also, clone libraries were generated in a smaller group (n = 10) that overlapped with but was not entirely a subset of the individuals selected for deep sequencing (n = 44). Therefore, differences in relative abundances may be related to methodological biases and/or different individuals in these subsets.
Small sequence differences between similar OTUs defined in clone libraries versus pyrosequencing may reflect actual differences between closely related strains or may result from different biases associated with the generation of consensus sequences using different software and data sets for clones (Gap4 using the full-length cpn60 UT) versus pyrosequencing reads (newbler using reads ranging from 150 to 400 bp). A closer look reveals that all OTUs found only in clone libraries are very close to OTUs found in reads. This question was formally addressed by comparing total branch length based on OTUs from reads (16.5 base substitutions per site or bsps), clones (5.1 bsps), and isolates (6.1 bsps). The total branch length when adding clone-only OTUs to read OTUs was 19.1 bsps. Therefore, the amount of unique branch length contributed by clone-only OTUs is 2.6 bsps.
The large number of OTUs with species level designations in the cpnDB contrasts with recent 16S rRNA gene-based molecular studies that largely describe the vaginal microbiota at the genus or higher taxonomic levels (29, 34, 46, 51). Our findings indicate that the higher resolution of the protein-encoding cpn60 UT allows greater separation between closely related organisms than other conventional targets do (49). We report 55 species in 29 genera from 200-bp assemblies representing just over 600,000 reads, compared with 23 species in 10 genera reported from over 12,000,000 72-bp Illumina reads of 16S rRNA gene v6 in a much larger group of African women (29).
The other studies report virtually no species level information. For example, all studies report that Prevotella was observed in most individuals; however, none report species level designations for this genus. In contrast, our study detected nine Prevotella species (P. amnii, P. bergensis, P. bivia, P. buccalis, P. corporis, P. disiens, P. melaninogenica, P. timonensis, and P. zoogleoformans) and three Porphyromonas species (P. asaccharolytica, P. gingivalis, and P. uenonis) in the phylum Bacteroidetes with an average of 96.8% identity.
The phylogeny of deep-sequencing OTUs shown in the context of clone libraries and isolate sequences demonstrates that the overall composition of the vaginal microbiota in this group is strikingly consistent with what has been observed in other groups of women worldwide (Fig. 1; see Table S1 in the supplemental material) (25, 46, 51). Since these women report a median of four commercial sex work clients per day (range, 1 to 10) and frequent postcoital douching (48), it is somewhat surprising that their vaginal microbiota should be so similar to that of other groups. This finding suggests a robustness of vaginal microbial communities despite frequent physical disturbance.
The hypothesis that HESN women have increased levels of specific types of Lactobacillus and/or reduced levels of specific types of BV-related organisms compared to those of HIV-N and HIV+ women was not supported. We originally estimated that a sample size of 15 individuals in each group would be sufficient to detect a 50% difference in the presence of a specific OTU; however, the high resolution of the present data set (number of OTUs defined) and the extensive variability observed between individuals make it difficult to draw firm conclusions based on such small numbers of individuals at a single time point. Longitudinal studies designed to determine the dynamics of the vaginal microbiota in relation to pro- and antiinflammatory mucosal factors may reveal distinct patterns in HESN individuals. The present study provides a solid framework on which to base future studies.
Our results suggest that HIV+ women in this cohort have a vaginal microbiota, including increased E. coli and reduced L. crispatus levels, distinct from that of HESN or HIV-N women. However, a recent 16S rRNA gene-based survey of the vaginal microbiota in HIV+ women in Tanzania did not detect any individuals with dominant E. coli (29), indicating that this finding may be a specific characteristic of HIV+ women in this cohort, since most also receive daily prophylaxis with the antibiotic trimethoprim-sulfamethoxazole (Septrin) (48).
Based on UniFrac PCA, we defined samples based on phylogenetic characteristics determined by deep sequencing, i.e., a “molecular definition” of BV. We found that samples with BV+ and BV− diagnoses by Nugent score were significantly different in terms of the richness, diversity, and abundance of Gardnerella, Bacteroidetes, Clostridiales, Lactobacillus, and Proteobacteria OTUs by deep sequencing, indicating an excellent correspondence between molecular and traditional definitions for these categories.
A cluster of samples with intermediate levels of Lactobacillus and dominant Gardnerella was identified (mBVI). These samples also had intermediate levels of Clostridiales, Bacteroidetes, and Proteobacteria abundance compared to those of BV+ and BV− samples. Although a discussion of BV dynamics remains speculative in the absence of longitudinal data, these samples hypothetically represent a transition stage between a Lactobacillus-dominated phenotype (BV−), characterized by a shift to Gardnerella dominance as levels of Lactobacillus fall, and gradual establishment of the Gardnerella-Prevotella-Clostridiales codominant phenotype (BV+). These findings are consistent with in vitro work suggesting that nutritional interactions between Gardnerella and Prevotella are likely to be involved in the establishment of BV (44).
By statistical analysis of the relative abundance of specific OTUs and k-means clustering techniques in the Genespring environment, we have defined several potential biomarkers of BV, including increased levels of Clostridiales clones NC030 and NC040, increased P. amnii, and reduced levels of L. iners. These OTUs are good candidates for rapid approaches to quantify fluctuations in specific bacterial populations associated with BV, as we have recently shown (17).
Surprisingly, several alpha- and betaproteobacterial OTUs, including the newly described species V. paradoxus, were found to be most abundant in BV− samples. Like many others, we have shown that increased Lactobacillus abundance is strongly tied to BV− status; however, no previous studies that we are aware of have described a non-Lactobacillus organism with increased abundance in BV− individuals. Further work is required to confirm this potentially important finding.
Understanding fluctuations in specific bacterial populations in the vagina and their influence on the susceptibility of the mucosal barrier to infection with HIV and other pathogens is critical to defining mechanisms underlying the well-known association between BV and HIV. The highly detailed species and strain level definition of the vaginal microbiota related to BV in this study provides a framework in which to track specific microbes in HIV-exposed individuals in order to address hypotheses about vaginal microbiology, mucosal resistance factors, immune quiescence, and resistance to HIV.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the members of the Majengo cohort for their ongoing participation in HIV-related studies, as well as the dedicated clinical and laboratory staffs in Nairobi and Winnipeg who make this research possible. Technical support in the Genomics Core was provided by B. Kaplan and E. Bellamy. Thanks to G. Reid for advice related to study design and to P. Schloss for guidance in applying mothur for ecological analyses.
Funding for this study was provided by the National Institutes of Health and the Bill & Melinda Gates Foundation. J.J.S. received grants from the Canadian Institutes of Health Research (CIHR) Doctoral Research Award and International Centre for Infectious Diseases/CIHR Training Program.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Ahmed N., et al. 2010. Suppression of human immunodeficiency virus type 1 replication in macrophages by commensal bacteria preferentially stimulating Toll-like receptor 4. J. Gen. Virol. 91:2804–2813 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Amsel R., et al. 1983. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74:14–22 [DOI] [PubMed] [Google Scholar]
- 4. Anesti V., et al. 2005. Isolation and molecular detection of methylotrophic bacteria occurring in the human mouth. Environ. Microbiol. 7:1227–1238 [DOI] [PubMed] [Google Scholar]
- 5. Atashili J., Poole C., Ndumbe P. M., Adimora A. A., Smith J. S. 2008. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS 22:1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ball T. B., et al. 2007. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS 21:1091–1101 [DOI] [PubMed] [Google Scholar]
- 7. Beigi R. H., Wiesenfeld H. C., Hillier S. L., Straw T., Krohn M. A. 2005. Factors associated with absence of H2O2-producing Lactobacillus among women with bacterial vaginosis. J. Infect. Dis. 191:924–929 [DOI] [PubMed] [Google Scholar]
- 8. Bonfield J. K., Smith K., Staden R. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boskey E. R., Telsch K. M., Whaley K. J., Moench T. R., Cone R. A. 1999. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect. Immun. 67:5170–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Braat H., et al. 2004. Lactobacillus rhamnosus induces peripheral hyporesponsiveness in stimulated CD4+ T cells via modulation of dendritic cell function. Am. J. Clin. Nutr. 80:1618–1625 [DOI] [PubMed] [Google Scholar]
- 11. Burgener A., et al. 2008. Identification of differentially expressed proteins in the cervical mucosa of HIV-1-resistant sex workers. J. Proteome Res. 7:4446–4454 [DOI] [PubMed] [Google Scholar]
- 12. Card C. M., et al. 2009. Decreased immune activation in resistance to HIV-1 infection is associated with an elevated frequency of CD4(+)CD25(+)FOXP3(+) regulatory T cells. J. Infect. Dis. 199:1318–1322 [DOI] [PubMed] [Google Scholar]
- 13. Cauci S., Monte R., Driussi S., Lanzafame P., Quadrifoglio F. 1998. Impairment of the mucosal immune system: IgA and IgM cleavage detected in vaginal washings of a subgroup of patients with bacterial vaginosis. J. Infect. Dis. 178:1698–1706 [DOI] [PubMed] [Google Scholar]
- 14. Dethlefsen L., McFall-Ngai M., Relman D. A. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donachie S. P., Foster J. S., Brown M. V. 2007. Culture clash: challenging the dogma of microbial diversity. ISME J. 1:97–99 [DOI] [PubMed] [Google Scholar]
- 16. Dumonceaux T. J., et al. 2006. Enumeration of specific bacterial populations in complex intestinal communities using quantitative PCR based on the chaperonin-60 target. J. Microbiol. Methods 64:46–62 [DOI] [PubMed] [Google Scholar]
- 17. Dumonceaux T. J., et al. 2009. Multiplex detection of bacteria associated with normal microbiota and with bacterial vaginosis in vaginal swabs by use of oligonucleotide-coupled fluorescent microspheres. J. Clin. Microbiol. 47:4067–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fleming D. T., Wasserheit J. N. 1999. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect. 75:3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fowke K. R., et al. 1996. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348:1347–1351 [DOI] [PubMed] [Google Scholar]
- 20. Goh S. H., et al. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldenberg R. L., Culhane J. F., Iams J. D., Romero R. 2008. Epidemiology and causes of preterm birth. Lancet 371:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gottlieb M. S., et al. 1981. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N. Engl. J. Med. 305:1425–1431 [DOI] [PubMed] [Google Scholar]
- 23. Haase A. T. 2005. Perils at mucosal front lines for HIV and SIV and their hosts. Nat. Rev. Immunol. 5:783–792 [DOI] [PubMed] [Google Scholar]
- 24. Harwich M. D., Jr, et al. 2010. Drawing the line between commensal and pathogenic Gardnerella vaginalis through genome analysis and virulence studies. BMC Genomics 11:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill J. E., et al. 2005. Characterization of vaginal microflora of healthy, nonpregnant women by chaperonin-60 sequence-based methods. Am. J. Obstet. Gynecol. 193:682–692 [DOI] [PubMed] [Google Scholar]
- 26. Hill J. E., et al. 2006. Identification of Campylobacter spp. and discrimination from Helicobacter and Arcobacter spp. by direct sequencing of PCR-amplified cpn60 sequences and comparison to cpnDB, a chaperonin reference sequence database. J. Med. Microbiol. 55:393–399 [DOI] [PubMed] [Google Scholar]
- 27. Hill J. E., Penny S. L., Crowell K. G., Goh S. H., Hemmingsen S. M. 2004. cpnDB: a chaperonin sequence database. Genome Res. 14:1669–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hillier S. L. 1998. The vaginal microbial ecosystem and resistance to HIV. AIDS Res. Hum. Retroviruses 14(Suppl. 1):S17–S21 [PubMed] [Google Scholar]
- 29. Hummelen R., et al. 2010. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 5:e12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iqbal S. M., et al. 2009. Elevated elafin/trappin-2 in the female genital tract is associated with protection against HIV acquisition. AIDS 23:1669–1677 [DOI] [PubMed] [Google Scholar]
- 31. Kaul R., et al. 2001. Late seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J. Clin. Invest. 107:341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsson P. G., et al. 2005. Bacterial vaginosis. Transmission, role in genital tract infection and pregnancy outcome: an enigma. APMIS 113:233–245 [DOI] [PubMed] [Google Scholar]
- 33. Lin P. W., et al. 2009. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic. Biol. Med. 47:1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ling Z., et al. 2010. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics 11:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livengood C. H. 2009. Bacterial vaginosis: an overview for 2009. Rev. Obstet. Gynecol. 2:28–37 [PMC free article] [PubMed] [Google Scholar]
- 36. Lozupone C., Hamady M., Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. MacDonald K. S., et al. 2000. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:1581–1589 [DOI] [PubMed] [Google Scholar]
- 39. Milne I., et al. 2010. Tablet—next generation sequence assembly visualization. Bioinformatics 26:401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitchell C. M., et al. 2008. Bacterial vaginosis, not HIV, is primarily responsible for increased vaginal concentrations of proinflammatory cytokines. AIDS Res. Hum. Retroviruses 24:667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nugent R. P., Krohn M. A., Hillier S. L. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pearson W. R., Lipman D. J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Piot P., et al. 1984. Biotypes of Gardnerella vaginalis. J. Clin. Microbiol. 20:677–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pybus V., Onderdonk A. B. 1997. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J. Infect. Dis. 175:406–413 [DOI] [PubMed] [Google Scholar]
- 45. Rabe L. K., Hillier S. L. 2003. Optimization of media for detection of hydrogen peroxide production by Lactobacillus species. J. Clin. Microbiol. 41:3260–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ravel J., et al. 2010. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. http://www.pnas.org/content/early/2010/06/02/1002611107.full.pdf [DOI] [PMC free article] [PubMed]
- 47. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 48. Schellenberg J. 2010. The microbiological context of HIV resistance. Ph.D. thesis. University of Manitoba, Winnipeg, Manitoba, Canada: http://hdl.handle.net/1993/4022 [Google Scholar]
- 49. Schellenberg J., et al. 2009. Pyrosequencing of the chaperonin-60 universal target as a tool for determining microbial community composition. Appl. Environ. Microbiol. 75:2889–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spear G. T., et al. 2008. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J. Infect. Dis. 198:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stock S. J., et al. 2009. Elafin (SKALP/Trappin-2/proteinase inhibitor-3) is produced by the cervix in pregnancy and cervicovaginal levels are diminished in bacterial vaginosis. Reprod. Sci. 16:1125–1134 [DOI] [PubMed] [Google Scholar]
- 53. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 54. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tien M. T., et al. 2006. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J. Immunol. 176:1228–1237 [DOI] [PubMed] [Google Scholar]
- 56. Turnbaugh P. J., et al. 2007. The human microbiome project. Nature 449:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Westrom L., Evaldson G., Holmes K. K. 1984. Taxonomy of vaginosis: bacterial vaginosis—a definition. Scand. J. Urol. Nephrol. Suppl. 86:259–2646598922 [Google Scholar]
- 58. Yeoman C. J., et al. 2010. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS One 5:e12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhu T., et al. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391:594–597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.