Abstract
A gene involved in the production of medium-chain α-olefins was identified in the cyanobacterium Synechococcus sp. strain PCC 7002. The gene encodes a large multidomain protein with homology to type I polyketide synthases, suggesting a route for hydrocarbon biosynthesis from fatty acids via an elongation decarboxylation mechanism.
INTRODUCTION
Biological hydrocarbons are a promising alternative to petroleum-based liquid transportation fuels as they would be compatible with current engines and distribution systems yet be sourced from renewable substrates. Although it has been known for decades that some species of bacteria, including Synechococcus sp. strain PCC 7002 (formerly Agmenellum quadruplicatum), can synthesize hydrocarbons (20), until recently, very little was known about their biosynthesis. Enzymes responsible for producing three types of hydrocarbons (alkanes, internal olefins, and α-olefins) have recently been identified. Cyanobacterial alkanes can be derived from fatty acids by decarbonylation of the corresponding aldehydes (16). Alkenes with internal double bonds can be generated from the head-to-head condensation of fatty acids; genes involved in this pathway have been described for Micrococcus luteus (2). Recently, a P450 fatty acid decarboxylase was reported to be involved in α-olefin biosynthesis in Jeotgalicoccus sp. (15). In this study, we analyzed the hydrocarbon profile of the cyanobacterium Synechococcus sp. PCC 7002 and demonstrated the involvement of a gene with modular organization, similar to a polyketide synthase, in the synthesis of medium-chain α- olefins. Feeding studies suggested that the putative enzyme used an elongation-decarboxylation mechanism to convert fatty acyl-acyl carrier proteins (fatty acyl-ACPs) to α-olefins.
PCC 7002 was reported to synthesize two C19 alkenes, but nothing was known about their structure (20). In order to characterize these compounds, cultures were grown photosynthetically (140 μE/m2/s) at 35°C in 100 ml of medium A (18) to an optical density at 730 nm (OD730) of 0.2, and cell pellets were subjected to lipid extraction and analysis by gas chromatography-mass spectrometry (GC-MS) (12). Two major peaks were observed at 14.87 min and 15.15 min (Fig. 1) whose mass spectra (see Fig. S2 in the supplemental material) were consistent with a 19:2 and a 19:1 hydrocarbon, respectively. In order to determine the position of the double bonds, the hydrocarbon mixture was derivatized with dimethyl disulfide (DMDS) (19) and analyzed by GC-MS. The spectrum for the DMDS adduct of the C19:1 hydrocarbon showed molecular ions corresponding to a terminal double bond (Fig. S3). The compound was confirmed as 1-nonadecene by comparison with a commercial standard. The spectrum for the DMDS adduct of the C19:2 hydrocarbon also showed ions consistent with a terminal double bond. To identify the position of the internal double bond, hydrocarbons were purified over silica gel and subjected to permanganate/periodate oxidation (8). The mass spectra of the major resulting peak were consistent with a 13-carbon dicarboxylic, dimethyl ester (Fig. S4), suggesting that the 19:2 species is 1,14-nonadecadiene, a compound previously observed in a distinct isolate of Synechococcus sp. (8).
Fig. 1.
Comparison of hydrocarbon extracts from the wild-type and mutant strains of PCC 7002. GC-MS signal was normalized to the height of an internal standard peak (hexadecane, 8.3 min). Two hydrocarbons, 1,14-nonadecadiene and 1-nonadecene, were identified in extracts of the wild type and the promoter replacement Φ(PpsbA-ols) mutant but not in extracts of the Δols mutant.
Analysis of the PCC 7002 fatty acid profile indicated that the largest species were C18 (13). Therefore, it is unlikely that a decarboxylase like the one described for Jeotgalicoccus sp. is involved in the biosynthesis of the C19 α-olefins. Biochemical characterization of lysates from the microalgae Botryococcus braunii suggested that α-olefins can also be formed from fatty acids by an elongation-decarboxylation mechanism (21), resulting in α-olefins one carbon larger than the fatty acid substrate. However, the enzymes involved in this conversion remain unknown. In search for additional genes involved in α-olefin formation, we considered the biosynthesis of curacin A, a natural product isolated from the marine cyanobacterium Lyngbya majuscula (4) that contains a terminal double bond. Curacin A is synthesized by enzymes encoded by a 64-kb gene cluster comprising nine polyketide synthases and one nonribosomal peptide synthase module. Biochemical characterization (9) showed that the last module, encoded by curM, is responsible for forming the terminal double bond.
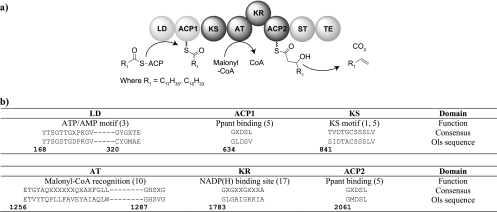
On the basis of these observations, we hypothesized that enzymes with homology to CurM could be involved in the biosynthesis of the α-olefins observed in PCC 7002. We used the basic local alignment search tool (BLAST) from NCBI to look for homologs to CurM in PCC 7002. This search identified an open reading frame encoding a protein with 45% amino acid sequence identity to CurM (SYNPCC7002_A1173, here referred to as the ols protein for olefin synthase). Several motif sequences commonly found in polyketide synthases were identified within ols (Fig. 2). Overall, ols encodes a protein with C-terminal domain architecture that is highly similar to CurM, including the polyketide elongation module and terminal olefin forming domains. Unlike for CurM, the N terminus of Ols contains two additional domains, which are predicted to comprise a loading module (Fig. 2). We hypothesize that substrates are loaded onto the ACP1 domain by the ATP consuming loading domain (LD). Once loaded, the central extension module (ketosynthase [KS], acyltransferase [AT], ketoreductase [KR], ACP) would add two carbons from malonyl-coenzyme A (CoA) (a malonyl-CoA recognition motif was found with in the AT domain) (Fig. 2a) to the acyl-substrate and reduce the β-keto group to a β-hydroxyl. The presence of a sulfotransferase (ST) domain adjacent to ACP2 suggests that it activates the β-hydroxyl group via sulfation. Activation is required to drive subsequent dehydration and decarboxylation reactions that could be catalyzed by the C-terminal thioesterase (TE) domain analogous to the final reactions performed by CurM.
Fig. 2.
(a) Domain organization and proposed mechanism of a putative olefin synthase encoded by ols. (b) Partial sequence alignments of Ols with consensus polyketide synthase domain motifs. Numbers correspond to the amino acid positions in Ols. References are in parentheses. Ppant, phosphopantetheine.
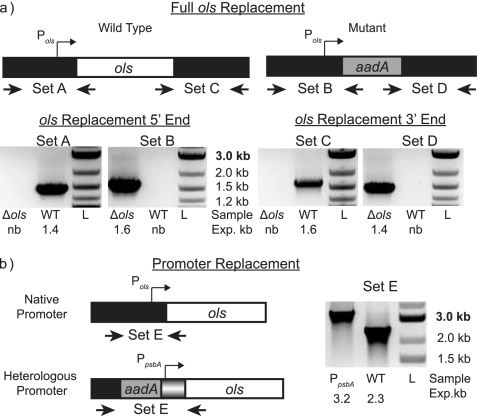
To confirm the involvement of ols in α-olefin biosynthesis, a fully segregated null mutant of the ols gene was made by homologous recombination of a linear DNA fragment containing a streptomycin resistance cassette flanked by 1,000 bases homologous to the regions flanking ols (7). Mutants were verified by PCR (Fig. 3). Table 1 gives the oligonucleotide sequences used in construction and verification. The mutant strain (Δols) was grown under conditions identical to those used for the wild-type strain and subjected to hydrocarbon analysis. Hydrocarbon extracts of Δols did not contain any detectable olefins (Fig. 1). Moreover, a deletion of only 1,000 bp at the 5′ end of the gene corresponding to the putative loading domain gave identical results (not shown). No significant differences were observed when the fatty acid profiles of the wild type and the mutant strain were compared (not shown).
Fig. 3.
Confirmation of PCC 7002 mutants by PCR. (a) Primers amplifying the 5′ and 3′ junctions of ols (sets A and C) and the expected integrated resistance cassette (sets B and D) were used to generate PCR products specific to each strain. Gels of each PCR confirm the replacement of ols with a streptomycin resistance cassette (aadA). (b) Primers flanking the promoter of ols were used to confirm the size of the genomic sequence on both the wild type and the Φ(PpsbA-ols) mutant. The larger size of mutant PCR product is due to the presence of an aadA expression cassette positioned upstream of PpsbA. nb, no band; exp, expected.
Table 1.
Oligonucleotides useda
| No. | Oligonucleotide name | Sequence |
|---|---|---|
| 1 | OFE7002-Sp-a2 | CCGTTCTGCAGCCTGTGAATGGAAATTCTGGACTCCGTATCC |
| 2 | OFE7002-Sp-a1 | CCAACCGGAGGTTGAAGCGGACTACA |
| 3 | OFE7002-Sp-BamHI-b2 | CCGTTGGATCCGCAAAGTGCAGTCCGAAAACCCGTAAATATTAGATCC |
| 4 | OFE7002-Sp-BamHI-b1 | GCCAAGTCAAAGGGTTTCTGGGCATGG |
| 5 | OFE7002-Sp-b2 | CCGTTGAATTCCAAGGGACAGAAACAACGGTGACCTTGG |
| 6 | OFE7002-Sp-b1 | GGGAAAACGACAACTGAGACCCACCAC |
| 7 | Prom-sw-a2 | CCAGAATTCCGGAGCTTCATCCTGGGGACAATGG |
| 8 | Prom-sw-a1 | GCTTTCAGCCCACCTGTTCCCAATATGC |
| 9 | Prom-sw-b1 | CCAAGGTCACCGTTGTTTCTGTCCCTTG |
| 10 | Prom-Amar-b2-1 | GAGACAGGATGAGGATCGTTTCGCATGGTTGGTCAATTTGCAAATTTCGTCGATCTGC |
| 11 | Prom-Amar-b2-2 | CTGTTGAATAACAAGGACGGATCTGATCAAGAGACAGGATGAGGATCGTTTCGCATG |
| 12 | Prom-Amar-b2-3 | GTTGACACGGGCGTATAAGACATGTTATACTGTTGAATAACAAGGACGGATCTGATCAAG |
| 13 | Prom-Amar-b2-4 | CCACTGCAGGATCTCAATGAATATTGGTTGACACGGGCGTATAAGACATGTTATACTG |
| 14 | Gaz7002-Seq2-Rv | CGTTGATCGCCTTTAGCCACC |
| 15 | aadA-Rv2 | GCAAGATAGCCAGATCAATGTCGATCGTG |
| 16 | Gaz7002-Seq11 | CCCAAAGACCTCTCGGCGTTC |
| 17 | aadA-Fw2 | GACATTCTTGCAGGTATCTTCGAGCCAGC |
| 18 | SYNPCC7002_A1173-RVT | TTATTGTGTTTTGGGTACAGG |
| 19 | petB-RVT | TTACAAAGGACCAGAAATACC |
| 20 | SYNPCC7002_A1173-RTF | TGGCATTAGCAGACGACGTTACCT |
| 21 | SYNPCC7002_A1173-RTR | TGGAGATCAGCAGGGCGGTTAAAT |
| 22 | petB-RT-Fw | GATTCGCAATGACCTTCTAC |
| 23 | petB-RT-Rv | CCAGTAATCCAAGTCAGCTC |
Restriction sites used in cloning are underlined. Oligonucleotides 1 to 4 were used for making the ols knockout cassette, oligonucleotides 1, 2, 5, and 6 for making the loading domain knockout cassette, oligonucleotides 10 to 13 for making the Amaranthus promoter (PpsbA) replacement cassette, oligonucleotides 7 to 9 and 13 for promoter replacement, oligonucleotides 14 to 17 for screening of mutant strains, and oligonucleotides 18 to 23 for quantitative reverse transcription-PCR (qRT-PCR).
To demonstrate a positive correlation between the ols gene and olefin production, the 250 bases immediately upstream of the ols coding sequence were replaced with the sequence that controls transcription of psbA in Amaranthus hybridus (6). A fully segregated mutant harboring the promoter replacement Φ(PpsbA-ols) was obtained and verified by PCR (Fig. 3). Hydrocarbon extracts from cultures of the mutant strains contained significantly increased titers of each olefin (Fig. 1). A 2-fold increase in 1-nonadecene production and a 5-fold increase in 1,14-nonadecadiene were observed in cultures grown at 35°C in medium A (Table 2). mRNA was extracted from each culture using the Trizol 95 method (14). Quantitative PCR of ols mRNA, using primers that amplified a short 104-bp segment at the 3′ end of ols, confirmed a 2.2-fold increase in mRNA in the promoter replacement mutant relative to the level for wild-type PCC 7002 (Table 2).
Table 2.
Olefin production and olefin synthase expressiona
| Description or genotype | Estimated C19:2 concn (μg/ml/OD730 unit)b | C19:1 concn (μg/ml/ OD730 unit) | RNA level relative to WT levelc |
|---|---|---|---|
| WT | 0.15 ± 0.06 | 1.60 ± 0.24 | 1.00 ± 0.10 |
| Δols | ND | ND | NA |
| Δols-LDd | ND | ND | 0.07 ± 0.01 |
| Φ(PpsbA-ols) | 0.75 ± 0.13 | 3.45 ± 0.71 | 2.20 ± 0.30 |
ND, not detected; NA, not applicable. Data represent averages of results from three biological replicates ± standard deviations.
Concentrations of 1,14-nonadecadiene were estimated from a dilution series of 1-nonadecene analytical standards.
ols RNA levels, determined by quantitative PCR (qPCR), were normalized to the amount of petB mRNA in each sample and compared to the wild-type-PCC7002 ratio.
Loading domain disruption mutant.
Further analysis of hydrocarbon extracts of the wild-type and mutant strains revealed trace amounts of two additional hydrocarbons (at 12.6 and 12.8 min) that also disappeared in extracts of the Δols strain and increased in extracts of the Φ(PpsbA-ols) mutant (Fig. 4). We identified the latter of the two compounds as 1-octadecene by its mass spectrum and by comparison with a commercial standard. The mass spectrum of the first compound was consistent with octadecadiene, but the compound was present in insufficient quantities to confirm its structure. On the basis of the proposed mechanism for α-olefin formation, we hypothesized that heptadecanoic acid (C17:0) might be the substrate for 1-octadecene formation. In order to test this hypothesis, we fed C17:0 to each of the three strains (final concentration, 0.1 mM) and cultures were grown to an OD730 of 0.3. We observed increases in the peak areas for 1-octadecene in both the wild-type and the Φ(PpsbA-ols) strains (not shown). These results, combined with the fact that fatty acids no larger than C18 have been observed in PCC 7002 (13), suggest that elongation of fatty acids is required for α-olefin formation. Interestingly, feeding of pentadecanoic acid (C15:0) did not result in the formation of 1-hexadecene but in an increase of 1-octadecene as well (Fig. 4). PCC 7002 contains an ortholog of an acyl-ACP synthetase responsible for activation of exogenous free fatty acids (11). Therefore, it is likely that an acyl-ACP is the direct substrate of the ols gene product. Further in vitro work will be required to confirm this hypothesis.
Fig. 4.
Comparison of hydrocarbon extracts from wild type (WT), ols deletion mutant (Δols), and Φ(PpsbA-ols) strains of PCC 7002 supplemented with pentadecanoic (C15:0) acid. The GC-MS signal was normalized to the height of an internal standard peak (hexadecane, 8.3 min). Supplementation of cultures with heptadecanoic (C17:0) resulted in similar traces (not shown). Supplementation of odd chain fatty acids resulted in increased production of 1-octadecene (C18:1) and a compound consistent with a doubly unsaturated 18-carbon hydrocarbon (C18:2). No peaks corresponding to a 1-hexadecene analytical standard were observed in any of the extracts.
In summary, we have identified a novel biological route for producing hydrocarbons in cyanobacteria. While the highest observed hydrocarbon titer, approximately 4.2 μg/ml/OD730 unit, was suboptimal for commercial production, increasing the supply of fatty acid substrates via metabolic engineering and enhancing the rate of carbon dioxide uptake may increase olefin titers. In addition, the combinatorial nature of polyketide synthesis raises the possibility of using CurM or Ols to produce a wide range of industrially relevant α-olefins. The N-terminal loading domain's substrate specificity could be altered to prefer shorter-chain-length fatty acids, thereby generating olefins with sizes similar to those of diesel constituents. Alternatively, the existing loading domain could be replaced with modules from other natural product pathways that act on short organic and amino acids to yield higher-value chemical building blocks.
Supplementary Material
Acknowledgments
We acknowledge Spencer Hoover, Ben Caes, Ron Raines, and John Yin for their contributions.
This work was supported with funding provided by the University of Wisconsin—Madison Graduate School and instrument time provided by the DOE Great Lakes Bioenergy Research Center (GLBRC; DOE Office of Science BER DE-FC02-07ER64494).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Aparicio J. F., et al. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9–16 [DOI] [PubMed] [Google Scholar]
- 2. Beller H. R., Goh E. B., Keasling J. D. 2010. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl. Environ. Microbiol. 76:1212–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Black P. N., Dirusso C. C., Metzger A. K., Heimert T. L. 1992. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme-A synthetase. J. Biol. Chem. 267:25513–25520 [PubMed] [Google Scholar]
- 4. Chang Z., et al. 2004. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 67:1356–1367 [DOI] [PubMed] [Google Scholar]
- 5. Donadio S., Katz L. 1992. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111:51–60 [DOI] [PubMed] [Google Scholar]
- 6. Elhai J. 1993. Strong and regulated promoters in the cyanobacterium Anabaena PCC 7120. FEMS Microbiol. Lett. 114:179–184 [DOI] [PubMed] [Google Scholar]
- 7. Frigaard N. U., Sakuragi Y., Bryant D. A. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum. Methods Mol. Biol. 274:325–340 [DOI] [PubMed] [Google Scholar]
- 8. Goodloe R. S., Light R. J. 1982. Structure and composition of hydrocarbons and fatty acids from a marine blue-green alga, Synechococcus sp. Biochim. Biophys. Acta 710:485–492 [Google Scholar]
- 9. Gu L. C., et al. 2009. Polyketide decarboxylative chain termination preceded by O-sulfonation in curacin A biosynthesis. J. Am. Chem. Soc. 131:16033–16035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haydock S. F., et al. 1995. Divergent sequence motifs correlated with the substrate-specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide syntheses. FEBS Lett. 374:246–248 [DOI] [PubMed] [Google Scholar]
- 11. Kaczmarzyk D., Fulda M. 2010. Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 152:1598–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lennen R. M., Braden D. J., West R. M., Dumesic J. A., Pfleger B. F. 2010. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol. Bioeng. 106:193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parker P. L., Van Baalen C., Maurer L. 1967. Fatty acids in 11 species of blue-green algae—geochemical significance. Science 155:707–708 [DOI] [PubMed] [Google Scholar]
- 14. Pinto F. L., Thapper A., Sontheim W., Lindblad P. 2009. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rude M. A., et al. 2011. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus sp. Appl. Environ. Microbiol. 77:1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schirmer A., Rude M. A., Li X., Popova E., del Cardayre S. B. 2010. Microbial biosynthesis of alkanes. Science 329:559–562 [DOI] [PubMed] [Google Scholar]
- 17. Scrutton N. S., Berry A., Perham R. N. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38–43 [DOI] [PubMed] [Google Scholar]
- 18. Stevens S. E., Jr., Patterson C. O., Myers J. 1973. Production of hydrogen-peroxide by blue-green-algae: a survey. J. Phycol. 9:427–430 [Google Scholar]
- 19. Vincent M., Guglielmetti G., Cassani G., Tonini C. 1987. Determination of double-bond position in diunsaturated compounds by mass spectrometry of dimethyl disulfide derivatives. Anal. Chem. 59:694–699 [Google Scholar]
- 20. Winters K., Parker P. L., Van Baalen C. 1969. Hydrocarbons of blue-green algae: geochemical significance. Science 163:467–468 [DOI] [PubMed] [Google Scholar]
- 21. Yong T. P. C., Largeau C., Casadevall E. 1986. Biosynthesis of non-isoprenoid hydrocarbons by the microalga Botryococcus braunii—evidences for an elongation-decarboxylation mechanism—activation of decarboxylation. Nouv. J. Chim. 10:701–707 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.