Abstract
We developed a new set of software tools that enable the speed and response kinetics of large numbers of tethered bacterial cells to be rapidly measured and analyzed. The software provides precision, accuracy, and a good signal-to-noise ratio combined with ease of data handling and processing. The software was tested on the single-cell chemosensory response kinetics of large numbers of Rhodobacter sphaeroides cells grown under either aerobic or photoheterotrophic conditions and either in chemostats or in batch cultures, allowing the effects of growth conditions on responses to be accurately measured. Aerobically and photoheterotrophically grown R. sphaeroides exhibited significantly different chemosensory response kinetics and cell-to-cell variability in their responses to 100 μM propionate. A greater proportion of the population of aerobically grown cells responded to a 100 μM step decrease in propionate; they adapted faster and showed less cell-to-cell variability than photosynthetic populations. Growth in chemostats did not significantly reduce the measured cell to cell variability but did change the adaptation kinetics for photoheterotrophically grown cells.
INTRODUCTION
The process by which bacteria bias their movement toward regions that contain higher concentrations of favorable chemicals and lower concentrations of unfavorable chemicals is known as chemotaxis (39). Motility and chemotaxis are widespread among bacterial species and are essential for many processes, such as the establishment of symbioses (14, 18, 19), biofilm formation (37), and virulence (12, 40). Due to their small size, bacteria use temporal sensing to bias their overall direction of movement (32). Bacteria swim by rotating semirigid, helical flagella and in a homogeneous environment change direction every few seconds. In the extensively studied enteric bacterium Escherichia coli, these changes are caused by transient changes in the direction of flagellar motor rotation, triggering a tumble. During these periods, bacteria reorient, and when swimming resumes it is usually in a new direction, so that, overall, their movement is random (38). In a nonhomogeneous environment, the reversal frequency is modulated by the chemotaxis pathway to bias the overall direction of swimming in a favorable direction. While chemotaxis in other bacteria is based on principles similar to those described in E. coli and most chemotactic bacteria have the components found in the E. coli single pathway, many bacteria have more than one pathway regulating their motors (13, 26, 28). One of the best-studied bacteria with a complex chemotaxis pathway is Rhodobacter sphaeroides (25, 26).
R. sphaeroides is a purple nonsulfur alphaproteobacterium that can grow using either aerobic or anaerobic respiration or photosynthesis and is a model organism for the study of complex chemotactic networks (16, 25, 26). This bacterium uses a single stop-start flagellar motor, stopping rotation rather than tumbling, and shows taxis to a wide range of stimuli, including sugars, light, oxygen, and organic acids, such as acetate and propionate. Interestingly, responses to certain stimuli such as oxygen and light depend on growth conditions (15, 27, 30), indicating that R. sphaeroides is able to tune its tactic responses to the environmental conditions.
Both aerobic and photoheterotrophic populations show chemotaxis, despite apparent differences in expression levels of the chemotaxis proteins. To understand the effect of these differences on the behavior of single cells, rather than populations, it is essential to be able to accurately analyze, on a single-cell level, the chemosensory response kinetics of large numbers of cells.
Many different assays have been developed to study bacterial chemotaxis. On the population level, capillary (2) and swim plate (1) assays have been widely used. At the single-cell level, three-dimensional tracking of a free swimming cell (6) and tracking of a bead attached to the flagellum filament using a quadrant photodiode (8) or back-focal-plane interferometry (31) allow quantitative data on a single cell to be obtained. To analyze large numbers of individuals from within a single population, analysis of tethered cell rotation (34) or recently developed microfluidic technologies (3, 4, 11) are the techniques of choice. Tethered cell analysis depends on attaching a bacterial cell to a microscope slide by its flagellum, usually using antiflagellar antibody. The behavior of the motor, and thus the response, is determined by tracking the rotation of the cell body in response to changing stimuli. Tracking the rotation of tethered cells allows single-cell analysis of multiple cells from within a population and has been a key technique in quantifying fundamental properties of the E. coli chemotaxis system such as tumbling frequency, run lengths or response kinetics and sensitivities (7, 9, 10, 32). In the late 1980s, this technique was successfully adapted for R. sphaeroides (24) and has yielded invaluable information on the R. sphaeroides chemotaxis system (17, 20, 27, 30, 33). However, quantitative measurements require multiple cells to be analyzed and the data on responses needs to be extracted rapidly and accurately across populations, and the above techniques were slow and labor-intensive.
In the present study we have developed software that allows the tracking of large numbers of tethered single cells, extracting reliable, simultaneous quantitative data on their response kinetics. This improved technique shows that R. sphaeroides cells grown under aerobic or photoheterotrophic conditions have different behaviors and variabilities in responses to stimuli, something not identified by other methods.
MATERIALS AND METHODS
Growth conditions.
R. sphaeroides WS8N (36) was grown in succinate medium (35) at 30°C and harvested in mid-exponential phase (optical density at 700 nm [OD700] between 0.45 and 0.55) when cells are very motile. This ensures limited self-shading in photosynthetic conditions and ensures oxygen saturation for aerobic batch cultures.
Batch cultures were grown either aerobically in the dark in 250-ml flasks containing 50 ml of medium shaken at 255 rpm or photoheterotrophically without shaking, in airtight 25-ml flasks illuminated with white light at low intensity (5 W/m2).
Chemostat cultures were grown in continuous mode in a 2.5-liter BioFlo 310 Fermentor from New-Brunswick Scientific (working volumes, 0.8 or 1.5 liter). Succinate medium was inoculated with stationary phase batch cultures and mixed at 80 rpm. For photosynthetic cultures, cells were initially grown in the dark without sparging to consume the oxygen in the medium. When the oxygen was depleted, the cells were illuminated with white light at low intensity (10 W/m2) without gas sparging. Aerobic cultures were grown without illumination and with air sparging at 5 standard liters per min. Once the cultures reached an OD700 of 0.45 ± 0.55, fresh succinate medium was fed into the culture vessels at dilution rates equivalent to the cell maximum growth rate of 0.11 ± 0.01 h−1 for photosynthetic cultures and 0.52 ± 0.05 h−1 for aerobic cultures. Samples were collected at steady state, during which the OD700 was between 0.45 and 0.55 in both cultures, the pH was constant at 7.29 ± 0.01 and 7.54 ± 0.02, and the dissolved oxygen in the medium was equal to 0.45% ± 0.25% and 20.50% ± 0.30% in photosynthetic and aerobic cultures, respectively. The light intensity was measured by using a Skye Instrument SKP200 light meter. Light intensities in photosynthetic-chemostat and batch conditions were chosen to have equivalent amounts of photosystem pigments synthesized in both conditions (measured by spectrophotometry).
Cell tethering.
Portions (1 ml) of cells in mid-exponential phase were harvested, washed, and resuspended in tethering buffer (10 mM Na-PIPES containing chloramphenicol at 30 μg/ml to prevent further protein synthesis). The cells were tethered by their flagella (without flagellum shearing, since R. sphaeroides is monoflagellate) onto a coverslip by adding 10 μl of cell suspension with 2 μl of 10,000-fold-diluted anti-flagellum antibody, which spontaneously adsorb to the coverslip glass. After 20 min of incubation in a humidity chamber, the coverslip was inverted onto a microscope flow chamber, and tethering buffer was flowed through. Cells were observed and recorded using ×40 magnification under phase contrast (Nikon Optiphot phase-contrast microscope). Solutions were flowed through the chamber at a rate of 0.12 ml/min. For each bacterial population studied, at least three biological replicates were analyzed.
RESULTS
Video capture and image analysis.
Tethered cells were recorded by using a digital DALSA Genie-HM640 camera, capable of high-speed image acquisition (up to 295 fps). A digital high-speed camera increased the acquisition rate over that of analogue cameras and improved rotation speed determination and eliminated possible stroboscopic effects. Movies were usually acquired and recorded at 100 fps with an exposure time of 6 ms. This avoids stroboscopic effects while keeping the size of each movie reasonable. Movies were archived on network storage.
New software, BRAS (for bacterial rotation analysis software), was written to analyze the movies and extract the motion of individual cells. This software allows analysis of the motion of an unlimited number of nonoverlapping single cells per field of view (Fig. 1A). Written in Python, this software relies on different open-source libraries: OpenCV (http://opencv.willowgarage.com/wiki/) for image extraction and manipulation, NumPy (http://numpy.scipy.org/) for data processing, and matplotlib (http://matplotlib.sourceforge.net/) for graphical outputs.
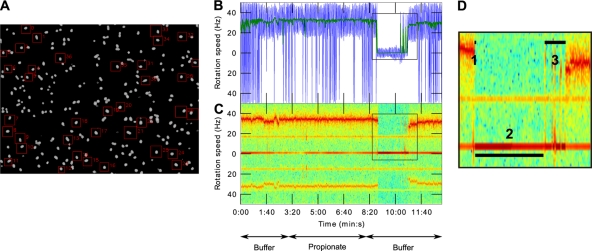
Fig. 1.
Single-cell level chemosensory response analysis using the improved computer tracking of tethered rotating cells. (A) Video frame of a microscope field of view (×40 magnification) of tethered bacteria whose motions are to be analyzed using BRAS. The regions used for motion analyses are delimited by red squares. (B and C) “Click & mean”-generated graphical representations of a single-cell rotation speed over time. Arrows indicate the solution in the microscope chamber. (B) Instantaneous speed measurements. Colors: blue, raw 100-Hz signal; green, moving average of the same signal using a Gaussian window of 1 s. (C) Frequency domain analysis of bacterial motion used to extract rotation speeds. The black squares highlight a chemosensory response to propionate removal. (D) Enlargement of the chemosensory response represented in panel C. The numbers show the different phases of a chemosensory response which can be measured using a “click & mean” interactive interface. Numbered segments: 1, duration of the stopping phase; 2, duration of the stopped phase; 3, duration of the recovery phase. The total chemosensory response duration equals 1+2+3.
BRAS works in two phases. The first is interactive and allows the user to choose an unlimited number of windows of interest (Fig. 1A). Each window defines a region containing the single cell to be analyzed. The size of the windows is variable and can be adapted to the size of the bacterium. The user sets a level threshold to remove the video background. The cells are thus the only objects visible in the processing windows. To allow for stage drift, the movie can be viewed at different time points, and the window size can be optimized. When the user has selected the bacteria for analysis, the noninteractive phase iterates over all the frames of the movie. For each time slice, the image is cut into smaller images defined by the windows chosen in the previous step. In each window the position of the center of mass is calculated. Finally, BRAS stores the coordinates of each bacterium, within their window coordinate system, for every frame.
BRAS has been designed to run on a normal desktop computer: it analyzes each frame independently, using little memory. The analysis time is determined by the data transfer rate: on a standard local hard drive, a 15-min movie (100 fps, 800 × 600 resolution) would be analyzed in ∼5 min, independently of the number of analyzed bacteria. A typical 15 min movie of 20 tethered bacteria to be analyzed would generate 20 MB of useful tracking data from a 25-GB video file.
Signal processing.
Two analysis methods were developed to extract the rotation speed from the position of the bacterium's center of mass. The first uses the coordinates to calculate the angle of the bacteria: θ = arctan(Y/X). This angle is then differentiated to obtain the rotation speed: f = (1/2π)(∂θ/∂t). This method may produce a noisy signal that can then be filtered by using a moving average technique: a convolution is performed between the signal and a Gaussian shape window. The second method transforms the time series of the two coordinates into a complex signal: S = X + jY then uses short-time Fourier transforms to generate a spectrogram of the complex signal. The spectrogram generated shows the change of the spectral components over time, i.e., the rotation speed of the analyzed bacteria. This analysis is much less noisy than phase difference methods and allows unambiguous determination of the rotation speed. In the experiments described here, the Fourier transforms were performed using 128 point windows.
Using these two methods, a graphical interface referred to as “click & mean” was developed to allow the output from the two methods to be presented and analyzed (Fig. 1B and C). Time of interest intervals can be graphically selected, and the average speeds as well as the direction of rotation over these intervals was determined by using either method (a positive speed corresponds to an anticlockwise rotation and a negative speed to a clockwise rotation). For the angle difference analysis, a mean of the rotation speed over the chosen interval is calculated, whereas for the spectral analysis a discrete Fourier transform over the chosen interval is performed. This set of analysis programs reduces the large video files to the relevant biological parameters and enables easier analysis of the bacterial behavior. The capacity for analysis of many bacteria from a single movie allows statistically robust studies to be performed both easily and relatively cheaply. All software and associated documentation can be downloaded (www.sysbio.ox.ac.uk).
Chemosensory response and speed analysis.
The developed software was tested on the responses of R. sphaeroides grown under different environmental conditions. To study chemotaxis responses, solutions were flowed through the microscope chamber, and changes in the tethered cell rotation were analyzed. The cells were initially equilibrated in tethering buffer to give background behavior and then exposed to 5 min of 100 μM propionate, followed by 5 min of tethering buffer, equivalent to a stepdown in attractant concentration. As shown in Fig. 1B and C, in the initial tethering buffer and in propionate, the rotational behavior is similar. The major chemosensory response in R. sphaeroides is to a stepdown in attractant concentration and, as expected, on propionate removal the cells stop rotating. They remain stopped for a time and then start to rotate again as they adapt to the stimulus. This sequence of events corresponds to a chemotactic response to a decrease in attractant concentration (Fig. 1B and C).
To test the ability of the software to accurately measure these responses, tethered populations were given a 100 μM step decrease in propionate concentration, and speed was measured by Fourier transformation analyses using “click and mean.” Bacteria were divided into two categories: (i) unresponsive cells showing no stop within 2 min of the step decrease in propionate concentration and (ii) responsive cells showing a stop within 2 min of the step decrease.
For responsive cells, cells are additionally classified as (i) cells adapting, i.e., cells returning to their initial pattern of rotation within 5 min of the step decrease in propionate and (ii) cells that did not adapt, i.e., cells that did not return to their initial pattern of rotation within 5 min.
For cells showing adaptation, the chemosensory response was further divided into three phases: (i) the stopping phase corresponding to the transition time between cell rotation and the stop, (ii) the stopped phase during which cells do not rotate, and (iii) the recovery phase corresponding to the time from the cells starting to rotate and a return to their initial pattern of rotation (Fig. 1D). The chemotaxis response duration is equal to the sum of these three phases (Fig. 1C). Dividing the response into three phases allows information to be extracted about the different stages of a response: the stopping phase is related to the sensing and transduction of the stimulus, whereas the stopped and recovery phases are related to the adaptation of the cells to the stimulus.
Each time trace, corresponding to a single bacterium's displacement over time, was individually processed by using “click & mean” to extract the instantaneous rotation speed of each bacterium using two independent methods described above. The interactive visualization interface of “click & mean” subsequently allows the selection and measurement of periods of interest during the responses with either the instantaneous rotation speed measurements or the frequency domain analysis graphical representations. In the present work, the latter representation was preferred since it generally gave the clearest results.
This new approach has several advantages. (i) It allows convenient data storage and handling. (ii) Data are easily processed, and an unlimited number of cells per field of view can be analyzed, increasing the sample size and thus the accuracy of the study. (iii) The camera's fast acquisition frame rate improves time resolution, increasing both precision of the chemotaxis response phase measurements and cell rotation speed determination. Rotation speed measurements obtained in the present study showed that tethered R. sphaeroides cells can rotate at up to 44 Hz, confirming the need for a high acquisition frame rate to limit potential stroboscopic effects. (iv) “Click & mean” allows a precise measurement of the duration of each chemotaxis response phase.
Comparison of “click & mean” with previous software.
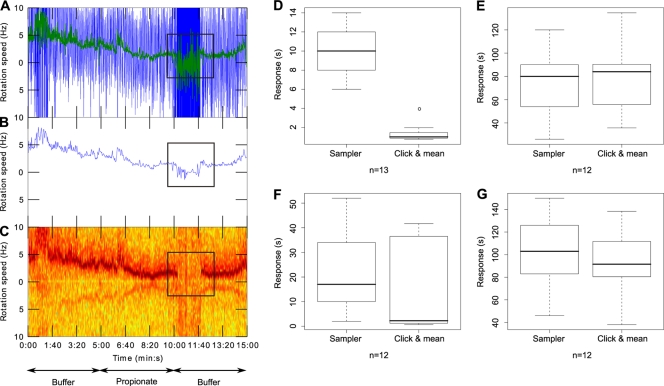
To test the software, we analyzed videos of tethered cells recorded with an analog camera and tracked with the previous “AROT7” image analysis software (Hobson Tracking System, Ltd., Sheffield, England) and the “Sampler” system (which uses a 100-point, 2-s, moving average to smooth the data) and compared this to “click & mean.” Processing the raw speed measurements (Fig. 2 A) with “Sampler” increases the signal/noise ratio but does not allow a precise determination of the chemosensory responses kinetics (Fig. 2B), whereas the “click & mean” frequency domain analysis extracts a cleaner rotation speed from the raw data (Fig. 2C). As a result, stopping- and recovery-phase durations, which are generally short, are overestimated when the previous “Sampler” system is used compared to the “click & mean” approach (Fig. 2D and F), unlike the longer stopped phase (Fig. 2E), and consequently the overall chemosensory responses are slightly shorter and show a lower spread when analyzed using “click & mean” versus “Sampler” (Fig. 2G).
Fig. 2.
Comparison of chemosensory responses analyzed with the “Sampler” software or with “click & mean.” Tethered cell rotation was recorded with an analog camera at 50 fps and processed with AROT7 software. Panels A, B, and C depict the findings for single-cell rotation speed over time. Arrows indicate the solution in the microscope chamber. Black squares highlight the chemosensory response to propionate removal. (A) Instantaneous speed measurements. Colors: blue, raw signal at 50 Hz; green, moving average of the raw signal using a window of 1 s. (B) Signal after “Sampler” processing (window averaging of 100 points). (C) Frequency domain analysis. (D, E, F, and G) Box plots of the different phases of the chemosensory responses and of the overall chemosensory responses obtained with the previous “Sampler” system versus the new “click & mean” system. For each box plot, the lower, middle, and upper horizontal lines of the box represent the first quartile, the median, and the third quartile, respectively. The lower and upper extremities of the dashed lines represent the lowest datum and the highest datum, respectively, which are no more than 1.5 times the interquartile range (difference between the third and the first quartiles) from the box. The circles represent data that are outliers. “n” represents the number of cells analyzed in each panel. The individual panels represent the stopping phase (phase 1) (D), the stopped phase (phase 2) (E), the recovery phase (phase 3) (F), and the overall chemosensory response (G).
Chemotactic responses of aerobic and photoheterotrophic cells.
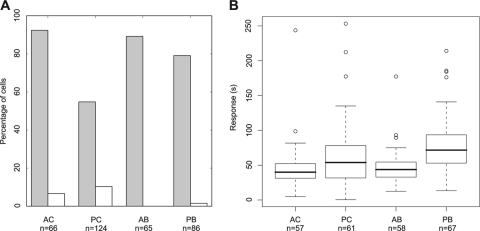
We used the new software to test the stepdown responses of large numbers of tethered R. sphaeroides grown under different conditions: (i) aerobic chemostat grown (AC), (ii) aerobic batch grown (AB), (iii) photoheterotrophic chemostat grown (PC), and (iv) photoheterotrophic batch grown (PB). About 90% of cells grown under aerobic conditions showed a stop response to propionate removal (92.4% for AC-grown cells and 89.2% for AB-grown cells), whereas only 55.8% of PC-grown cells and 76.1% of PB-grown cells responded. Among the responsive cells, the large majority of stopping cells adapted, although for AC-, PC-, and PB-grown cells, a small proportion of cells stopped but did not adapt within the time frame of the experiment (Fig. 3 A).
Fig. 3.
Chemosensory responses of bacterial populations grown in AC (aerobic chemostat), PC (photoheterotrophic chemostat), AB (aerobic batch), and PB (photoheterotrophic batch) conditions. “n” is the number of cells analyzed for each condition. (A) Total percentage of responsive cells (gray bars) and percentage of responsive cells not adapting (white bars); (B) box plots of the chemosensory response durations of the responsive adapting cells. For each box plot, the lower, middle, and upper horizontal lines of the box represent the first quartile, the median, and the third quartile, respectively. The lower and upper extremities of the dashed lines represent the lowest datum and the highest datum, respectively, which are no more than 1.5 times the interquartile range (difference between the third and the first quartiles) from the box. The circles represent data that are outliers.
Comparison of aerobically grown cells with photoheterotrophically grown cells showed that cell-to-cell variability in the chemosensory responses for aerobically grown cells was ∼2-fold lower than for photoheterotrophically grown cells (Fig. 3B). Aerobically grown cells also had shorter chemosensory responses than photoheterotrophically grown cells (Fig. 3B), with PB-versus-AB and PC-versus-AC populations being significantly different (Wilcoxon rank-sum tests, P < 0.05), showing a shorter adaptation time when cells are grown aerobically.
Effect of culturing method on responses.
Bacteria grown under aerobic conditions (AC or AB) showed similar chemosensory response durations (Wilcoxon rank-sum test, P > 0.05) and spreads (Fig. 3B). This indicates that aerobic growth in a chemostat or in batch culture does not affect the overall chemosensory response durations or cell-to-cell variability. Photoheterotrophically grown cells showed similar chemosensory response spreads for PC- and PB-grown cells (Fig. 3B). However, PB-grown cells showed a significantly longer chemosensory response than PC-grown cells (Wilcoxon rank-sum test, P < 0.05) (Fig. 3B). This suggests that batch-growth conditions do not increase cell-to-cell variability under photoheterotrophic conditions but significantly increase the duration of the chemosensory response.
Rotation rates and adaptation time.
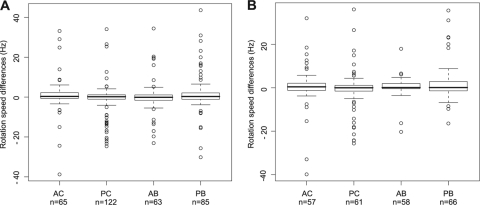
The method used here to study chemosensory responses does not allow determination of the absolute flagellar motor speed of rotation since parameters such as the length of the cell influence the speed of rotation of tethered cells. However, given that each individual bacterium is considered separately in this analysis, we investigated whether the rotation speed of an individual cell varies upon addition of attractant. When we compared the speed of rotation in propionate with the speed in the first tethering buffer for each individual cell (Fig. 1C and Fig. 4 A), we found that R. sphaeroides cells rotated at a constant speed and hence did not show chemokinesis to the attractant propionate. Moreover, comparison of each individual cell speed of rotation in propionate with the second tethering buffer indicates that adapting cells returned to their basal rotation rate after the stimulus, suggesting exact adaptation to the stimulus under all four growth conditions (Fig. 4B).
Fig. 4.
Box plots representing differences in speeds of rotation in propionate and tethering buffer of single cells grown in AC (aerobic chemostat), PC (photoheterotrophic chemostat), AB (aerobic batch), and PB (photoheterotrophic batch) conditions. For each box plot, the lower, middle, and upper horizontal lines of the box represent the first quartile, the median, and the third quartile, respectively. The lower and upper extremities of the dashed lines represent the lowest datum and the highest datum, respectively, which are no more than 1.5 times the interquartile range (difference between the third and the first quartiles) from the box. The circles represent data that are outliers. “n” represents the number of cells analyzed for each condition. (A) Differences between single-cell rotation speeds in propionate and in the first tethering buffer flow; (B) differences between single-cell rotation speeds in propionate and in the second tethering buffer flow.
DISCUSSION
In the present study, computer tracking of tethered rotating cells was improved and used to accurately analyze the chemosensory response kinetics of large numbers of individual R. sphaeroides cells within a population.
Previous tethered cell analysis methods were unable to measure large numbers of cells and also had problems regarding precision, accuracy, signal-to-noise ratio, and data handling and processing. The use of a fast digital camera allows easier data handling and processing and also increases the acquisition frame rate, improving the precision of measurement of chemosensory response durations and rotation speeds. The new image analysis software BRAS enables easy and fast processing of an unlimited number of individual cells from the movies, considerably increasing the overall number of cells studied in a single experiment. This software is also able to track different shapes of cells: aerobic R. sphaeroides cells are rod-shaped, whereas photosynthetic grown cells are much rounder and smaller. The new “click & mean” software also allows reliable measurements, based on frequency domain analysis, of tethered single-cell rotation speeds (and, if required, of the direction of cell rotation) with a high signal-to-noise ratio. This improved computer tracking of tethered cells thus allows (i) the processing of large numbers of cells, (ii) reliable and precise quantitative analysis of chemosensory responses, and (iii) permits the use of simple and affordable materials, with software freely available online.
Although the current filtering used by “click & mean” means that stops shorter than 100 ms will not be measured, this could be achievable by modifying the filters applied to the raw data generated by “BRAS.” The new software was tested on the single-cell chemosensory response kinetics of large numbers of R. sphaeroides cells grown under aerobic or photosynthetic conditions, either in chemostats or in batch, and showed that R. sphaeroides responds to a step decrease in attractant but does not exhibit chemokinesis. This finding correlates with results obtained in a previous work on R. sphaeroides flagellar motor using bead assays and BFP interferometry (23) and represents a good validation of our improved technique.
Our results show that the chemosensory response kinetics and cell-to-cell variability in responses are affected by the physiology of the cells, although growing cells under highly controlled chemostat conditions or in batch conditions does not significantly influence cell-to-cell variability. Aerobically grown cells showed greater homogeneity than photosynthetically grown cells to a 100 μM step decrease in propionate, with only one-half to two-thirds of photosynthetically grown cells showing a chemotaxis response. Aerobically grown cells also adapt faster than photosynthetically grown cells. Detailed analyses of the chemosensory responses kinetics for aerobic and photosynthetic growth conditions showed significantly more cell-to-cell variability in photosynthetic populations.
Adaptation to the persistence of a stimulus depends on the adaptation proteins, CheB and CheR. Previous studies in E. coli have shown that variations in bacterial chemotaxis behavior are related to cellular levels of CheB and CheR (5, 21). R. sphaeroides has two adaptation systems, and their expression levels vary under different growth conditions (29, 33). The different ratios of adaptation proteins in aerobic versus photoheterotrophic growth conditions could therefore explain the faster adaptation period measured in aerobically versus photoheterotrophically grown cells. Interestingly, however, the different ratios of adaptation proteins in these two growth conditions do not affect exact adaptation. Indeed, a large majority of both aerobically and photosynthetically grown cells show exact adaptation to the stimulus. These findings are consistent with results obtained in E. coli by Alon et al. (5), who showed that adaptation times but not the precision of adaptation vary with protein concentrations. In addition, our results, obtained on a single-cell level, correlate with recent Park et al. (22) findings which indicated that exact adaptation in E. coli occurs at a single-cell level, as well as at a population level (5).
Thus, the improved motion analysis system described here allows us to enhance our understanding of R. sphaeroides complex chemotaxis system by measuring, in large numbers of single cells, fundamental properties of this system, such as adaptation kinetics and robustness or cell-to-cell variability. Given the features of this new analysis system, similar types of studies could easily be performed on other chemotactic bacteria.
ACKNOWLEDGMENT
This research was funded by the UK Biotechnology and Biological Sciences Research Council.
Footnotes
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Adler J. 1966. Chemotaxis in bacteria. Science 153:708–716 [DOI] [PubMed] [Google Scholar]
- 2. Adler J. 1969. Chemoreceptors in bacteria. Science 166:1588–1597 [DOI] [PubMed] [Google Scholar]
- 3. Ahmed T., Shimizu T. S., Stocker R. 2010. Microfluidics for bacterial chemotaxis. Integr. Biol. (Camb.) 2:604–629 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed T., Stocker R. 2008. Experimental verification of the behavioral foundation of bacterial transport parameters using microfluidics. Biophys. J. 95:4481–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alon U., Surette M. G., Barkai N., Leibler S. 1999. Robustness in bacterial chemotaxis. Nature 397:168–171 [DOI] [PubMed] [Google Scholar]
- 6. Berg H. C., Brown D. A. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239:500–504 [DOI] [PubMed] [Google Scholar]
- 7. Berg H. C., Tedesco P. M. 1975. Transient response to chemotactic stimuli in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 72:3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berry R. M., Armitage J. P. 2000. Response kinetics of tethered Rhodobacter sphaeroides to changes in light intensity. Biophys. J. 78:1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Block S. M., Segall J. E., Berg H. C. 1982. Impulse responses in bacterial chemotaxis. Cell 31:215–226 [DOI] [PubMed] [Google Scholar]
- 10. Block S. M., Segall J. E., Berg H. C. 1983. Adaptation kinetics in bacterial chemotaxis. J. Bacteriol. 154:312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Englert D. L., Manson M. D., Jayaraman A. 2009. Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl. Environ. Microbiol. 75:4557–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garvis S., et al. 2009. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog. 5:e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geer L. Y., Domrachev M., Lipman D. J., Bryant S. H. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greer-Phillips S. E., Stephens B. B., Alexandre G. 2004. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J. Bacteriol. 186:6595–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamblin P. A., Maguire B. A., Grishanin R. N., Armitage J. P. 1997. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol. Microbiol. 26:1083–1096 [DOI] [PubMed] [Google Scholar]
- 16. Mackenzie C., et al. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 61:283–307 [DOI] [PubMed] [Google Scholar]
- 17. Martin A. C., Wadhams G. H., Armitage J. P. 2001. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol. Microbiol. 40:1261–1272 [DOI] [PubMed] [Google Scholar]
- 18. Miller L. D., Yost C. K., Hynes M. F., Alexandre G. 2007. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 63:348–362 [DOI] [PubMed] [Google Scholar]
- 19. Millikan D. S., Ruby E. G. 2003. FlrA, a σ54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J. Bacteriol. 185:3547–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Packer H. L., Armitage J. P. 2000. Behavioral responses of Rhodobacter sphaeroides to linear gradients of the nutrients succinate and acetate. Appl. Environ. Microbiol. 66:5186–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park H., Guet C. C., Emonet T., Cluzel P. 2010. Fine-tuning of chemotactic response in Escherichia coli determined by high-throughput capillary assay. Curr. Microbiol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park H., et al. 2010. Interdependence of behavioral variability and response to small stimuli in bacteria. Nature 468:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pilizota T., et al. 2009. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 106:11582–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poole P. S., Sinclair D. R., Armitage J. P. 1988. Real time computer tracking of free-swimming and tethered rotating cells. Anal. Biochem. 175:52–58 [DOI] [PubMed] [Google Scholar]
- 25. Porter S. L., Wadhams G. H., Armitage J. P. 2008. Rhodobacter sphaeroides: complexity in chemotactic signaling. Trends Microbiol. 16:251–260 [DOI] [PubMed] [Google Scholar]
- 26. Porter S. L., Wadhams G. H., Armitage J. P. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 27. Porter S. L., Warren A. V., Martin A. C., Armitage J. P. 2002. The third chemotaxis locus of Rhodobacter sphaeroides is essential for chemotaxis. Mol. Microbiol. 46:1081–1094 [DOI] [PubMed] [Google Scholar]
- 28. Pruitt K. D., Tatusova T., Maglott D. R. 2007. NCBI reference sequences (refseq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roh J. H., Smith W. E., Kaplan S. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1 redox active gene expression profile. J. Biol. Chem. 279:9146–9155 [DOI] [PubMed] [Google Scholar]
- 30. Romagnoli S., Packer H. L., Armitage J. P. 2002. Tactic responses to oxygen in the phototrophic bacterium Rhodobacter sphaeroides WS8N. J. Bacteriol. 184:5590–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ryu W. S., Berry R. M., Berg H. C. 2000. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403:444–447 [DOI] [PubMed] [Google Scholar]
- 32. Segall J. E., Block S. M., Berg H. C. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 83:8987–8991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shah D. S., Porter S. L., Martin A. C., Hamblin P. A., Armitage J. P. 2000. Fine tuning bacterial chemotaxis: analysis of Rhodobacter sphaeroides behavior under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes. EMBO J. 19:4601–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Silverman M., Simon M. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249:73–74 [DOI] [PubMed] [Google Scholar]
- 35. Sistrom W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22:778–785 [DOI] [PubMed] [Google Scholar]
- 36. Sockett R. E., Foster J. C. A., Armitage J. P. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473–479 [Google Scholar]
- 37. Stoodley P., Sauer K., Davies D. G., Costerton J. W. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209 [DOI] [PubMed] [Google Scholar]
- 38. Turner L., Ryu W. S., Berg H. C. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wadhams G. H., Armitage J. P. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 40. Williams S. M., et al. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]