Abstract
Cronobacter (formerly Enterobacter sakazakii) is a recently defined genus consisting of six species, C. sakazakii, C. malonaticus, C. dublinensis, C. muytjensii, C. turicensis, and Cronobacter genomospecies 1. In this study, MboII restriction fragment length polymorphism (RFLP) patterns of O-antigen gene clusters, located between galF and gnd, were used to identify serotypes in Cronobacter spp. Seven O-antigen RFLP clusters were generated, including three C. sakazakii clusters, previously identified as serotypes O1, O2, and O3. The O-antigen regions of six strains with unique RFLP patterns, including two C. sakazakii strains, two C. malonaticus strains, one C. turicensis strain, and one C. muytjensii strain, revealed three O-antigen gene clusters shared among Cronobacter species. PCR assays were developed, targeting the wzx O-antigen polymerase gene, and used to screen 231 Cronobacter strains to determine the frequency of these newly identified serotypes.
INTRODUCTION
Cronobacter spp. are a group of emerging opportunistic food-borne pathogens that causes rare cases of neonatal meningitis, septicemia, and enterocolitis (4, 9, 24, 32, 46). Typical vehicles of infection are temperature-abused, contaminated infant formula and dried food products, and these organisms have been cultured from a variety of food production environments (13, 17, 20). The Cronobacter genus consists of six species, Cronobacter sakazakii, C. malonaticus, C. muytjensii, C. turicensis, C. dublinensis (three subspecies, Cronobacter dublinensis subsp. dublinensis, C. dublinensis subsp. lausannensis, and C. dublinensis subsp. lactaridi), and Cronobacter genomospecies 1, that are distinguishable with the use of biochemical and molecular techniques (19, 41).
The O antigen is a highly variable constituent of the lipopolysaccharide (LPS) of Gram-negative bacteria. It consists of oligosaccharide repeats ranging in size from 2 to 8 sugar residues, and the variability comes from the different sugars present, their specific order in the O-antigen structure, and the chemical linkages between sugar residues (48). In the Enterobacteriaceae, O-antigen gene clusters are primarily located between galF and gnd, contain from 6 to 19 genes, and are usually 6 to 20 kb in length (39). O-antigen regions contain three classes of genes: (i) those encoding enzymes specific to biosynthetic pathways of nucleotide sugars, (ii) glycosyl transferase genes that provide unique linkages between sugar residues, and (iii) O-antigen-processing genes, namely, wzx and wzy, required for assembly and transport of the polysaccharide repeats (39). The conserved yet divergent nature of the glycosyl transferase and O-antigen-processing genes has been utilized in developing PCR assays for serotyping.
Serotyping assays based on PCR specific to O-antigen genes have become acceptable methods for typing Escherichia coli, Salmonella, Shigella, and many other Gram-negative bacteria, and this method was successful in identifying C. sakazakii serotypes O1 and O2 (25, 26, 31, 49). In a recent study, serological typing was used to identify five additional C. sakazakii serotypes, bringing the total number of serotypes for this species to seven (42). Other information available about Cronobacter O antigens comes from chemical characterization studies that have identified the repeat polysaccharide sugar composition of five C. sakazakii isolates, one C. malonaticus isolate, and one C. muytjensii isolate (1, 2, 7, 27–30). Two of the C. sakazakii O-antigen polysaccharide structures were from C. sakazakii O1 and O2 strains, and the structural analyses were in agreement with the O-antigen gene cluster results (1, 2, 31). In addition, there is a third C. sakazakii O-polysaccharide structure that has the same composition but a different pentasaccharide arrangement from the analyzed O1 structure, suggesting a serological relationship (2, 27).
Traditional serological typing is expensive, time-consuming, and labor-intensive. Additionally, commercial antisera are not always available, as is the case with Cronobacter spp., making PCR assays that target serotype-specific genes useful alternatives to serological typing. In this study, we sequenced and characterized additional O-antigen gene clusters in Cronobacter spp. and developed PCR assays specific to these new serotypes.
MATERIALS AND METHODS
Bacterial strains used in this study.
A collection of 231 strains representing the six Cronobacter species was used in this study. Species designations were determined according to the classification scheme suggested by Iversen et al. (19), using the rpoB PCR method developed by Stoop et al. (41). The collection represents isolates from clinical, food, and environmental sources from diverse geographic locations. All strain information and results were submitted to the Pathogen Annotated Tracking Resource Network (PATRN), located at http://www.patrn.net/ (43), and is accessible to users after a free registration process. Enterobacter helveticus and Citrobacter sedlakii, which are closely related to Cronobacter, were negative-control strains for the O-antigen-specific PCR screening. The six Cronobacter strains used for O-antigen sequencing in this study are listed in Table 1.
Table 1.
Cronobacter strains used for O-antigen sequencing
| Strain name | Synonym(s) | Species | Country | Sample type(s) |
|---|---|---|---|---|
| LMG 23827 | z3032, E866, 3032, E3032, DSM 18703, CI3032 | C. turicensis | Switzerland | Blood, neonatal meningitis |
| E615 | CI615 | C. malonaticus | Czech Republic | Clinical |
| CDC 1059-77 | C. sakazakii | United States | Unknown | |
| 2156 | 2005-14-08 | C. sakazakii | United States | Blood |
| E769 | CI769 | C. muytjensii | Denmark | Milk powder |
| LMG 23826 | E825, CI825, CDC 1058-77, API 76-2121, DSM 18702 | C. malonaticus | United States | Breast abscess |
Frozen bacterial cultures were stored at −80°C in Trypticase soy broth (BBL, Cockeysville, MD) supplemented with 1% NaCl (TSBS) and 50% glycerol. For propagation, frozen cultures were plated on Trypticase soy agar (TSA; BBL) supplemented with 1% NaCl (TSAS) or Luria-Bertani (LB) agar (LBA; BBL) and the plates incubated for 16 to 18 h at 37°C.
PCR of Cronobacter O-antigen gene clusters.
Bacterial genomic DNA was purified from log-phase broth cultures grown in LB at 37°C using a Qiagen DNeasy blood and tissue kit (Qiagen, Inc., Valencia, CA). PCR primers specific to the galF and gnd genes of C. sakazakii ATCC BAA-894 (Table 2) were used in long PCR with AccuPrime Taq high-fidelity DNA polymerase (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's instructions (23, 31). The PCR cycling conditions consisted of an initial denaturing step at 94°C for 30 s, followed by 35 cycles of 94°C for 10 s, 65°C for 35 s, and 68°C for 15 min, followed by a final 10-min extension step at 72°C.
Table 2.
PCR primers used in this study
| Target gene | Serogroup(s) | Primer pair | Primer sequence | Amplicon size (bp) | Source or reference |
|---|---|---|---|---|---|
| wehC | C. sakazakii O1 | EsLPS1F | CACGTTCGCCCTGCAAAAAT | 341 | 31 |
| EsLPS1R | GCAAGCGGCCAGACTGGATA | ||||
| wehI | C. sakazakii O2 | EsLPS2F | TCCTGCATTTGTGGATTTTGC | 329 | 31 |
| EsLPS2R | AACGCATTGCGCTTGAGAAA | ||||
| wzx | C. turicnesis O1 and C. malonaticus O1 | z3032-wzxF5 | AGGGGCACGGCTTAGTTCTGG | 323 | This study |
| z3032-wzxR4 | CCCGCTTGCCCTTCACCTAAC | ||||
| wzx | C. sakazakii O3 and C. muytjensii O1 | 2156-wzxF1 | TGGCTGTCATGGTTTTCTTGC | 258 | This study |
| 2156-wzxR1 | TAGTTGGCACCATCAACGCC | ||||
| wzx | C. malonaticus O2 | E825-wzxF3 | TGGCCCTTGTTAGCAAGACGTTTC | 394 | This study |
| E825-wzxR2 | ATCCACATGCCGTCCTTCATCTGT | ||||
| galFa | EsgalF-F2 | TACCCACTCCTCCAAGAACG | ∼8,000–14,000 | 31 | |
| gnda | gndSeqR1 | TTTGTCACGAGAGCGGTTGAATAC | This study |
Primers for amplification of O-antigen regions.
Restriction fragment length polymorphism (RFLP) analysis of O-antigen amplicons by use of MboII.
O-antigen PCR amplicons (1.5 μg) were digested with MboII (New England BioLabs, Ipswich, MA) according to the manufacturer's instructions. Restriction digests were subjected to gel electrophoresis using 1.5% agarose gels and were visualized with ethidium bromide. TIFF images were imported into Bionumerics software (Applied Maths, Inc., Austin, TX), and dendrograms were generated using the Dice similarity coefficient and the unweighted-pair group method with arithmetic means (UPGMA). Band tolerance and optimization coefficient settings of 1.5% were applied. In silico analysis of MboII restriction sites of O-antigen amplicons was performed with the NEBcutter program (47).
Subcloning and DNA sequencing of Cronobacter O-antigen amplicons.
O-antigen amplicons from C. sakazakii 2156, C. malonaticus LMG 23826, and C. turicensis LMG 23827 isolates were subcloned using a TOPO shotgun subcloning kit (Invitrogen, Carlsbad, CA). Long PCR amplicons were generated in triplicate, pooled, and subcloned according to the manufacturer's instructions. Individual clones were screened using PCR with the T7 and M13 universal primers supplied in the kit to verify the presence of an insert. Positive clones were submitted to Macrogen (Rockville, MD) for sequencing using the T7 and M13 universal primers. The O-antigen regions of C. sakazakii CDC 1059-77, C. malonaticus E615, and C. muytjensii E769 were sequenced using the primer walking method (14).
Sequence analysis of putative O-antigen gene clusters.
Sequencing data were assembled in Bionumerics, and BLAST analysis was used to search available databases for amino acid similarities. The Pfam database was used to search for conserved protein domains; cluster and phylogenetic analyses were done with the MEGA4 program (12, 44). Evolutionary relationships of concatenated wzx and wzy nucleotide sequences were determined with MEGA4 (44) using the neighbor-joining method (37), with distances computed using the maximum composite likelihood method (45). Transmembrane-spanning domains were identified using the THMMER program (22).
PCR screening of Cronobacter genomic DNA for serotype identification.
PCR primers specific to the wzx genes of C. turicensis LMG 23827, C. sakazakii 2156, and C. malonaticus LMG 23826 were designed from the O-antigen regions sequenced in this study (Table 2). These PCR primers and those previously published by Mullane et al. (31) to detect C. sakazakii serotypes O1 and O2 were used to screen a collection of 231 Cronobacter strains. The PCR cycling conditions for all reactions included an initial denaturation step at 95°C for 2 min, followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 5 min.
Nucleotide sequence accession numbers.
The nucleotide sequences for C. turicensis LMG 23827, C. malonaticus E615, C. sakazakii 2156, C. sakazakii CDC 1059-77, C. muytjensii E769, and C. malonaticus LMG 23826 were deposited in GenBank under accession numbers HQ646166, HQ646167, HQ646168, HQ646169, HQ646170, and HQ646171, respectively.
RESULTS
RFLP analysis of Cronobacter O-antigen amplicons with MboII restriction digestion.
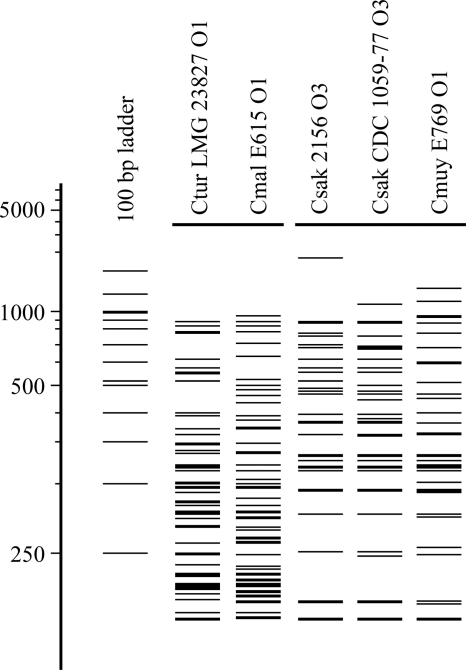
RFLP analysis of 44 Cronobacter strains discriminated seven species-specific Cronobacter clusters (Fig. 1). Two of the C. sakazakii clusters correspond to those identified as C. sakazakii serotypes O1 and O2 by Mullane et al. (31), and a third cluster, consisting of two distinct RFLP patterns, is similar to a C. sakazakii O3 cluster identified by Sun et al. (personal communication) (42). The C. malonaticus strains clustered into two RFLP groups, and there is a single cluster of two C. muytjensii strains (Fig. 1). Finally, the RFLP pattern of the single C. turicensis strain is distinct from those of all of the other strains.
Fig. 1.
Cluster analysis of MboII digests of Cronobacter O-antigen gene clusters digested with MboII. The dendrogram was generated using Bionumerics software, with an 80% similarity value used to distinguish serogroup-specific clusters. Asterisks (*) indicate the Cronobacter strains used for O-antigen sequencing in this study.
These data prompted us to sequence the O-antigen regions from six strains with distinct RFLP patterns in order to define new serotypes for this genus. These included C. sakazakii 2156 and C. sakazakii CDC 1059-77 (from the same cluster but with distinct RFLP patterns), C. malonaticus E615, C. malonaticus LMG 23826, C. muytjensii E769, and C. turicensis LMG 23827 (Fig. 1). Open reading frames (ORFs) were identified and assigned functions based on comparisons with current databases and are summarized in Table 3. Gene names for serotype-specific amplicons were assigned according to the bacterial polysaccharide gene nomenclature scheme described by Reeves et al. (36).
Table 3.
ORFs in new Cronobacter gene clustersa
| Gene cluster group and ORF | Gene | Position (nt) | Size (aa) | Species or strain with similar proteinb (GenBank accession no.) | % identity/% similarity/size (aa) of gap (no. of overlapping amino acids)c | Putative function |
|---|---|---|---|---|---|---|
| C. malonaticus O1 and C. turicensis O1 gene clusters | ||||||
| 1 | wffW | 1048–1443 | 131 | (ACA24847, ACA24835) | 94/97 (131) | Glycerol-3-phosphate cytidyltransferase |
| 2 | wffX | 1440–2600 | 386 | (ACA24848, ACA24836) | 83/92 (386) | Glycerophosphotransferase |
| 3 | wffY | 2597–3694 | 365 | (ACA24849, ACA24837) | 78/90 (365) | Glycosyl transferase group 1 |
| 4 | wffZ | 3684–4739 | 351 | (ACA24850, ACA24838) | 79/90 (351) | Glycosyl transferase group 1 |
| 5 | wzx | 4742–6142 | 466 | (ACA24851, ACA24839) | 83/93 (466) | O-antigen flipase |
| 6 | wzy | 6145–7302 | 385 | (ACA24852, ACA24840) | 67/82 (385) | O-antigen polymerase |
| 7 | wfgA | 7290–8408 | 372 | (ACA24853, ACA24841) | 81/91 (369) | Glycosyl transferase group 1 |
| 8 | fnlA | 8345–9469 | 374 | (ACA24854, ACA24842) | 99/99 (344) | 4,6-Dehydratase, 3-and 5-epimerase |
| 9 | fnlB | 9487–10575 | 362 | (ACA24855, ACA24843) | 85/93 (347) | Epimerase |
| 10 | fnlC | 10575–11705 | 376 | (ACA24856, ACA24844) | 94/97 (376) | C-2 epimerase |
| 11 | wbuB | 11705–12913 | 402 | (ACA24857, ACA24845) | 76/86 (398) | Glycosyl transferase group 1 |
| 12 | wbuC | 12904–13308 | 134 | (ACA24858, ACA24846) | 71/88 (131) | Unknown protein function |
| C. sakazakii O3 and C. muytjensii O1 gene clusters | ||||||
| 1 | rmlB | 1119–2201 | 360 | RmlB, Cronobacter sakazakii O2 (ABX51885) | 99/99 (360) | dTDP-d-glucose-4,6-dehydratase |
| 2 | rmlD | 2198–3100 | 300 | Cronobacter sakazakii O2 (ABX51886) | 98/99 (300) | dTDP-6-deoxy-l-mannose dehydrogenase |
| 3 | rmlA | 3150–4028 | 292 | Cronobacter sakazakii O2 (ABX51887) | 94/98 (292) | Glucose-1-phosphate thymidylyltransferase |
| 4 | rmlC | 4032–4586 | 184 | E. coli (ZP_07502864) | 72/83 (175) | Putative dTDP-6-deoxy-d-glucose-3,5 epimerase |
| 5 | fdtA | 4583–4999 | 139 | Pectobacterium carotovorum (ACT12347) | 76/93 (127) | WxcM domain protein isomerase |
| 6 | fdtC | 4923–5435 | 170 | Salmonella enterica (ADI39339) | 75/83 (144) | dTDP-d-Fuc3N acetyltransferase |
| 7 | fdtB | 5437–6543 | 368 | Shewanella baltica OS195 (ZP_07069428) | 70/82 (365) | DegT/DnrJ/EryC1/StrS aminotransferase |
| 8 | wzx | 6540–7811 | 423 | Pectobacterium carotovorum (ZP_03826804) | 52/71 (413) | O-antigen flipase |
| 9 | wdaN | 7801–9228 | 475 | E. coli O71 (ADI77032) | 39/59 (422) | Glycosyltransferase |
| 10 | wzy | 9277–10548 | 423 | E. coli (ADI77033) | 23/44 (412) | O-antigen polymerase |
| 11 | wehJ | 10556–11605 | 349 | Pseudomonas aeruginosa (AAM27646) | 41/59 (342) | Glycosyl transferase family 2 |
| 12 | wehK | 11608–12693 | 361 | Flavobacteria bacterium (EAZ96080) | 26/47 (378) | Glycosyl transferases group 1 |
| 13 | wehL | 12695–13465 | 256 | Cronobacter sakazakii O1 (ABX51882) | 66/80 (246) | Glycosyl transferase family 2 |
| C. malonaticus O2 gene cluster | ||||||
| 1 | wehM | 1009–1791 | 260 | Fusobacterium mortiferum (EEO36671) | 31/48/32 (265) | Glycosyl transferase family 2 |
| 2 | wehN | 1793–3034 | 413 | Marinomonas sp. (EAQ66012) | 33/51/19 (346) | Capsule biosynthesis protein |
| 3 | wzx | 3027–4232 | 401 | Geobacillus kaustophilus (BAD77602) | 23/47/18 (400) | O-antigen flipase |
| 4 | wzy | 4222–5448 | 408 | Proteus mirabilis (ADL32282) | 24/44/39 (286) | O-antigen polymerase |
| 5 | wehO | 5450–6289 | 279 | Actinobacillus minor (EEF16045) | 32/51/8 (275) | Glycosyl transferase family 14; Core-2/I-Branching enzyme |
| 6 | wehP | 6286–7122 | 278 | E. coli (AAY28251) | 51/66/0 (268) | Glycosyl transferase family 2 |
| 7 | wehQ | 7147–7824 | 225 | Erwinia sp. (ADP10993) | 41/61/5 (173) | Acyltransferase 3 |
| 8 | wehR | 7856–8188 | 110 | Burkholderia phymatum (ACC71456) | 37/60/8 (77) | Acyltransferase 3 |
nt, nucleotides; aa, amino acids.
The GenBank accession numbers for the C. malonaticus O1 and C. turicensis O1 genes are all from E. coli O29 (GenBank accession no. EU294173) and S. dysenteriae (GenBank accession no. EU294172) predicted proteins.
Size of gap is included for genes in the O5 gene cluster.
Nucleotide sequence analysis and identification of new Cronobacter serotypes.
PCR products using primers specific to the galF and gnd genes of C. turicensis LMG 23827, C. malonaticus E615, C. sakazakii 2156 and CDC 1059-77, C. muytjensii E769, and C. malonaticus LMG 23826 resulted in sequences of 13,995, 13,499, 13,741, 13,687, 13,688, and 8,415 bp, respectively. BLAST analysis revealed three unique O-antigen gene clusters among these six strains (Fig. 2).
Fig. 2.
O-antigen gene clusters of Cronobacter serotypes. (A) Predicted genes for the C. turicensis O1 and C. malonaticus O1 serotypes derived from the sequences of C. turicensis LMG 23827 and C. malonaticus E615. (B) Predicted genes for the C. sakazakii O3 and C. muytjensii O1 serotypes derived from the sequences of C. sakazakii 2156, C. sakazakii CDC 1059-77, and C. muytjensii E769. (C) Predicted genes for the C. malonaticus O2 serotype derived from the sequence of C. malonaticus LMG 23826.
The nucleotide sequences of C. turicensis LMG 23827 and C. malonaticus E615 (excluding galF and gnd) are 91% homologous and were designated C. turicensis serotype O1 and C. malonaticus O1, respectively. In silico digestion with MboII and BLAST analysis of the two gene clusters showed that despite vastly different RFLP patterns at the species level (Fig. 3), the predicted ORFs from these O-antigen regions encode the same proteins (Fig. 2 and Table 3). Furthermore, the predicted proteins in the C. turicensis and C. malonaticus O1 gene clusters are highly similar to those of Shigella dysenteriae D11 and E. coli 029 (71 to 91% identities); therefore, the nomenclature of the S. dysenteriae D11 and E. coli 029 (D11/O29) gene clusters was used in naming the genes contained in these two new Cronobacter O-antigen regions (Fig. 2 and Table 3) (25, 33).
Fig. 3.
In silico analysis, showing the variability in MboII restriction patterns of serogroups sequenced in this study using the NEBcutter program.
The O-antigen gene clusters of C. sakazakii 2156 and CDC 1059-77 and C. muytjensii E769, designated C. sakazakii serotype O3 and C. muytjensii O1, respectively, carry 13 ORFs (Fig. 2). BLAST analysis revealed 99% nucleotide homology between the two C. sakazakii gene clusters and 91% homology between the C. sakazakii and C. muytjensii gene clusters. Even though the MboII RFLP patterns of the O-antigen amplicons from these three strains differed considerably (Fig. 1), in silico and BLAST analysis revealed that this variability is due to differences in the location of MboII sites that do not change the predicted translation of proteins in these gene clusters (Fig. 3 and Table 3).
The O-antigen gene cluster of C. malonaticus LMG 23826 consists of eight ORFs and was designated C. malonaticus serotype O2 (Fig. 2). The predicted proteins encoded by genes in the C. malonaticus O2 gene cluster possessed the lowest similarities to currently known O-antigen region proteins, with all of the amino acid alignments having several gaps (Table 3). The exception to this observation is the predicted product of ORF 5, which is 51% similar to the Core-2/I-Branching family of enzymes (Table 3).
Nucleotide sugar synthesis pathway genes.
Five genes encode putative nucleotide sugar synthesis pathway enzymes with 83 to 99% identities to D11/O29 homologous gene products in the C. turicensis O1 and C. malonaticus O1 gene clusters (Fig. 2 and Table 3). These include WffW and WffX, which are part of a CDP-glycerol nucleotide sugar synthesis pathway that produces a glycerol-3-phosphate moiety, and the FnlABC proteins, known to produce 2-acetamido-2,6-dideoxy-hexose (UDP-l-FucNAc) in the D11/O29 O antigens (25, 33, 39).
The C. sakazakii O3 and C. muytjensii O1 gene clusters encode putative enzymes involved in two nucleotide sugar synthesis pathways (Fig. 2 and Table 3). The RmlBDAC proteins are known to function in a dTDP-sugar biosynthesis pathway that produces dTDP-l-rhamnose (39). ORFs 5, 6, and 7 share significant identity with FdtA, FdtC, and FdtB (76, 75, and 76%), respectively, a group of enzymes required for the synthesis of the dTDP-sugar 3-acetamido-3,6-dideoxy-d-galactose (dTDP-d-Fucp3NAc) (34, 39). They also share 73, 73, and 63% identities with QdtA, QdtC, and QdtB, respectively, of E. coli O71 and Salmonella enterica O28, which, based on studies with Thermoanaerobacterium thermosaccharolyticum, are part of a biosynthetic pathway that includes the RmlA and RmlB proteins. The product of this pathway is dTDP-3-acetamido-3,6-dideoxy-α-d-glucose (dTDP-α-d-Qui3NAc) (16, 35). ORFs 5, 6, and 7 of the C. sakazakii O3 and C. muytjensii O1 O-antigen regions were named fdtA, fdtC, and fdtB, respectively, because of their slightly higher similarities, with the caveat that these gene names may change if structural data that reveal the sugars present in these O-antigen gene clusters become available (Table 3).
Surprisingly, none of the putative proteins encoded by the ORFs in the C. malonaticus O2 gene cluster are common to the nucleotide sugar synthesis pathways typically found in O-antigen regions, suggesting that genes outside the O-antigen region encode the O repeat sugars in this serotype (39).
Sugar transferase genes.
Glycosyl transferase genes have been categorized into 26 families based on amino acid sequence similarities (3). The C. turicensis O1 and C. malonaticus O1 gene clusters encode four putative glycosyl transferase family 1 proteins that share 78, 79, 81, and 76% identity with WffY, WffZ, WfgA, and WbuB, respectively, in the D11/O29 O-antigen regions (Fig. 2 and Table 3). Four genes in the C. sakazakii O3 and C. muytjensii O1 gene clusters encode predicted glycosyl transferase proteins, including one family 1 protein and two family 2 proteins. The predicted product of the fourth putative glycosyl transferase, encoded by ORF 9 of C. sakazakii O3 and C. muytjensii O1, is similar to WdaN, a glycosyl transferase recently identified in E. coli O71 and S. enterica O28, which does not share conserved domains with any other of the glycosyl transferase families (Fig. 2 and Table 3) (16). There are three predicted glycosyl transferase proteins in the C. malonaticus O2 gene cluster (two family 2 proteins and a third protein, which has conserved domains relative to glycosyl transferases in family 14/Core-2/I-Branching enzymes) (Table 3).
O-antigen-processing genes.
Putative Wzx (O-antigen flippase) and Wzy (O-antigen polymerase) proteins were identified in all of the Cronobacter O-antigen gene clusters sequenced in this study (Fig. 2 and Table 3). O-antigen-processing proteins possess several well-pronounced transmembrane-spanning domains (6, 8). TMHMM analysis of the serotype-specific Cronobacter Wzx proteins revealed 11 to 14 transmembrane-spanning domains. The Wzy proteins were predicted to have 10 or 11 transmembrane-spanning domains.
Other genes in Cronobacter O-antigen gene clusters.
The last predicted ORF in the C. turicensis O1 and C. malonaticus O1 gene clusters, ORF 12, shares 71% identity with WbuC of D11/O29, which has no known function (Fig. 2 and Table 3). This gene is also present in the O-antigen gene clusters of E. coli O26, O145, and O98 (5, 10, 11).
The putative ORF 2 protein of the C. malonaticus O2 gene cluster shares 33% identity with a predicted capsule polysaccharide biosynthesis protein of Pfam PF01943 (Table 3) (12). The conserved domains of the proteins in the PF01943 family are thought to be involved in the proper translocation and surface expression of capsule polymers (12). ORFs 7 and 8 of the C. malonaticus O2 gene cluster carry predicted acyltransferase genes associated with exopolysaccharide biosynthesis (Table 3). ORF 7 is 41% identical to the amino terminus of a predicted acyltransferase 3 in Erwinia sp. strain Ejp617, and the putative product of ORF 8 is 34% identical to the carboxyl terminus of a predicted acyltransferase 3 protein in Burkholderia phymatum (Table 3). Multiple rounds of sequencing through both DNA strands in this region confirmed that the two ORFs are in fact separate genes. The partial identities of ORFs 7 and 8 to opposite ends of the same type of protein suggest the presence of a frameshift mutation that resulted in the splitting of this coding region into 2 genes.
Comparison of the C. turicensis LMG 23827 sequence from this study to the published sequence of C. turicensis z3032.
Our work with the O-antigen region of C. turicensis LMG 23827 coincided with the release of the whole-genome sequence from the synonymous C. turicensis strain, z3032, by Stephan et al. (GenBank accession no. NC_013282) (40). A comparison of the nucleotide sequence from the O-antigen gene cluster of LMG 23827 obtained in our study to that of z3032 revealed three nucleotide differences in z3032, including an extra thymine in the wzx gene, an extra thymine in the wzy gene, and a missing adenine in the fnlA gene. Each of these nucleotide differences result in frameshift mutations that affect the predicted translations of ORFs 5 to 12 of the z3032 O-antigen locus. We believe that the sequence generated in this study reconciles these discrepancies.
PCR assays developed to detect Cronobacter serotypes.
In a previous study, Mullane et al. (31) identified serotypes O1 and O2 of C. sakazakii using PCR primers specific to the O1 wehC gene and the O2 wehI gene. Analysis of the six Cronobacter wzx genes sequenced in this study led to the design of three O-antigen cluster-specific PCR primer pairs, which identify five new Cronobacter serotypes. Genomic DNA from 231 Cronobacter isolates in our collection was screened with these primer pairs, and 32% and 31% of our strains were identified as C. sakazakii serotypes O1 and O2, respectively (Table 4). Furthermore, all of the C. sakazakii serotype O1 and O2 strains cluster into two clades by MboII RFLP fingerprinting (Fig. 1).
Table 4.
Distribution of Cronobacter serogroups among 231 strains
| Species | No. of strains |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| C. sakazakii O1a | C. sakazakii O2b | C. sakazakii O3c | C. malonaticus O1c | C. malonaticus O2c | C. muytjensii O1c | C. turicensis O1c | NDd | Total | |
| C. dublinensis | 6 | 6 | |||||||
| C. malonaticus | 4 | 21 | 25 | ||||||
| C. muytjensii | 2 | 10 | 12 | ||||||
| C. sakazakii | 74 | 71 | 10 | 25 | 180 | ||||
| C. turicensis | 1 | 5 | 6 | ||||||
| Cronobacter genomospecies 1 | 2 | 2 | |||||||
| Serogroup total (%) | 32.0 | 30.7 | 4.3 | 1.7 | 9.1 | 0.9 | 0.4 | 20.8 | |
Cronobacter O1 serogroup PCR is specific to the wehC gene.
Cronobacter O2 serogroup PCR is specific to the wehI gene.
Cronobacter serogroup PCR is specific to the wzx gene.
ND, not determined (negative with Cronobacter PCR assays tested in this study).
Four C. malonaticus isolates were identified by the same wzx PCR primer set as the one used for the C. turicensis isolate from which the wzx PCR primers were designed, and these groups of isolates were designated C. malonaticus serotype O1 and C. turicensis serotype O1, respectively (Table 4). Three of the four C. malonaticus O1 strains form one RFLP clade, which has a different fingerprint from the C. turicensis O1 strain (Fig. 1). Additionally, 10 C. sakazakii and 2 C. muytjensii strains, which form two clades, were identified by the same wzx primer pair (Fig. 1 and Table 4). A third wzx primer pair identified a C. malonaticus serotype O2 clade, consisting of 21 (9%) isolates (Fig. 1 and Table 4). Twenty-one percent of the isolates in our collection were negative by the Cronobacter serotype-specific PCR assays developed to date, including all of the C. dublinensis and C. genomospecies 1 strains (Table 4). C. sakazakii has the most diverse serotype profile of all the species in our collection, with 32% (74) of the strains positive by the O1 PCR assay, 31% (71) positive by the O2 PCR assay, 4% (10) positive by the O3 PCR assay, and 14% (25) not determined (ND) (Table 4). Similarly, most of the C. muytjensii and C. turicensis strains tested in this study were negative by all of the serotype PCR assays, suggesting that there are additional serotypes within the Cronobacter genus. C. malonaticus is the only species in which all isolates in our collection were assigned to a serotype (Table 4).
DISCUSSION
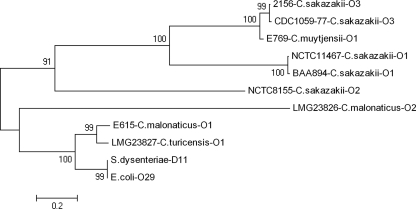
The O-antigen region is one of the most variable regions in the membranes of Gram-negative bacteria, and this variability is routinely used to discriminate serotypes within species of bacteria. Within O-antigen gene clusters, the wzx and wzy genes have the most diverse nucleotide sequences, which has led to their wide use in serotype-specific PCR assays. The use of RFLP fingerprinting with MboII, combined with DNA sequence analysis of Cronobacter O-antigen regions, allowed us to develop serotype-specific PCR assays from Cronobacter wzx gene sequences, revealing similarities between C. malonaticus O1 and C. turicensis O1 as well as C. sakazakii O3 and C. muytjensii O1 strains, which had different RFLP fingerprints. Additionally, the C. malonaticus O1 and C. turicensis O1 O-antigen gene clusters are highly similar to those from S. dysenteriae D11 and E. coli O29. Analysis of the nucleotide sequences of concatenated wzx and wzy genes from Cronobacter spp. and D11/O29 confirmed these phylogenetic relationships (Fig. 4). In all cases, Cronobacter wzx and wzy sequences from different species with similar O-antigen regions clustered together, and as expected, C. malonaticus O1 and C. turicensis O1 wzx and wzy genes clustered with those from the O-antigen regions of D11/O29. Furthermore, O-antigen sequences from the same species clustered together, a pattern that was not apparent from the clustering of the RFLP fingerprints (Fig. 4).
Fig. 4.
Evolutionary relationships of concatenated wzx and wzy nucleotide sequences from Cronobacter species serotypes, E. coli O29 and S. dysenteriae D11. Relationships were analyzed with MEGA4 using the neighbor-joining method, with distances computed using the maximum composite likelihood method. Numbers on the branches denote bootstrap percentages from 1,000 bootstrap replicates, with a cutoff value of 50%. The GenBank accession numbers for NCTC 11467, BAA 894, NCTC 8155, S. dysenteriae D11, and E. coli O29 are EU076545, NC_009778, EU076546, EU294172, and EU294173, respectively.
The sequence similarities between C. malonaticus O1, C. turicensis O1, and O29/D11 suggests similar ancestries for the origination of the O-antigen regions from these four strains (25, 33). Liu et al. determined that 18 Shigella type strains share their O-antigen gene clusters with an E. coli gene cluster (25). Evolutionary relationships have also been suggested for the origins of O-antigen gene clusters in E. coli and Salmonella. Some examples include E. coli O103 and Salmonella O55, E. coli O157 and Salmonella O30, E. coli O55 and Salmonella O50, E. coli O77 and Salmonella O:6,14, and E. coli O71 and S. enterica O28, where all of the pairs of gene clusters share genetic and structural similarities (16, 26, 38, 49). The presence of wbuC in the C. malonaticus O1 and C. turicensis O1 gene clusters also suggests evolutionary ties to E. coli. In E. coli O29, O26, and O145, S. dysenteriae D11, C. malonaticus O1, and C. turicensis O1, wbuC is downstream of the genes fnlA, fnlB, fnlC, and wbuB (Fig. 2), which encode the proteins for the putative or, in some instances, known synthesis of UDP-l-FucNAc (10, 11, 25). In E. coli O98, wbuC is downstream of a similar group of genes, comprising fnlA, qnlA, qnlB, and wbwW, which synthesize UDP-l-QuiNAc, and interestingly, the E. coli O98 gene cluster was acquired by Yersinia kristensenii O11 through lateral gene transfer without the wbuC gene (5). The function of WbuC is not known, and although it has been proposed to be a nonfunctional remnant gene product in E. coli O26 and O145 (10, 11), Cunneen and Reeves (5) suggested that its conservation in E. coli implies that wbuC must have some function specific to E. coli and now possibly in C. malonaticus and C. turicensis. The C. malonaticus O1 and C. turicensis O1 gene clusters are another example of the existence of related ancestors responsible for the evolution of antigenic variation among Enterobacteriaceae. It is likely that lateral gene transfer by homologous recombination between species is responsible for gene sharing since most of the clusters are located between galF and gnd genes. Additionally, it is probable that if it were available, C. malonaticus O1 and C. turicensis O1 antisera would cross-react between strains, as was the case with pairs of E. coli and Shigella strains, including D11/O29, which have identical or closely related O-antigen structures (25).
Our analysis of the C. sakazakii O3 and C. muytjensii O1 gene clusters revealed additional similarities among the O-antigen regions from different Cronobacter species. Initial screening with the serotype-specific PCR assays developed by Mullane et al. (31) followed by cluster analysis of MboII RFLP fingerprints revealed a clade of C. sakazakii strains that were negative by both the C. sakazakii O1 and the C. sakazakii O2 assays (Fig. 1). We sequenced the O-antigen regions of C. sakazakii 2156 and CDC 1059-77 because although they were in the same clade, they had different RFLP fingerprints (Fig. 1). Surprisingly, the wzx-specific PCR assay designed from these sequences was positive for two C. muytjensii isolates that form their own RFLP clade, in addition to all members of the subgroups of the C. sakazakii O3 antigen clade (Fig. 1). The C. sakazakii O3 and C. muytjensii O1 O-antigen gene clusters contain the rmlBDAC genes, a highly conserved and well characterized set of genes known to encode the proteins for the production of l-rhamnose in the O-antigen regions of E. coli and S. enterica (39). The same genes, in the same order, are present at the 5′ end of the C. sakazakii O2 gene cluster, and it was recently reported that l-rhamnose is present in the O polysaccharide of C. sakazakii O2 by chemical composition analysis (1, 31). It is reasonable to assume that the C. sakazakii O3 and C. muytjensii O1 O polysaccharides also contain l-rhamnose sugar residues.
In many O-antigen gene clusters, the rmlA and rmlB genes provide an intermediate sugar that serves as a branching point for other dTDP-sugar biosynthesis pathways (39). Two examples of this are in the S-layer glycoprotein of Aneurinibacillus thermoaerophilus, where studies have shown that FdtA, FdtB, and FdtC use the rmlAB branch point product to synthesize dTDP-d-Fucp3NAc, and in Thermoanaerobacterium thermosaccharolyticum, where QdtA, QdtB, and QdtC are part of a biosynthetic pathway that includes RmlA and RmlB to produce dTDP-3-acetamido-3,6-dideoxy-α-d-glucose (dTDP-d-Qui3Nac) (34, 35). ORFs 5, 6, and 7 of the C. sakazakii O3 and C. muytjensii O1 regions were named FdtA, FdtC, and FdtB, respectively, because of their similarities to proteins predicted to synthesize dTDP-d-Fuc3Nac (Table 3). However, these ORFs are also similar to the E. coli O71 QdtA, QdtC, and QdtB proteins, which are part of the sugar synthesis pathway for dTDP-d-Qui3NAC (16). A recent structural analysis study of the O polysaccharide from a C. sakazakii O1 isolate showed that it contains dTDP-d-Qui3NAC as part of the repeat sugars, whereas the molecular analysis of Mullane et al. (31) identified genes in the O1 cluster as fdtA, fdtB, and fdtC. The results of the structural analysis suggest that the fdt gene names originally given by Mullane et al. (31) be changed to qdt to account for this finding (2). Without knowing the chemical composition of the C. sakazakii O3 and C. muytjensii O1 O polysaccharide, it is difficult to definitively assign gene names; therefore, we suggest the provisional names of fdtA, fdtC, and fdtB until structural data are available to confirm the sugars present in this O-antigen region.
The C. malonaticus O2 serotype accounts for 21% of our strain collection. Analysis of the O-antigen region showed that there are no genes for nucleotide sugar synthesis predicted from the sequence data, which accounts for the shorter length of this gene cluster (∼8 kb, compared to ∼12 to 14 kb for the other Cronobacter O-antigen regions). There are at least 69 sugars known to be components of the repeat polysaccharide structures in Gram-negative bacteria, and some, such as glucose and galactose, provide housekeeping functions, and therefore, their genes are not duplicated in the O-antigen region (39). For example, the O-polysaccharide structure of C. malonaticus 3267 isolated from powdered infant formula in Australia is known, and the polymer consists of a branched pentasaccharide composed of d-glucose (d-Glc), d-galactose (d-Gal), 2-amino-2-deoxy-d-glucose (d-GlcN), 2-amino-2-deoxy-d-galactose (d-GalN), and 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) (29). All of these sugars provide other functions to bacterial cells, and therefore, it is plausible that their genes are not part of the O-antigen gene clusters. The presence of numerous glucose and galactose sugars in the O polysaccharide of C. malonaticus 3267 supports the absence of nucleotide sugar synthesis pathway genes in our sequence of the C. malonaticus O2 gene cluster, as they are probably carried elsewhere in the genome. Further, Kdo, a predicted protein in the C. malonaticus O-antigen region and a known component of the inner core oligosaccharide of LPS, is rarely found in O-antigen polysaccharide structures, with some exceptions, such as the O structure of Providencia alcalifaciens O36 (15, 21). It was not determined in this study whether the putative acyltransferase 3 genes carried by ORFs 7 and 8 of the C. malonaticus O2 gene cluster are functional; however, the structural analysis of C. malonaticus 3267 did not identify any O-acetylated residues in the O-antigen repeat unit, which suggests that the enzyme is inactive (29). Structural analysis of C. malonaticus LMG 23826 and genetic characterization of C. malonaticus 3267 are necessary for further investigation of these hypotheses.
In conclusion, we have identified five new serotypes of Cronobacter spp., using PCR assays with primers specific to the wzx genes of the O-antigen locus. The assays reported here will be valuable tools for identifying and typing Cronobacter species in clinical, environmental, and food samples. Cluster analysis of the RFLP patterns from isolates in our strain collection showed that there are additional serotypes in all of the Cronobacter species except C. malonaticus (data not shown). Additional studies are needed to define the serotypes for these strains.
ACKNOWLEDGMENTS
We thank Atin R. Datta, Barbara A. McCardell, and Marianna D. Solomotis for reviewing the manuscript.
K. G. Jarvis, L. Hu, and C. J. Grim are Oak Ridge Institute for Science and Education fellows, and we thank the Department of Energy for their support.
Footnotes
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Arbatsky N. P., et al. 2010. Structure of the O-polysaccharide of Cronobacter sakazakii O2 with a randomly O-acetylated l-rhamnose residue. Carbohydr. Res. 345:2090–2094doi:10.1016/j.carres.2010.07.014 [DOI] [PubMed] [Google Scholar]
- 2. Arbatsky N. P., et al. 2010. Structure of the O-polysaccharide of Cronobacter sakazakii O1 containing 3-(N-acetyl-l-alanyl)amino-3,6-dideoxy-d-glucose. Carbohydr. Res. 345:2095–2098doi:10.1016/j.carres.2010.07.013 [DOI] [PubMed] [Google Scholar]
- 3. Campbell J. A., Davies G. J., Bulone V., Henrissat B. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326(Pt. 3):929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caubilla-Barron J., et al. 2007. Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J. Clin. Microbiol. 45:3979–3985doi:10.1128/JCM.01075-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cunneen M. M., Reeves P. R. 2007. The Yersinia kristensenii O11 O-antigen gene cluster was acquired by lateral gene transfer and incorporated at a novel chromosomal locus. Mol. Biol. Evol. 24:1355–1365doi:10.1093/molbev/msm058 [DOI] [PubMed] [Google Scholar]
- 6. Cunneen M. M., Reeves P. R. 2008. Membrane topology of the Salmonella enterica serovar Typhimurium group B O-antigen translocase Wzx. FEMS Microbiol. Lett. 287:76–84doi:10.1111/j.1574-6968.2008.01295.x [DOI] [PubMed] [Google Scholar]
- 7. Czerwicka M., et al. 2010. Structure of the O-polysaccharide isolated from Cronobacter sakazakii 767. Carbohydr. Res. 345:908–913 doi:10.1016/j.carres.2010.01.020 [DOI] [PubMed] [Google Scholar]
- 8. Daniels C., Vindurampulle C., Morona R. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28:1211–1222 [DOI] [PubMed] [Google Scholar]
- 9. Drudy D., et al. 2006. Characterization of a collection of Enterobacter sakazakii isolates from environmental and food sources. Int. J. Food Microbiol. 110:127–134 doi:10.1016/j.ijfoodmicro.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 10. D'Souza J. M., Wang L., Reeves P. 2002. Sequence of the Escherichia coli O26 O antigen gene cluster and identification of O26 specific genes. Gene 297:123–127 [DOI] [PubMed] [Google Scholar]
- 11. Feng L., et al. 2005. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O antigen and development of an O145 serogroup-specific PCR assay. J. Bacteriol. 187:758–764 doi:10.1128/JB.187.2.758-764.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finn R. D., et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–D288 doi:10.1093/nar/gkm960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedemann M. 2007. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int. J. Food Microbiol. 116:1–10 doi:10.1016/j.ijfoodmicro.2006.12.018 [DOI] [PubMed] [Google Scholar]
- 14. Giesecke H., Obermaier B., Domdey H., Neubert W. J. 1992. Rapid sequencing of the Sendai virus 6.8 kb large (L) gene through primer walking with an automated DNA sequencer. J. Virol. Methods 38:47–60 doi:10.1016/0166-0934(92)90168-D [DOI] [PubMed] [Google Scholar]
- 15. Holst O. 2007. The structures of core regions from enterobacterial lipopolysaccharides—an update. FEMS Microbiol. Lett. 271:3–11 doi:10.1111/j.1574-6968.2007.00708.x [DOI] [PubMed] [Google Scholar]
- 16. Hu B., et al. 2010. Structural and genetic evidence for the close relationship between Escherichia coli O71 and Salmonella enterica O28 O-antigens. FEMS Immunol. Med. Microbiol. 59:161–169 doi:10.1111/j.1574-695X.2010.00676.x [DOI] [PubMed] [Google Scholar]
- 17. Iversen C., Druggan P., Forsythe S. 2004. A selective differential medium for Enterobacter sakazakii, a preliminary study. Int. J. Food Microbiol. 96:133–139 doi:10.1016/j.ijfoodmicro.2004.01.024 [DOI] [PubMed] [Google Scholar]
- 18. Iversen C., et al. 2007. Identification of “Cronobacter” spp. (Enterobacter sakazakii). J. Clin. Microbiol. 45:3814–3816 doi:10.1128/JCM.01026-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iversen C., et al. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 58:1442–1447 doi:10.1099/ijs.0.65577-0 [DOI] [PubMed] [Google Scholar]
- 20. Kim K., et al. 2008. Prevalence and genetic diversity of Enterobacter sakazakii in ingredients of infant foods. Int. J. Food Microbiol. 122:196–203 doi:10.1016/j.ijfoodmicro.2007.11.072 [DOI] [PubMed] [Google Scholar]
- 21. Kocharova N. A., et al. 2007. The structure of the O-polysaccharide from the lipopolysaccharide of Providencia alcalifaciens O36 containing 3-deoxy-d-manno-oct-2-ulosonic acid. Carbohydr. Res. 342:665–670 doi:10.1016/j.carres.2006.06.003 [DOI] [PubMed] [Google Scholar]
- 22. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 doi:10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 23. Kucerova E., et al. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556 doi:10.1371/journal.pone.0009556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lai K. K. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113–122 [DOI] [PubMed] [Google Scholar]
- 25. Liu B., et al. 2008. Structure and genetics of Shigella O antigens. FEMS Microbiol. Rev. 32:627–653 doi:10.1111/j.1574-6976.2008.00114.x [DOI] [PubMed] [Google Scholar]
- 26. Liu B., et al. 2010. Genetic and structural relationships of Salmonella O55 and Escherichia coli O103 O-antigens and identification of a 3-hydroxybutanoyltransferase gene involved in the synthesis of a Fuc3N derivative. Glycobiology 20:679–688 doi:10.1093/glycob/cwq015 [DOI] [PubMed] [Google Scholar]
- 27. MacLean L. L., Pagotto F., Farber J. M., Perry M. B. 2009. Structure of the antigenic repeating pentasaccharide unit of the LPS O-polysaccharide of Cronobacter sakazakii implicated in the Tennessee outbreak. Biochem. Cell Biol. 87:459–465 doi:10.1139/o09-004 [DOI] [PubMed] [Google Scholar]
- 28. MacLean L. L., Pagotto F., Farber J. M., Perry M. B. 2009. The structure of the O-antigen in the endotoxin of the emerging food pathogen Cronobacter (Enterobacter) muytjensii strain 3270. Carbohydr. Res. 344:667–671 doi:10.1016/j.carres.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 29. MacLean L. L., Vinogradov E., Pagotto F., Farber J. M., Perry M. B. 2009. Characterization of the O-antigen in the lipopolysaccharide of Cronobacter (Enterobacter) malonaticus 3267. Biochem. Cell Biol. 87:927–932 doi:10.1139/o09-059 [DOI] [PubMed] [Google Scholar]
- 30. MacLean L. L., Vinogradov E., Pagotto F., Farber J. M., Perry M. B. 2010. The structure of the O-antigen of Cronobacter sakazakii HPB 2855 isolate involved in a neonatal infection. Carbohydr. Res. 345:1932–1937 doi:10.1016/j.carres.2010.06.020 [DOI] [PubMed] [Google Scholar]
- 31. Mullane N., et al. 2008. Molecular analysis of the Enterobacter sakazakii O-antigen gene locus. Appl. Environ. Microbiol. 74:3783–3794 doi:10.1128/AEM.02302-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muytjens H. L., et al. 1983. Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J. Clin. Microbiol. 18:115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perepelov A. V., et al. 2006. Structure of a teichoic acid-like O-polysaccharide of Escherichia coli O29. Carbohydr. Res. 341:2176–2180 doi:10.1016/j.carres.2006.05.016 [DOI] [PubMed] [Google Scholar]
- 34. Pfoestl A., Hofinger A., Kosma P., Messner P. 2003. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 278:26410–26417 doi:10.1074/jbc.M300858200 [DOI] [PubMed] [Google Scholar]
- 35. Pfostl A., et al. 2008. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-glucose. Biochem. J. 410:187–194 doi:10.1042/BJ20071044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reeves P. R., et al. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495–503 [DOI] [PubMed] [Google Scholar]
- 37. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 38. Samuel G., Hogbin J. P., Wang L., Reeves P. R. 2004. Relationships of the Escherichia coli O157, O111, and O55 O-antigen gene clusters with those of Salmonella enterica and Citrobacter freundii, which express identical O antigens. J. Bacteriol. 186:6536–6543 doi:10.1128/JB.186.19.6536-6543.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samuel G., Reeves P. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519 [DOI] [PubMed] [Google Scholar]
- 40. Stephan R., Lehner A., Tischler P., Rattei T. 2011. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J. Bacteriol. 193:309–310 doi:10.1128/JB.01162-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoop B., Lehner A., Iversen C., Fanning S., Stephan R. 2009. Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter. Int. J. Food Microbiol. 136:165–168 doi:10.1016/j.ijfoodmicro.2009.04.023 [DOI] [PubMed] [Google Scholar]
- 42. Sun Y., et al. 2011. Development of an O-antigen serotyping scheme for Cronobacter sakazakii. Appl. Environ. Microbiol. 77:2209–2214 doi:10.1128/AEM.02229-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tall B. D., et al. 2010. Development and application of a Pathogen Annotated Tracking Resource Network (PATRN) system to aid analysis of foodborne pathogens, abstr. R-2447. Abstr. 110th Gen. Meet. Am., Soc. Microbiol. American Society for Microbiology, Washington, DC [Google Scholar]
- 44. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 doi:10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 45. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 doi:10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Acker J., et al. 2001. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39:293–297 doi:10.1128/JCM.39.1.293-297.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vincze T., Posfai J., Roberts R. J. 2003. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 31:3688–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang L., Wang Q., Reeves P. R. 2010. The variation of O antigens in gram-negative bacteria. Subcell. Biochem. 53:123–152 doi:10.1007/978-90-481-9078-2_6 [DOI] [PubMed] [Google Scholar]
- 49. Wang W., et al. 2007. A group of Escherichia coli and Salmonella enterica O antigens sharing a common backbone structure. Microbiology 153:2159–2167 doi:10.1099/mic.0.2007/004192-0 [DOI] [PubMed] [Google Scholar]