Abstract
Nine approaches to recover viral RNA from environmental silty sediments were newly developed and compared to quantify RNA viruses in sediments using molecular methods. Four of the nine approaches employed direct procedures for extracting RNA from sediments (direct methods), and the remaining five approaches used indirect methods wherein viral particles were recovered before RNA extraction. A direct method using an SDS buffer with EDTA to lyse viral capsids in sediments, phenol-chloroform-isoamyl alcohol to extract RNA, isopropanol to concentrate RNA, and magnetic beads to purify RNA resulted in the highest rate of recovery (geometric mean of 11%, with a geometric standard deviation of 0.02; n = 7) of poliovirus 1 (PV1) inoculated in an environmental sediment sample. The direct method exhibiting the highest rate of PV1 recovery was applied to environmental sediment samples. One hundred eight sediment samples were collected from the Takagi River, Miyagi, Japan, and its estuary from November 2007 to April 2009, and the genomic RNAs of enterovirus and human norovirus in these samples were quantified by reverse transcription (RT)-quantitative PCR (qPCR). The human norovirus genome was detected in one sample collected at the bay, although its concentration was below the quantification limit. Meanwhile, the enterovirus genome was detected in two samples at the river mouth and river at concentrations of 8.6 × 102 and 2.4 × 102 copies/g (wet weight), respectively. This is the first report to obtain quantitative data for a human pathogenic virus in a river and in estuarine sediments using RT-qPCR.
INTRODUCTION
Bacterial (Mycobacterium avium [29] and Clostridium botulinum type E [19]), protozoan (Cryptosporidium species [22]), and viral (enterovirus [EV] [5, 13, 15], hepatitis A virus [HAV] [6, 14], and rotavirus [6]) pathogens have been detected in environmental sediments. Whittington et al. reported previously that M. avium in sediments from a dam lake survived 12 to 26 weeks longer than it did in a water column (29). Moreover, the persistence of viral pathogens in environmental sediments has been reported. Smith et al. demonstrated previously that poliovirus 1 (PV1), coxsackieviruses B3 and A9, and echovirus 1 survived significantly longer when associated with marine sediments (23). A 3-log reduction in the infectivity of PV1 was observed in 14 days in seawater having marine sediments, whereas such a reduction was observed in 4 days in seawater without sediments (23). These results suggest that environmental sediments have a protective effect on pathogens (1, 20), and the association of pathogens with environmental sediments cannot be ignored when considering the fate of pathogens in water environments (22).
When storms, tides, or strong winds cause sediment resuspension, pathogens in sediments are also resuspended, resulting in high pathogen levels in the water column. Dorner et al. indicated previously the importance of the resuspension of microorganisms from stream sediments rather than land-based sources according to the hydrological simulation of Escherichia coli during storm events (4). The quantitative detection of pathogens in environmental sediments is thus crucial for assessing the health effects of exposure to pathogen-contaminated sediments or pathogen-resuspended water. However, the quantification of pathogens in sediments using molecular methods such as quantitative PCR (qPCR) has been difficult because of the presence of inhibitory substances such as humic substances that affect the efficiency of genome extraction and enzymatic genome amplification (24, 25, 28, 30). Particularly, the preparation of viral RNA from environmental sediments is even more difficult because viral particles are not completely different from those inhibitory substances in terms of physical characteristics such as molecular weight and isoelectric point. Furthermore, the loss of viral RNA due to adsorption to soil particles and degradation by RNase occurs easily.
Sample preparation methods to recover viruses from sediments, particularly molecular detection methods, are still under development. Conventional methods of preparing samples from sediments, usually consisting of the dispersion of sediments in buffer solutions to elute viral particles, centrifugation to remove sediments, and concentration of the supernatant, were developed on the premise that cell culture-based plaque assays can be used to detect viruses (5, 13, 15). Previously, Green and Lewis employed reverse transcription (RT)-PCR to detect HAV and rotavirus in concentrated samples using a conventional sample preparation method for virus detection in cell cultures (6); however, the efficiency of the recovery of viral RNA was not investigated. Haramoto et al. detected F-specific RNA phages in river sediments by combining culturing and qPCR methods (10), but to our knowledge, there is no report of quantitative data for human pathogenic enteric viral RNA in a river and in estuarine sediments obtained by RT-qPCR.
In this study, nine procedures to recover viral RNA from environmental sediments were newly developed and compared in terms of efficiency and robustness of recovery. The nine procedures employed in this study can be divided into two approaches, i.e., direct and indirect methods of viral RNA recovery. For direct methods, viral RNA was extracted directly from sediment samples, whereas viral particles were eluted from sediment samples before RNA extraction by indirect methods. The recovery rate was evaluated by using PV1 inoculated into sediment samples. The procedure exhibiting the highest and most stable recovery rate was used for viral RNA recovery from sediment samples collected from the Takagi River, Miyagi, Japan, and its estuary, and genomes of EV and human norovirus (HuNoV) were quantified by RT-qPCR.
MATERIALS AND METHODS
Environmental sediment samples.
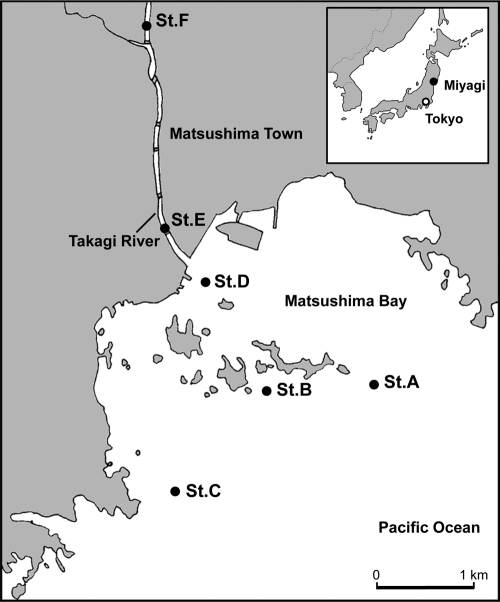
Sediment samples were collected monthly from the Takagi River, Miyagi, Japan, and its estuary during ebb tide. Figure 1 illustrates the sampling sites: sites A, B, and C were located in the bay; site D was located at the river mouth; and sites E and F were located in the downstream river. There is a small dam to control the flow of the river just below site F. Samples were obtained by using an Ekman-Birge-type bottom sampler with a square area of 15 cm by 15 cm; the top 1-cm layer was collected. All sediment samples in this study consisted mainly of silt and clay in the range of 67% to 85% and 8% to 13%, respectively (16). The water content and ignition loss (organic matter content calculated from the weight loss at 600°C for 1 h in an electric furnace) of the samples ranged from 62% to 81% and 2.9% to 3.8%, respectively (17). Sediment samples were transported to the laboratory in sterile containers on ice and stored at −20°C until analysis. One hundred eight samples were collected from the six sampling sites between November 2007 and April 2009.
Fig. 1.
Sampling sites (St.) at the Takagi River and its estuary region. (Adapted from reference 17 with permission.)
Indirect methods to recover viral RNA from sediments.
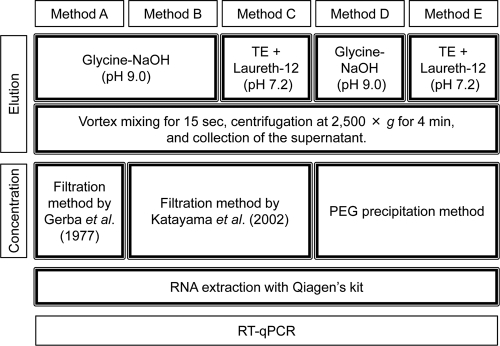
Sediment samples collected at site F were used to compare viral RNA recovery rates because sediment characteristics such as particle size distribution and water content were similar among sampling sites. All indirect methods tested in this study were modified on the basis of an approach developed previously by Gerba et al. (5); this approach does not use beef extract, a possible inhibitor of viral genome detection (21). Indirect methods consisted of three steps: the elution of viral particles by dispersing sediments in buffer solutions and centrifugation to remove the sediments (elution step), concentration of the eluted viruses in the supernatant (concentration step), and viral RNA extraction from the virus concentrate (extraction step), using a QIAamp viral RNA minikit (Qiagen, Tokyo, Japan). Detailed elution and concentration procedures for each indirect method are described below and summarized in Fig. 2.
Fig. 2.
Summary of the elution and concentration steps for each indirect method.
(i) Method A.
For method A, glycine-NaOH buffer (pH 9.0) was used as the virus elution buffer instead of the original buffer at pH 11.5, and a 15-s vortex was employed instead of a 10-min agitation with a shake table (5). The modified procedure was as follows. Five grams of sediment sample under wet conditions was placed into a 50-ml centrifuge tube, and 15 ml of 0.25 M glycine-NaOH buffer (pH 9.0) containing 0.05 M EDTA was added. The tube was vortexed for 15 s and centrifuged at 2,500 × g for 4 min to remove the sediments. The supernatant was collected, and its pH was adjusted to 3.5 by using 1 M glycine-HCl buffer (pH 2.0). Aluminum chloride (1 M) was then added to yield a final concentration of 0.06 M, and the solution was passed through a type HA membrane filter (0.45-μm pore size and 90-mm diameter; Millipore, Tokyo, Japan). Viruses were eluted from the filter by the passage of 10 ml of 0.25 M glycine-NaOH buffer (pH 11.5), and the eluate was immediately neutralized by the addition of 1 M glycine-HCl buffer (pH 2.0).
(ii) Method B.
Method B was the same as method A, with one modification: the negatively charged membrane filtration method (12) was employed in the concentration step. After centrifugation at 2,500 × g for 4 min in the elution step, magnesium chloride (0.25 M) was added to the supernatant at a final concentration of 0.1 M, and the solution was passed through an HA membrane filter (0.45-μm pore size and 90-mm diameter; Millipore). Subsequently, 200 ml of 0.5 mM H2SO4 (pH 3.0) was passed through the filter, followed by 10 ml of 1 mM NaOH (pH 11.0) to elute viral particles. The filtrate was recovered in a tube containing 100 μl of 50 mM H2SO4 and 100 μl of 100× Tris-EDTA (TE) buffer for neutralization.
(iii) Method C.
Method C was the same as method B, with one modification: TE buffer (pH 7.2) containing Laureth-12 (Kanto Chemical Co., Tokyo, Japan) was used instead of glycine-NaOH buffer (pH 9.0) in the elution step to prevent the elution of humic substances from the sediments. The elution buffer consisted of 0.1% (wt/vol) Laureth-12, 10 mM Tris, 1 mM EDTA, and 0.015% (vol/vol) Antifoam Y-30 (Sigma-Aldrich Co., Tokyo, Japan), which is in accordance with Method 1622 of the U.S. EPA (26). The negatively charged membrane filtration method (12) was employed in the concentration step as described above for method B.
(iv) Method D.
Method D was the same as method B except that instead of the membrane filtration method, the polyethylene glycol (PEG) precipitation method was applied in the concentration step. After centrifugation at 2,500 × g for 4 min in the elution step, an equal volume of a PEG solution containing 16% (wt/vol) PEG 6000 (Kanto Chemical Co.) and 4.7% (wt/vol) NaCl was added to the supernatant. The suspension was mixed vigorously and incubated overnight at 4°C. After centrifugation at 9,000 × g for 30 min at 4°C, the supernatant was discarded. The pellet was suspended in 1 ml of deionized distilled water (DDW) with a vortex mixer and centrifuged at 9,000 × g for 10 min at 4°C. The supernatant was collected as a virus concentrate.
(v) Method E.
For method E, TE buffer (pH 7.2) containing Laureth-12 was used in the elution step instead of the glycine-NaOH buffer (pH 9.0) used in method D to prevent the elution of humic substances from the sediments. The PEG precipitation method was applied in the concentration step as described above for method D.
Direct methods to recover viral RNA from sediments.
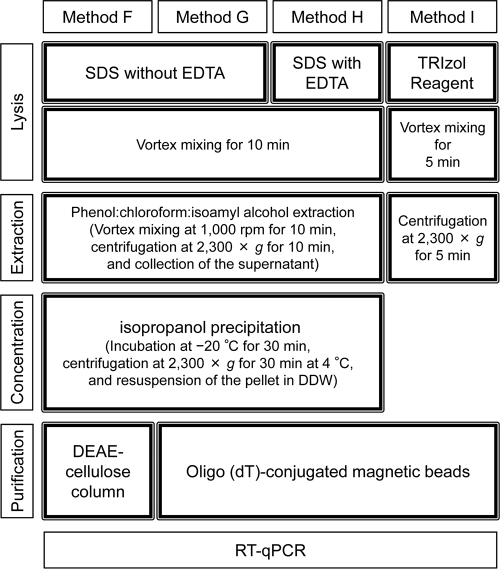
Direct methods consisted of four steps: lysis of viral capsids in sediments (lysis step), separation of viral RNA from sediments by centrifugation (extraction step), RNA concentration from the supernatant (concentration step), and purification of the RNA concentrate (purification step). Details of each of the above-mentioned procedures are summarized in Fig. 3 and described below.
Fig. 3.
Summary of the lysis, extraction, concentration, and purification steps for each direct method.
(i) Method F.
For method F, 5 g of wet sediment was placed into a 50-ml centrifuge tube. Five milliliters of 0.3 M sodium phosphate buffer (pH 5.8) was added to the sediment sample, and the sample was suspended using a vortex mixer. Five milliliters of lysis buffer (0.5 M Tris [pH 8.0], 0.1 M NaCl, 2% SDS, 8 mg skim milk/g sediment) was added to the suspension, and the tubes were processed in a Multi Vortex-Genie (SI-M286; Scientific Industries, New York, NY) at 1,000 rpm for 10 min. The lysis buffer was modified from that developed previously by Ikeda et al. (8), wherein EDTA was excluded to curb the elution of humic substances from sediments. Twelve milliliters of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol, molecular biology grade; Invitrogen, CA) was added, and the tubes were vortexed again at 1,000 rpm for 10 min. The tubes were centrifuged at 2,300 × g for 10 min at room temperature. The supernatant (upper aqueous phase of 12 ml) was collected and transferred into a clean 50-ml centrifuge tube. Twelve milliliters of isopropanol (molecular biology grade; Wako Pure Chemical Industries, Osaka, Japan) was added, and the sample was mixed vigorously with a vortex mixer. The resulting solution was incubated at −20°C for 30 min, and the tubes were centrifuged at 2,300 × g for 30 min at 4°C. The supernatant was decanted, and the tubes were inverted on a paper towel for 5 min. One milliliter of DDW (DNase/RNase free; Invitrogen) was added to the tubes, and the pellet was resuspended by placing the tubes into a heat block at 55°C for 5 min. Viral RNA was purified by using a DEAE-cellulose column according to the method described previously by Ikeda et al. (8). Briefly, the DEAE-cellulose resin (molecular biology grade; Wako Pure Chemical Industries) was suspended in 20 volumes of TE buffer (pH 8.0) containing 0.6 M NaCl. After the resin settled, the supernatant was discarded. Twenty volumes of TE buffer (pH 8.0) containing 0.6 M NaCl were added again, and the washing step was repeated. The equilibrated resin was stored at 4°C. Two milliliters of the resin was poured into the barrel of a 5-ml syringe (Terumo Co., Tokyo, Japan) plugged with a Millex syringe filter (5-μm pore size and 25-mm diameter; Millipore). The column was equilibrated with 2 ml of TE buffer (pH 8.0) containing 0.1 M NaCl (8). Two hundred microliters of TE buffer (pH 8.0) containing 0.2 M NaCl was added to an equal volume of extracted viral RNA and loaded onto the DEAE-cellulose column. After washing the column with 2 ml of TE buffer (pH 8.0) containing 0.4 M NaCl, viral RNA was eluted with 1 ml of TE buffer (pH 8.0) containing NaCl. The NaCl concentration in TE buffer for RNA elution was decided on the basis of the concentration of viral RNA in each eluted fraction (0.1 to 1 M), which was measured in advance.
(ii) Method G.
Method G was the same as method F except that magnetic beads were used in the RNA purification step instead of the DEAE-cellulose column. Briefly, after isopropanol precipitation in the concentration step, 200 μl of the concentrated viral RNA was purified by using FastTrack MAG Micro mRNA isolation kits (Invitrogen) according to the manufacturer's instructions.
(iii) Method H.
For method H, EDTA was added to the lysis buffer used in methods F and G at a final concentration of 0.1 M (8). It was expected that EDTA would dissolve multivalent cations in sediments that otherwise contribute to the adsorption of viral particles onto solids (7, 21) or increase the enzymatic activity of some bacterial RNases (3). The extraction, concentration, and purification steps were the same as those described above for method G.
(iv) Method I.
For method I, guanidine isothiocyanate was used in the RNA extraction buffer instead of SDS buffers. Briefly, 5 g of sediment (wet weight) was placed into a 50-ml centrifuge tube. Ten milliliters of TRIzol reagent (a monophasic solution of phenol and guanidine isothiocyanate; Invitrogen) was added to the tube, and the mixture was processed in the Multi Vortex-Genie at 1,000 rpm for 5 min. The tubes were centrifuged at 2,300 × g for 5 min at room temperature, and the supernatant was collected. Two hundred microliters of the supernatant containing viral RNA was purified by using FastTrack MAG Micro mRNA isolation kits as described above for methods G and H.
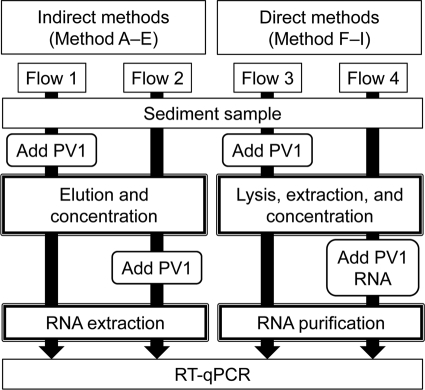
Recovery of PV1 inoculated into sediments.
Figure 4 illustrates an experimental flow for evaluating the rate of recovery of PV1 from sediments. PV1 was prepared by using the BGM kidney cell line and according to procedures described previously by Sano et al. (21). In the case of indirect methods (methods A to E), 1 μl (1.1 × 108 copies; standard deviation [SD] = 5.8 × 106 copies) of PV1 was inoculated into sediment samples, and the tubes were vortexed for 30 s. Methods A to E were applied for the recovery of the inoculated PV1 particles from sediment samples, and total rates of recovery of PV1 were calculated by using experimental flow 1 (Fig. 4). On the other hand, in experimental flow 2, sediment samples were first treated with methods A to E without PV1 inoculation, and 1 μl (1.1 × 108 copies; SD = 5.8 × 106) of PV1 was then added to each virus concentrate (Fig. 4) to evaluate the rates of recovery of the PV1 genome in the RNA extraction and RT-qPCR steps of the indirect methods. Based on the recovery rates with experimental flows 1 and 2, the recovery rates for the elution and concentration steps were calculated.
Fig. 4.
Experimental flows to evaluate the rates of recovery of inoculated PV1 from a sediment sample using indirect and direct methods.
In the case of direct methods (methods F to I), 1 μl (1.1 × 108 copies; SD = 5.8 × 106) of PV1 was inoculated into sediment samples, which were processed using methods F to I, and the total recovery rates were calculated (experimental flow 3) (Fig. 4). On the other hand, for experimental flow 4 (Fig. 4), sediment samples were processed without PV1 inoculation, and 1 μl (1.7 × 106 copies; SD = 8.1 × 105) of genomic RNA prepared from PV1 was added to the solution before the purification step to evaluate the recovery rates for the purification and RT-qPCR steps. Based on the recovery rates with experimental flows 3 and 4, the recovery rates for the lysis, extraction, and concentration steps were calculated.
Quantification of viral RNA.
cDNA was obtained from 5 μl of extracted RNA through reverse transcription using the First Strand cDNA synthesis kit for RT-PCR (Roche, Tokyo, Japan). The cDNA concentration of EV (including PV1) was determined by using qPCR with the LightCycler ST300 instrument (Roche) according to a method described previously by Monpoeho et al. (18). Each 20-μl PCR mixture contained 5 μl of cDNA, 4 μl of LightCycler TaqMan Master (Roche), 750 nM each primer, and 200 nM TaqMan probe (listed in Table 1). PCR conditions consisted of a denaturing step at 95°C for 10 min, followed by 50 cycles of 95°C for 15 s, annealing at 60°C for 20 s, and extension at 72°C for 11 s. The cDNA concentrations of HuNoV genogroups I and II were quantified by using qPCR with a CFX96 real-time system (Bio-Rad Laboratories, Tokyo, Japan) according to a method described previously by Kageyama et al. (11). Each 20-μl PCR mixture contained 5 μl of cDNA, 10 μl of iQ Supermix (Bio-Rad Laboratories), 400 nM each primer, 300 nM RING1(a)-TP, and 100 nM RING1(b)-TP for HuNoV GI or 300 nM RING2-TP for HuNoV GII (Table 1). PCR conditions consisted of a denaturing step at 95°C for 10 min, followed by 50 cycles of 95°C for 15 s, annealing at 56°C for 20 s, and extension at 72°C for 20 s. Based on the standard curve that was made by a 10-fold serial dilution of plasmid DNA (101 to 106 copies), the quantification limit was approximately 10 copies/PCR tube.
Table 1.
Primer and probe sequences for the detection of EV and HuNoV
| Virus | Primer or probe | Sequence (5′–3′)a | Reference |
|---|---|---|---|
| Enterovirus (poliovirus) | Ev2 (forward) | CCCCTGAATGCGGCTAATC | 18 |
| Ev1 (reverse) | GATTGTCACCATAAGCAGC | ||
| Ev-probe | FAM-TGGGAGGGCGATCGCAATCT-TAMRA | ||
| Human norovirus genogroup I | COG1-F | CGYTGGATGCGNTTYCATGA | 11 |
| COG1-R | CTTAGACGCCATCATCATTYAC | ||
| RING1(a)-TP | FAM-AGATYGCGATCYCCTGTCCA-TAMRA | ||
| RING1(b)-TP | FAM-AGATCGCGGTCTCCTGTCCA-TAMRA | ||
| Human norovirus genogroup II | COG2-F | CARGARBCNATGTTYAGRTGGATGAG | 11 |
| COG2-R | TCGACGCCATCTTCATTCACA | ||
| RING2-TP | FAM-TGGGAGGGCGATCGCAATCT-TAMRA |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
RESULTS AND DISCUSSION
PV1 recovery using indirect methods.
Table 2 shows the rates of recovery of inoculated PV1 from a sediment sample using indirect methods (methods A to E). PV1 was not recovered from any samples tested (n = 3) using method D. Because a large inhibitory effect on the steps of RNA extraction and RT-qPCR was observed when method D was used, the combination of virus elution with glycine-NaOH buffer and virus concentration with PEG precipitation appeared ineffective in removing substances that inhibited the molecular detection of viral RNA. Meanwhile, the rates of recovery of PV1 added to concentrates (experimental flow 2) (Fig. 4) were greater than 100% with other methods (methods A to C and E), indicating that the concentrates obtained using indirect methods did not include any substances that inhibited RNA extraction and RT-qPCR. Therefore, the recovery rates with the elution and concentration steps were estimated to match the total recovery rates for all methods excluding method D (Table 2).
Table 2.
Rates of recovery of inoculated PV1 from a sediment sample using indirect methodsa
| Method | Elution buffer | Concn method (reference) | Total (flow 1) |
Elution and concnb recovery rate (%) | Extraction and RT-qPCR (flow 2) |
No. of samples | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of copies detected/no. of inoculated copies | Recovery rate (%) |
No. of copies detected/no. of inoculated copies | Recovery rate (%) |
|||||||
| GM | GSD | GM | GSD | |||||||
| A | Glycine-NaOH | Filtration (5) | 1.9 × 106/1.1 × 108 | 1.8 | 0.17 | 1.8 | 1.7 × 108/1.1 × 108 | 165 | 0.06 | 3 |
| B | Glycine-NaOH | Filtration (12) | 5.7 × 106/1.1 × 108 | 5.4 | 0.53 | 5.4 | 1.2 × 108/1.1 × 108 | 112 | 0.20 | 9 |
| C | TE + Laureth-12 | Filtration (12) | 6.5 × 105/1.1 × 108 | 0.61 | 0.11 | 0.61 | 1.4 × 108/1.1 × 108 | 135 | 0.05 | 3 |
| D | Glycine-NaOH | PEG precipitation | 0/1.1 × 108 | — | — | Unknown | 0/1.1 × 108 | — | — | 3 |
| E | TE + Laureth-12 | PEG precipitation | 1.9 × 105/1.1 × 108 | 0.18 | 0.11 | 0.18 | 1.1 × 108/1.1 × 108 | 106 | 0.02 | 3 |
GM, geometric mean; GSD, geometric standard deviation; —, not detected.
Recovery rates in the elution and concentration steps were calculated on the basis of the assumption that the recovery rate for the extraction and RT-qPCR steps was 100%.
The geometric mean (GM) of the total PV1 recovery rate with method A was 1.8% (geometric standard deviation [GSD] = 0.17; n = 3). This recovery rate was much lower than that reported previously by Gerba et al., wherein 50% recovery was achieved (5). One reason for this difference in the recovery rate would be the differing sediment compositions. Gerba et al. used sediment samples consisting of organic mud and sand (5), whereas the sediment sample used in this study (from site F) was composed mainly of silt (67%) and clay (13%). As indicated previously by Johnson et al., the rate of virus recovery from sediments may depend on the particle size distribution of the sediment, particularly the composition of silt and clay (9). Johnson et al. tested some elution buffers for the recovery of PV1 from a variety of sediments, and the mean recovery rates were 3.2% and 0.9% from sediments containing 4.6% and 17.4% clay, respectively (9). The latter recovery rate and clay composition reported by Johnson et al. are comparable to those observed in this study, although those authors quantified PV1 using a cell culture-based plaque assay.
The GM of the rates of recovery of PV1 with method B was 5.4% (GSD = 0.53; n = 9), which was 3-fold higher than that with method A. This means that the negatively charged membrane filtration method (12) significantly improved PV1 recovery. However, the elution buffer consisting of TE plus Laureth-12 was ineffective even when used along with the negatively charged membrane filtration method; only 0.61% of inoculated PV1 was recovered (method C) (GSD = 0.11; n = 3) (Table 2). This buffer containing TE plus Laureth-12 also produced poor PV1 recovery when the concentration procedure was changed to PEG precipitation: only 0.18% of the inoculated PV1 was recovered (method E) (GSD = 0.11; n = 3) (Table 2). Laureth-12, a surfactant, is an important component of the buffer used to elute Cryptosporidium oocysts and Giardia cysts from membrane surfaces (26, 27). This surfactant may not be effective for eluting viral particles from environmental silty sediments. Another possibility is that the interfacial activity of Laureth-12 affected the adsorption of viral particles onto the negatively charged membrane. Consequently, the combination of glycine-NaOH buffer and the negatively charged membrane (method B) produced the highest recovery rate (5.4%) among indirect methods.
Indirect methods have advantages in the processability of relatively larger volumes of samples because the supernatant obtained in the elution step can be processed using various concentration methods for water samples, such as ultracentrifugation. Indirect methods would achieve a higher recovery efficiency if viral particles could be recovered in the elution and concentration steps without concentrating inhibitory substances.
PV1 recovery using direct methods.
Table 3 shows the PV1 recovery rates for the four direct methods (methods F to I). Naked genomic RNA of PV1 was added to the extracts from sediment samples to evaluate the inhibitory effects of coeluted substances on the purification and RT-qPCR steps. The PV1 recovery rate of method F, wherein SDS buffer and a DEAE-cellulose column were used for viral capsid lysis and RNA purification, respectively, was extremely low (GM = 0.09%; GSD = 0.004; n = 3). Because the recovery rate for the purification step using the DEAE-cellulose column and RT-qPCR was 15% (Table 3), the recovery rate for the lysis, extraction, and concentration steps was calculated to be 0.6%. This low recovery efficiency was improved approximately 10-fold when magnetic beads were employed for RNA purification: 0.77% of genomic RNA was recovered (method G) (GSD = 0.03; n = 3) (Table 3). This improvement of viral RNA recovery is owing to the increase in efficient recovery during the RNA purification and RT-qPCR steps (52%). This result indicates that magnetic beads, which can specifically capture RNA with poly(A) tails, were effective for purifying the positive-sense RNA genomes of viruses extracted from sediment samples.
Table 3.
Rates of recovery of inoculated PV1 from a sediment sample using direct methodsa
| Method | Lysis buffer | Purification method | Total (flow 3) |
Lysis, extraction, and concn recovery rate (%)b | Purification and RT-qPCR (flow 4) |
No. of samples | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of copies detected/no. of inoculated copies | Recovery rate (%) |
No. of copies detected/no. of inoculated copies | Recovery rate (%) |
|||||||
| GM | GSD | GM | GSD | |||||||
| F | SDS without EDTA | DEAE-cellulose | 9.9 × 104/1.1 × 108 | 0.09 | 0.004 | 0.6 | 2.6 × 105/1.7 × 106 | 15 | 0.18 | 3 |
| G | SDS without EDTA | Magnetic beads | 8.2 × 105/1.1 × 108 | 0.77 | 0.03 | 1.5 | 8.9 × 105/1.7 × 106 | 52 | 0.04 | 3 |
| H | SDS with EDTA | Magnetic beads | 1.2 × 107/1.1 × 108 | 11 | 0.02 | 41 | 4.7 × 105/1.7 × 106 | 27 | 0.10 | 7 |
| I | TRIzol reagent | Magnetic beads | 1.0 × 105/1.1 × 108 | 0.10 | 0.24 | 6.7 | 2.5 × 104/1.7 × 106 | 1.5 | 0.11 | 3 |
GM, geometric mean; GSD, geometric standard deviation.
Recovery rates for the lysis, extraction, and concentration steps were calculated from those of flow 3 and flow 4.
However, this effective purification of viral RNA with magnetic beads was not compatible with the TRIzol-based RNA extraction method, for which the recovery rate of PV1 was 0.10% (method I) (GSD = 0.24; n = 3) (Table 3). The RNA extraction efficiency with TRIzol itself was 6.7%, whereas the recovery rate for the purification and RT-qPCR steps was 1.5%. This means that the eluate obtained by TRIzol-based extraction includes substances that inhibit RNA purification with magnetic beads or that TRIzol itself affects the interaction of viral RNA with magnetic beads. On the other hand, the combination of RNA extraction using an SDS buffer with EDTA and RNA purification with magnetic beads exhibited the highest and most stable recovery rate (method H) (11%, with a GSD of 0.02) (Table 3). Because the recovery rate for the purification and RT-qPCR steps was 27% (Table 3), the rate of PV1 recovery for the lysis, extraction, and concentration steps was calculated to be 41%. These results indicate that the addition of EDTA to the SDS buffer dramatically improved viral RNA recovery from sediments. Because some bacterial RNases require divalent metal ion cofactors to maintain biological activity (3), the chelation of multivalent cations with EDTA may prevent the degradation of viral RNA even after extraction from sediments.
Quantitative detection of EV and HuNoV in environmental sediment samples.
One hundred eight sediment samples were collected from the Takagi River estuary from November 2007 to April 2009. Viral RNA was recovered from all samples using method H because this method resulted in the highest and most stable rate of recovery of inoculated PV1 among the nine approaches tested. A viral genome originating from HuNoV GII was detected in only one sample from site A, located at the bay, although its concentration was below the quantification limit (approximately 10 copies/PCR tube). Meanwhile, two samples that were positive for EV were obtained from site D in December 2008 and site E in February 2009. The concentrations of EV genomic RNAs in these positive samples were 8.6 × 102 copies/g (wet weight) (at site D) and 2.4 × 102 copies/g (wet weight) (at site E). To the best of our knowledge, this is the first report to obtain quantitative data for EVs in environmental sediment samples using RT-qPCR. In our previous study, total coliforms were detected from the sediment samples collected at sites A to F at concentrations of 7.3 × 102 to 7.5 × 104 CFU/100 g (dry weight) (16). In addition, human-specific Bacteroides-Prevotella 16S rRNA genetic markers were detected (17). These results suggested that the sediment samples were contaminated by human feces. However, the rates of detection of human pathogenic viruses were reasonably low. Although molecular detection methods such as PCR have been developed and widely used, particularly for noncultivable viruses (e.g., HuNoV), methods of preparing samples from sediments that are compatible with molecular detection methods have not been established. It was reported previously that humic substances coeluted from soil and sediments inhibit nucleic acid extraction (30), hybridization (24), and Taq DNA polymerase in PCRs (25, 28). The results obtained in this study suggested that the composition of multivalent cations in RNA extracts from sediments is also important for stably obtaining excellent rates of recovery of viral RNA.
In this study, nine approaches to recover viral RNA from environmental silty sediments were newly developed and compared to quantify human pathogenic viruses in sediments using RT-qPCR. The direct RNA extraction using an SDS buffer including EDTA to lyse viral capsids and magnetic beads to purify RNA (method H) exhibited the best rate of recovery of inoculated PV1, and method H was effective for quantifying viral RNA in environmental sediments using the molecular method.
Further discussion regarding the inhibitory control in molecular detection will be required to acquire correctly quantified values of virus concentrations in sediments. A mutant strain of mengovirus belonging to the family Picornaviridae has been successfully employed as a control for recovering HAV particles from clinical and shellfish samples (2). This mengovirus mutant could be applied as an inhibitory control for the methods developed in this study for extracting viral RNA from environmental sediments. The inhibition of RT and qPCR steps should be also monitored, and RNA transcripts obtained by in vitro transcription can be used for this purpose (2). An appropriate setup of inhibitory controls in quantifying enteric viral RNA in environmental sediments is crucial for future studies.
ACKNOWLEDGMENTS
This work was supported in part by the Japan Society for the Promotion of Science through grants-in-aid for scientific research (S, 19106009) and young scientists (startup, 20860010) as well as JSPS research fellowships for young scientists (19-5067).
Footnotes
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Alm E. W., Burke J., Spain A. 2003. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 37:3978–3982 [DOI] [PubMed] [Google Scholar]
- 2. Costafreda M. I., Bosch A., Pinto R. M. 2006. Development, evaluation, and standardization of a real-time reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72:3846–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuzic S., Hartmann R. K. 2005. Studies on Escherichia coli RNase P RNA with Zn2+ as the catalytic cofactor. Nucleic Acids Res. 33:2464–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dorner S. M., Anderson W. B., Slawson R. M., Kouwen N., Huck P. M. 2006. Hydrologic modeling of pathogen fate and transport. Environ. Sci. Technol. 40:4746–4753 [DOI] [PubMed] [Google Scholar]
- 5. Gerba C. P., Smith E. M., Melnick J. L. 1977. Development of a quantitative method for detecting enteroviruses in estuarine sediments. Appl. Environ. Microbiol. 34:158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Green D. H., Lewis G. D. 1999. Comparative detection of enteric viruses in wastewaters, sediments and oysters by reverse transcription-PCR and cell culture. Water Res. 33:1195–1200 [Google Scholar]
- 7. Gutierrez L., Mylon S. E., Nash B., Nguyen T. H. 2010. Deposition and aggregation kinetics of rotavirus in divalent cation solutions. Environ. Sci. Technol. 44:4552–4557 [DOI] [PubMed] [Google Scholar]
- 8. Ikeda S., Watanabe K. N., Minamisawa K., Ytow N. 2004. Evaluation of soil DNA from arable land in Japan using a modified direct-extraction method. Microbes Environ. 19:301–309 [Google Scholar]
- 9. Johnson R. A., Ellender R. D., Tsai S. C. 1984. Elution of enteric viruses from Mississippi estuarine sediments with lecithin-supplemented eluents. Appl. Environ. Microbiol. 48:581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haramoto E., et al. 2009. Application of real-time PCR assays to genotyping of F-specific phages in river water and sediments in Japan. Water Res. 43:3759–3764 [DOI] [PubMed] [Google Scholar]
- 11. Kageyama T., et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katayama H., Shimasaki A., Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LaBelle R. L., et al. 1980. Relationships between environmental factors, bacterial indicators, and the occurrence of enteric viruses in estuarine sediments. Appl. Environ. Microbiol. 39:588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Le Guyader F., Dubois E., Menard D., Pommepuy M. 1994. Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl. Environ. Microbiol. 60:3665–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lewis G. D., Loutit M. W., Austin F. J. 1985. A method for detecting human enteroviruses in aquatic sediments. J. Virol. Methods 10:153–162 [DOI] [PubMed] [Google Scholar]
- 16. Miura T., Masago Y., Chan Y. M., Imai T., Omura T. 2009. Detection of bacteria and enteric viruses from river and estuarine sediment. J. Water Environ. Technol. 7:307–316 [Google Scholar]
- 17. Miura T., Chan Y. M., Masago Y., Omura T. 2010. Evaluation of detection methods targeting host-specific Bacteroides spp. as a microbial source tracking marker, p. 13–19 In Fukushi K., Kurisu F., Oguma K., Furumai H., Fontanos P. (ed.), Southeast Asian water environment 4 . IWA Publishing, London, United Kingdom [Google Scholar]
- 18. Monpoeho S., et al. 2000. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. Biotechniques 29:88–93 [DOI] [PubMed] [Google Scholar]
- 19. Perez-Fuenteaja A., et al. 2006. Influence of limnological conditions on Clostridium botulinum type E presence in Eastern Lake Erie sediments (Great Lake, USA). Hydrobiologia 563:189–200 [Google Scholar]
- 20. Salvo V. S., Fabiano M. 2007. Mycological assessment of sediments in Ligurian beaches in the Northwestern Mediterranean: pathogens and opportunistic pathogens. J. Environ. Manage. 83:365–369 [DOI] [PubMed] [Google Scholar]
- 21. Sano D., Fukushi K., Yoshida Y., Omura T. 2003. Detection of enteric viruses in municipal sewage sludge by a combination of the enzymatic virus elution method and RT-PCR. Water Res. 37:3490–3498 [DOI] [PubMed] [Google Scholar]
- 22. Searcy K. E., Packman A. I., Atwill E. R., Harter T. 2006. Deposition of Cryptosporidium oocysts in streambeds. Appl. Environ. Microbiol. 72:1810–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith E. M., Gerba C. P., Melnick J. L. 1978. Role of sediment in the persistence of enteroviruses in the estuarine environment. Appl. Environ. Microbiol. 35:685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tebbe C. C., Vahjen W. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsai Y.-L., Olson B. H. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. U.S. EPA. 2005. Method 1622: Cryptosporidium in water by filtration/IMS/FA. U.S. EPA, Washington, DC.
- 27. U.S. EPA. 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. U.S. EPA, Washington, DC.
- 28. Watson R. J., Blackwell B. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46:633–642 [DOI] [PubMed] [Google Scholar]
- 29. Whittington R. J., Marsh I. B., Reddacliff L. A. 2005. Survival of Mycobacterium avium subsp. paratuberculosis in dam water and sediment. Appl. Environ. Microbiol. 71:5304–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou J. Z., Bruns M. A., Tiedje J. M. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]