Abstract
Streptococcus parasanguinis is among the most successful colonizers of the human body. Strain FW213 harbors a 7.0-kb cryptic plasmid, pFW213, with a copy number at 5 to 10 per chromosome. Sequence and functional analyses of pFW213 revealed that the open reading frame (ORF) encoding the replication protein (Rep) is essential for the replication of pFW213, and the putative plasmid addiction system (RelB and RelE) and an ORF (ORF6) with no known function are required for its stability. The minimal replicon of pFW213 contains the rep gene and its 5′-flanking 390-bp region. Within the minimal replicon, an A/T-rich region followed by 5 contiguous 22-bp repeats was located 5′ of the ATG of rep. No single-stranded replication intermediates were detected in the derivatives of pFW213, suggesting that pFW213 replicates via the theta replication mechanism. The minimal replicon was unstable in streptococcal hosts without selection, but the stability was greatly enhanced in derivatives containing the intact relBE genes. A Streptococcus-Escherichia coli shuttle vector, pCG1, was constructed with the pFW213 replicon. Plasmid pCG1 features a multiple cloning region and a spectinomycin resistance determinant that is expressed in both Streptococcus spp. and E. coli. Various streptococcal DNA fragments were cloned in pCG1, and the recombinant constructs were stably maintained in the streptococcal hosts. Since pCG1 is compatible with the most widely used streptococcal replicon, pVA380-1, pCG1 will provide a much needed tool allowing the cloning of two genes that work in concert in the same host.
INTRODUCTION
Plasmids are autonomously replicating extrachromosomal elements that generally do not carry genes essential for host cell survival. Knowledge of the basic replicon of naturally occurring plasmids is the foundation for building cloning, expression, and shuttle vectors. Most of the plasmids of Gram-negative bacterial origin replicate via theta replication, whereas most of the plasmids of Gram-positive bacterial origin replicate via rolling circle replication (RCR); exceptions exist in both cases (9). Generally, vectors derived from theta replication replicons have larger cloning size capacities and are more stably maintained than those of RCR replicons (24, 34, 40). Cloning of DNA fragments into RCR plasmids from Staphylococcus aureus leads to the formation of high-molecular-weight plasmid multimers, whereas DNA fragments of the same size inserted into the theta-replicating plasmid pAMβ1 remain segregation stable, indicating that the instability of the recombinant plasmids is a function of the RCR replication (18). The replication protein (Rep) of RCR plasmids possesses nicking-closing activity that recognizes and nicks a specific sequence within the double-stranded origin (dso). Therefore, it is hypothesized that the Reps of RCR plasmids could nick at sites with similar target sequences and, thus, generate various deletion derivatives during replication (2, 30).
The most commonly used replicons in the genetic analysis of oral streptococci are pAMβ1 and pVA380-1. Plasmid pAMβ1, which was originally isolated from Enterococcus faecalis, is a broad-host-range, conjugative, erythromycin (Em) resistance gene-carrying plasmid (7). The transmission of this plasmid between E. faecalis, Lactococcus lactis, and Lactobacillus spp. by conjugation is well documented (32, 36, 38). Plasmid pAMβ-1 replicates via the theta-type mechanism and, like most conjugative plasmids, is large (26.5 kb). Therefore, the use of pAMβ-1 as a cloning vector is limited. On the other hand, plasmid pVA380-1 is a 4.2-kb RCR plasmid isolated from Streptococcus ferus (29). This plasmid replicates in a broad range of Gram-positive bacterial hosts (11, 25). Similar to many RCR plasmids, pVA380-1 also contains a mob gene, which encodes a protein responsible for its mobilization. The mobilization of derivatives of pVA380-1 by a conjugative plasmid is an effective way to introduce foreign DNA into nontransformable streptococcal hosts (4, 22). Currently, most of the available streptococcal cloning and Streptococcus-Escherichia coli shuttle vectors are constructed based on the pVA380-1 replicon (11, 25, 27, 28). Given that plasmids of the same replication family are incompatible, this lack of diversity poses a limitation in genetic studies. The identification of other replicons able to function efficiently and stably in oral streptococci would allow the construction of cloning vectors and shuttle vectors that would be compatible with pVA380-1.
The human isolate Streptococcus parasanguinis FW213 (8) is a primary colonizer of the tooth surface and an opportunistic pathogen for subacute endocarditis (5). In an attempt to identify replicons for developing genetic tools that function in streptococci, we analyzed the cryptic plasmid pFW213 found in S. parasanguinis FW213 and utilized it to construct the shuttle vector described in this study.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. parasanguinis FW213, Streptococcus gordonii CH1, Streptococcus mutans GS5, Streptococcus sanguinis SK36, and their derivatives were grown in Todd-Hewitt (TH; Difco) broth or on agar plates at 37°C in a 5% CO2 atmosphere. Where indicated, Em at 5 μg ml−1, kanamycin (Km) at 250 μg ml−1, or spectinomycin (Sp) at 250 μg ml−1 was included in the growth medium (35). Recombinant E. coli strains were grown at 37°C with aeration in LB medium containing ampicillin (Ap) at 100 μg ml−1, Em at 200 μg ml−1, Km at 50 μg ml−1, Sp at 50 μg ml−1, or chloramphenicol (Cm) at 25 μg ml−1 as needed.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Phenotypea | Description | Source or reference |
|---|---|---|---|
| Streptococcal strains | |||
| S. parasanguinis FW213 | Wild-type strain harboring pFW213 | 7 | |
| S. gordonii CH1 | Plasmid-free, naturally competent streptococcal host | D. J. LeBlanc | |
| S. mutans GS5 | Plasmid-free, naturally competent streptococcal host | H. K. Kuramitsu | |
| S. sanguinis SK36 | Plasmid-free, naturally competent streptococcal host | T. Kitten | |
| E. coli plasmids | |||
| pCTL2 | Kmr | pDL290 harboring an internal fragment of pFW213 (nt 2996 to 5589) at SacI and NsiI sites | This study |
| pCTL4 | Kmr | pDL290 harboring an internal fragment of pFW213 (nt 1064 to 3621) at EcoRI site | This study |
| pDL290 | Kmr | pSC101 replicon-based E. coli vector | D. J. LeBlanc |
| pSU21 | Cmr | p15A replicon-based E. coli vector | 3 |
| Streptococcal plasmids | |||
| pCG1 | Spr | Streptococcus-E. coli shuttle vector composed of p15A origin, pFW213 basic replicon, and a spe encoding Sp resistance in both E. coli and streptococci | This study |
| pCTL14 | Spr | pFW213::spe at Klenow fragment-treated ClaI site | This study |
| pCTLr2 | Spr | Derivative of pCTL14, containing pFW213 nt 3800 to 5129 | This study |
| pCTLr3 | Spr | Derivative of pCTL14, containing pFW213 nt 3800 to 6180 | This study |
| pCTLr4 | Spr | Derivative of pCTL14, containing pFW213 nt 3800 to 6897 | This study |
| pCTLr5 | Spr | Derivative of pCTL14, containing pFW213 nt 3800 to 500 | This study |
| pDL276 | Kmr | pVA380-1 replicon-based Streptococcus-E. coli shuttle vector | 11 |
| pFW213 | Cryptic | Naturally occurring plasmid in S. parasanguinis FW213 | This study |
| pT12 | Spr | ExoIII deletion derivative of pCTL14, containing pFW213 nt 900 to 5700 | This study |
| pT16R | Spr | ExoIII deletion derivative of pCTL14, containing pFW213 nt 3800 to 900 | This study |
Cm, chloramphenicol; Km, kanamycin; Sp, spectinomycin; r, resistant.
Standard molecular manipulations.
Plasmid DNA was isolated from streptococcal strains by the method of Anderson and McKay (1). For large-scale preparation, plasmid DNA was purified further by centrifugation to equilibrium in cesium chloride-ethidium bromide (35). Plasmid DNA was introduced into S. parasanguinis FW213 by electroporation (6) and into S. gordonii CH1, S. mutans GS5, and S. sanguinis SK36 by natural transformation (23).
Total RNA was isolated from mid-exponential-phase cultures (optical density at 600 nm, ≈0.6) of S. parasanguinis FW213 by the method of Chen et al. (6) and further purified by using an RNeasy minikit (Qiagen). For reverse transcription-PCR (RT-PCR), the first-strand cDNA was synthesized from 10 μg of total cellular RNA with random hexamer primers. The cDNA was then amplified by PCR at high stringency (primer annealing set at 58°C) with primers specific for each open reading frame (ORF).
Restriction endonuclease and DNA-modifying enzymes were purchased from New England BioLabs (United States). Taq and Phusion DNA polymerase were purchased from Toyobo (Japan) and Finnzymes (Finland), respectively. Primers used in this study are listed in Table 2.
Table 2.
Primers used in this study
| Primer | Sequencea | Purpose |
|---|---|---|
| pFW213_ORF1_AS | 5′-GGTTGCTCAAATCGCCTTG | Expression analysis of ORF1 |
| pFW213_ORF1_S | 5′-GCTACATCCCAACGCATG | |
| pFW213_ORF2_S | 5′-GGCAGAATGGCCTCCCTAC | Expression analysis of ORF2 |
| pFW213_ORF2_AS | 5′-GACCGTGAGAATACGACGC | |
| pFW213_ORF3_S | 5′-GGTGCGTTATTCGCGCCG | Expression analysis of ORF3 |
| pFW213_ORF3_AS | 5′-GCTATTCCACGCCAACTATTG | |
| pFW213_ORF4_S | 5′-CTGGCATGACAAATATACGTC | Expression analysis of ORF4 |
| pFW213_ORF4_AS | 5′-GAGGAGCTCCATCTTCGTC | |
| pFW213_ORF5_S | 5′-GGTGATGTCTTTACGGTTC | Expression analysis of ORF5 |
| pFW213_ORF5_AS | 5′-GGTCCAGCATCTTCAGAG | |
| pFW213_ORF6_S | 5′ CGGGGTGAAAGTTTTGACTG | Expression analysis of ORF6 |
| pFW213_ORF6_AS | 5′-CAAAATTCCACCATCTCTTCG | |
| pFW213_ORF7_S | 5′-TTGATACCTGAAGCACAAGATGAT | Expression analysis of ORF7 |
| pFW213_ORF7_AS | 5′-TCACTTCTCTTACCAATAGCAATGA | |
| pFW213_ORF8_S | 5′-TTAAAACAGGAAGTCGGTGAGG | Expression analysis of ORF8 |
| pFW213_ORF8_AS | 5′-CCAAACCTTCACGCCAATTT | |
| pSU21AS | 5′-TTTTAAGGCAGTTATTGGTGCCT | To generate psu21 internal fragment |
| pSU21S | 5′-CATTCGCCATTCAGGCTGCG | |
| SpecPvuIS | 5′-GGATCCAAGCTTCGATCGTTCGAA | To generate pCTLr2, pCTLr3, pCTLr4, and pCTLr5 |
| Replicon2 | 5′-GATATTAGAATAGAGCGATCGAGCCATT | |
| Replicon3 | 5′-CCCCTAAAGCGATCGAAAGCTGGATGAC | |
| Replicon4 | 5′-CCGTACCGATCGCTGAAAATAGACCAG | |
| Replicon5 | 5′-GTTAGTACCTCCGATCGAAAAATTACAC | |
| secA2_S | 5′-AAAGGTGTTGCAGAATTAGGCGGC | qPCR for secA2 |
| secA2_AS | 5′-TCGTCCTCTCAACTGCCAGTCAAT | |
| gly_S | 5′-GTTGAAAGCGTGCGAACCCAGATA | qPCR for gly |
| gly_AS | 5′-CTTGTGGCACCAATTCCCTCGTTT | |
| galT2_S | 5′-GGAATACCTTCGCCCTTGTTTGGA | qPCR for galT2 |
| galT2_AS | 5′-AATTCCTTGCTGCACCAACTCCAC | |
| qPCR_S | 5′-TGGTTGGCATCCGTCTATCCCTAA | qPCR for pFW213 |
| qPCR_AS | 5′-TAAGAGGAATGCTCTCATGGTGGC |
Introduced restriction sequences are underlined.
Relative plasmid copy number determination.
Primers for quantitative real-time PCR (qPCR) were designed to have a predicted melting temperature of about 60°C and to generate products of approximately 100 bp in length. qPCRs were conducted using iQ SYBR green supermix (Bio-Rad). Serial dilutions (10−1 to 10−4) of total cellular DNA (10 μg) isolated from S. parasanguinis (containing native pFW213) were analyzed by qPCR using primer pairs specific for the rep of pFW213, the chromosome-borne secA2 (GenBank sequence accession number AY338765), gly (39), and galT2 (39). The copy numbers were calculated as the mean threshold cycle (CT) values of the amplicons of the chromosomal genes (single-copy reference) compared to the amplicon of the plasmid-borne rep using the formula 2ΔCT, where ΔCT is the difference between the threshold cycle number of the reference gene and that of the rep reaction.
Southern DNA-DNA hybridization and single-stranded DNA (ssDNA) detection.
Total cellular DNA was isolated from recombinant S. parasanguinis strains according to the method of Anderson and McKay (1) as modified by LeBlanc et al. (25). Four micrograms of isolated DNA was treated with RNase A and then divided into two aliquots. One aliquot was treated with S1 nuclease, and the other was not treated. Both S1 nuclease-treated and untreated samples were separated on 0.8% Tris-acetate-EDTA (TAE) agarose gels by electrophoresis and transferred to Hybond N+ membranes (Amersham) (37), with or without prior denaturation. Southern DNA-DNA hybridization was performed using a digoxigenin (DIG) DNA labeling and detection kit (Roche) according to the manufacturer's instructions.
Plasmid construction, sequencing analysis, and characterization of minimal replicon.
On the basis of the restriction endonuclease site map, two internal fragments of pFW213 were subcloned onto pDL290 (D. J. LeBlanc, personal communication) at the compatible sites to generate plasmids pCTL2 and pCTL4 for sequencing. Plasmid pCTL2 contains the SacI-NsiI fragment (nucleotides [nt] 2996 to 5589) of pFW213, and pCTL4 contains the EcoRI-EcoRI fragment (nt 1064 to 3261) (Fig. 1). The sequence of the region between these two clones (nt 5589 to 1064) was completed by primer walking. Automated DNA sequencing was done by Tri Biotech, Inc. (Taiwan), and the complete sequence of pFW213 was obtained from both directions. All sequence data assembly and analysis were performed using the Vector NTI software package and The European Molecular Biology Open Software Suite (EMBOSS). Database searches were performed using BLAST, located at the NCBI website.
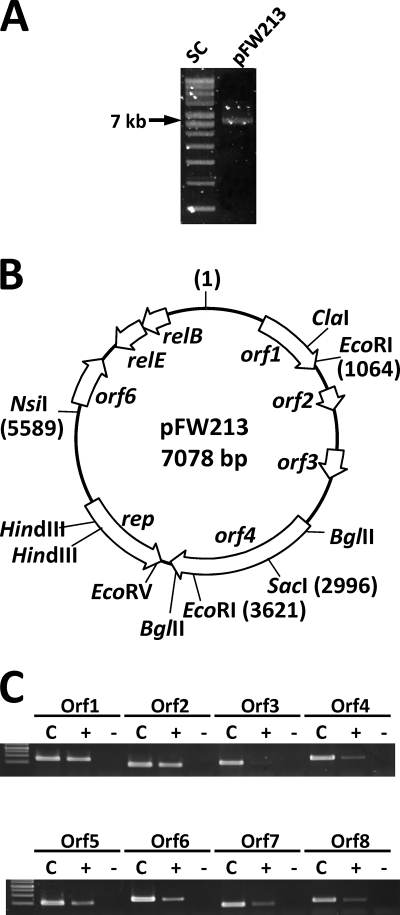
Fig. 1.
Plasmid pFW213. (A) Agarose gel electrophoresis of cesium chloride gradient-purified pFW213. SC, supercoiled DNA ladder. (B) Physical and genetic map of pFW213. Relative sizes, directions of transcription, and assigned designations are indicated. The assigned first base and positions of restriction sites used in subcloning pFW213 are listed in parentheses. (C) RT-PCR analysis of ORFs in pFW213. Two percent of the total cDNA generated from RT was amplified with specific primers, and 10% of the PCR products was run on a 1.8% TAE gel. C, products generated from total cellular DNA; +, reverse transcriptase was included in the reaction mixture; −, control groups without reverse transcriptase. A 1-kb DNA ladder was used as the marker.
A DNA fragment containing the Sp resistance gene (spe) (26) flanked by BamHI-PvuI and ApaI-NcoI sites was generated by PCR and cloned into the Klenow fragment-treated ClaI site of plasmid pFW213 to facilitate exonuclease III (ExoIII) analysis. The resulting plasmid (pCTL14) was digested with either BamHI-PvuI or ApaI-NcoI to allow for nested deletions generated by ExoIII from opposite ends of pFW213 (20). The resulting products were self-ligated and introduced into S. gordonii CH1. The properties of the plasmids isolated from Sp-resistant (Spr) transformants were verified by plasmid isolation and restriction endonuclease digestion. To define the minimal replicon of pFW213, fragments covering different lengths of the plasmid were generated by PCR using plasmid pT16R (Table 1) as the template. PCR products were digested, self-ligated, and transformed into S. gordonii CH1. The presence of desired recombinant plasmids in Spr transformants was confirmed as described above. All recombinant plasmids were then introduced into S. parasanguinis by electroporation and confirmed as described above.
Plasmid stability.
Derivatives of pFW213 were tested for stability in the streptococcal host. Briefly, overnight cultures of selected clones were grown in TH broth containing Sp to ensure the presence of the respective plasmid. The culture was then diluted at 10−4 in fresh TH broth without antibiotics and incubated for 16 h (13 generations) for a total of 10 days (117 generations). Isolated colonies of each subculture were obtained by serial dilution and plating on TH agar, and the percentage of Spr isolates at each time point was determined by inoculating 300 random colonies onto TH agar plates with and without Sp. The plasmid stability was calculated as the percentage of clones in the population that maintained the test plasmid.
Incompatibility assay.
The compatibility of pFW213 and pVA380-1 replicons was determined according to the method of Miki et al. (31), with minor modifications. The pVA380-1 replicon-based pDL276 (harboring kan) (11) was introduced into a pCTL14 (harboring spe)-containing S. parasanguinis strain, and transformants that were resistant to both Sp and Km were selected. The presence of both plasmids was confirmed as described above. Two Kmr and Spr double resistance colonies were suspended in 10 mM sodium phosphate buffer (pH 7), serially diluted, and then plated onto plain TH agar. After 16 h of incubation without selection (approximate 20 generations), 100 colonies were picked and patched onto TH agar plates. The patches were tested for the resistances conferred by either pDL276 (Kmr) or pCTL14 (Spr) by replica plating to TH agar containing Km or Sp, respectively. The process was repeated for 4 passages.
Construction of the shuttle cloning vector pCG1.
A DNA fragment containing the p15A origin of replication and the β-galactosidase gene (lacZ) of E. coli was amplified from pSU21 (3) by PCR using Phusion DNA polymerase with primer pair psu21S-psu21AS. The PCR product was then cloned into pCTLr2 at the T4 polymerase-treated PvuI site. The ligation mixture was used to transform E. coli DH10B, and Spr blue colonies on LB agar with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and Sp were selected. Recombinant plasmids were verified by plasmid isolation and restriction enzyme analysis, and the correct clone was further confirmed by sequence analysis. The resulting plasmid was designated pCG1.
Nucleotide sequence accession number.
The complete sequence of pFW213 has been deposited in GenBank with the accession number NC_012642.1.
RESULTS AND DISCUSSION
Identification and sequence analysis of pFW213.
One plasmid, designated pFW213, was detected in S. parasanguinis FW213 by agarose gel electrophoresis of the gradient-purified plasmid preparation (Fig. 1A). Sequence analysis revealed circular molecules of 7,078 bp with an overall 35% G+C, which is lower than the average G+C content (approximately 41%) of the known ORFs in S. parasanguinis FW213. The copy number of pFW213 in S. parasanguinis was estimated at 5 to 10 per chromosome by real-time qPCR analysis. Annotation of the sequence identified 8 ORFs greater than 50 amino acids (aa) in length. Among the predicted ORFs, 5 were transcribed from one orientation and 3 were transcribed from the opposite orientation (Fig. 1B). Specific mRNA was readily detected in all but ORF3 by RT-PCR; only a very faint band was detected for ORF3 under the experimental conditions used (Fig. 1C). The basic properties and promoter predictions of these ORFs are listed in Table 3.
Table 3.
Properties of ORFs identified in pFW213
| ORF | Position (nt) | Protein size (aa/kDa) | RBSa | Putative promoter sequenceb | Translation start codon | Protein with greatest similarityc | GenBank accession no.d |
|---|---|---|---|---|---|---|---|
| 1 | 502–1107 | 202/22.19 | AGAGG (8) | TGTGTTATAAT (70) | ATG | Conserved uncharacterized protein in S. sanguinis SK36 (202/202) | YP_001035954 |
| 2 | 1336–1524 | 63/7.13 | AGGGG (8) | TGGACG-N17-TATATC (74) | ATG | No significant similarity | NA |
| 3e | 1871–2125 | 85/9.88 | GGCAA (9) | TTGAGA-N15-TATTTT (136) | GTG | No significant similarity | NA |
| 4 | 2553–3838 | 432/51.5 | GAGGAA (8) | TGGTCA-N18-TTTAAA (118) | ATG | Mob of plasmid pBMYdx (392/426) | NP_981974 |
| 5 | 3928–4737 | 270/31.4 | AAAGG (8) | TTGACT-N18-TAGAAT (36) | ATG | RepB of plasmid pLME300 (242/259) | NP_783833.1 |
| 6 | 5615–6058 | 148/17.35 | None | TTCACA-N16-TAAACT (56) | GTG | No significant similarity | NA |
| 7 | 6206–6508 | 101/11.6 | AGAGG (4) | NA | TTG | RelE in S. downei F0415 (96/99) | EFQ56598.1 |
| 8 | 6502–6747 | 82/9.5 | GAGG (8) | TTGAAG-N18-TATAAT (26) | ATG | RelB in S. downei F0415 (77/79) | EFQ56528.1 |
The distance in nucleotides from the RBS to the translation start site is listed in parentheses.
The distance in nucleotides from the −10 element to the translation start site is listed in parentheses. NA, not available. N indicates G, A, T, or C.
The target that the ORF shares the highest homology to. Numbers in parentheses are the length of the alignment/target size, in amino acids.
GenBank sequence accession number of the locus listed under Similarity. NA, not available.
No significant product was detected by RT-PCR.
The deduced amino acid sequence of ORF1 was 94% identical to that of a hypothetical protein (SSA_2031) of S. sanguinis SK36. A high degree of homology (95% identity) was also observed with the C-terminal portion of the putative inorganic pyrophosphatase/exopolyphosphatase (YP_003363493) deduced from the newly sequenced Rothia mucilaginosa DY-18 genome. A transmembrane domain (aa 65 to 87) was suggested (http://www.cbs.dtu.dk/services/TMHMM/); however, no functional data are currently available for either protein. We also observed a putative rho-independent terminator (ΔGo = −12.19 kcal mol−1) located 19 bp 3′ of the stop codon of ORF1. Furthermore, no contiguous transcript was detected between ORF1 and ORF2 by RT-PCR, confirming that ORF1 is expressed as a single gene.
When performing BLAST analysis against the entire nonredundant GenBank database, no homology to ORF2 and ORF3 was found, albeit both ORFs contain ribosome binding sites (RBS) 5′ of the predicted translation start codon. Although the PCR product specific for ORF3 was not readily detected (Fig. 1C), we could not rule out the possibility that ORF3 was expressed at an extremely low level. On the other hand, the predicted protein encoded by ORF4 shared 35% and 31% identity with the Mob proteins of plasmid pBMYdx from Bacillus mycoides (10) and plasmid p9785s from Lactobacillus johnsonii FI9785 (21), respectively. A sequence identity of 32% was also observed with the putative Mob of pTX14-1 from Bacillus thuringiensis subsp. israelensis (GenBank sequence accession number AAB16962). These Mob proteins belong to the MOBV family of the conjugative transfer systems (14), among which the Mob protein of streptococcal plasmid pMV158 is the best studied. When ORF4 was compared to the members of pMV158 superfamily relaxases, the consensus sequences of the highly conserved motifs I (HXXR), II (NXXL), and III (HXDEXXPHXH) (13) were also found (data not shown). Although the 5′-flanking region of ORF2 shared 54% nucleotide identity with the oriT of pMV158, the proposed nick site and flanking sequences (13) were not found in pFW213. With an intact mob, pMV158 can be mobilized by a coresident conjugative plasmid (35). Multiple attempts failed to demonstrate mobilization activity of pFW213 in the presence of the conjugative plasmid pAMβ1 (data not shown). Thus, the function of the pFW213 ORF4 remains unclear.
ORF4 and ORF5 are separated by 89 bp and transcribed convergently. An inverted repeat (ΔGo = −10.96 kcal mol−1) which may act as the transcriptional terminator for ORF4 and ORF5 was located 25-bp 3′ of ORF4. Of note, this secondary structure is flanked by 5′-GACTTG (in the ORF4 orientation) and 5′-AATAAA (in the ORF5 orientation); thus, this shared terminator may have different strengths for ORFs 4 and 5. The highest homology (52% identity) was observed between the deduced amino acid sequence of ORF5 and the predicted theta replication protein RepB of pLME300 from Lactobacillus fermentum ROT1 (15). Identities of 52%, 46%, and 28% were also observed with the RepB proteins of pPLA4 (ABG23030.1) from Lactobacillus plantarum, of pYIT356 (YP_077168.1) from Lactobacillus casei, and of pKC5b (NP_862331.1) from L. fermentum KC5b (GenBank sequence accession numbers in parentheses). Among these three plasmids, the replication mode has been studied experimentally only in pKC5b, which replicates via the theta replication mode (34). As commonly seen in oriV of lactobacillus plasmids, including pLME300, pYIT356, and pKC5b, characteristics of iteron-based plasmids, such as a stretch of A/T-rich sequence and copies of about 20-bp direct-repeat (DR) sequences, are observed 5′ of the respective rep on each plasmid. Similarly, a 23-bp A/T-rich region and 5 copies of 22-bp DR sequences [5′-(G/A)(A/T)TGACTATTTTGTACCCCATT] were also found in the 5′-flanking region of ORF5 in pFW213 (Fig. 2).
Fig. 2.
Nucleotide sequence analysis of the putative ori of pFW213. The A/T-rich region is boxed. The DR sequence is overlined. The predicted RBS is italic and overlined, and the translation start codon of rep is indicated by a bent arrow.
ORF6 did not share any homology with ORFs in the database; neither could a putative RBS be found 5′ of the predicted translation start site. Thus, whether a protein is made from ORF6 is unclear. The translation start site of ORF7 is located within the 3′ end of ORF8. A contiguous transcript was identified from the midpoint of ORF7 to the end of ORF8 by RT-PCR analysis (data not shown), confirming that these ORFs are cotranscribed as an operon. A stem-loop structure (ΔGo = −10.96 kcal mol−1) was located 38 bp 3′ of ORF7. ORF7 and ORF8 shared 59% and 47% identity with the addiction module toxin of the RelE family and the antitoxin of the RelB family, respectively, of Streptococcus downei F0415. Of note, homologs of this relBE locus have been reported in several microbes via genome projects recently. Based on these sequence homologies, ORFs 5, 7, and 8 were designated rep, relE, and relB.
Based on the BLAST results for pFW213 and the variations in G+C content of its ORFs (from 47.61% of ORF2 to 32.65% of ORF6), it is likely that the formation of pFW213 is the result of multiple acquisitions of DNA fragments from various microbes via horizontal gene transfer. In addition to the coding region of ORF1, the 21-bp regions 5′ to ORF1 of pFW213 and to SSA_2031 of S. sanguinis SK36 are identical; however, further flanking sequences in the two species are not related. Notably, we were unable to clone the NsiI-EcoRI fragment (nt 5589 to 1064) of pFW213 in E. coli, even on the low-copy-number cloning vector pDL290, suggesting that the expression of ORF6, relBE, and/or ORF1 is lethal to E. coli.
Identification of the minimal replicon of pFW213.
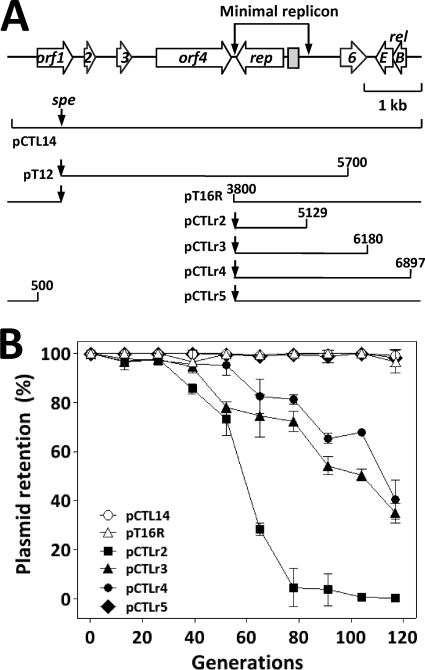
The two smallest ExoIII derivatives of pFW213 that could be established in S. gordonii CH1 were pT12 and pT16R (Fig. 3A). The overlapping region of these two plasmids contained the entire rep and its 1.2-kb 5′-flanking region (Fig. 3A). To further define the minimal replicon, DNA fragments of various lengths were generated by PCR using pT16R as a template. Under selective conditions, the smallest clone, pCTLr2, harbored the intact rep, all 5 copies of the DR sequence, and the A/T-rich region, thus defining the minimal replicon of pFW213.
Fig. 3.
The minimal replicon of pFW213. (A) Schematic diagram of pFW213. The relative location and the direction of transcription of each ORF in pFW213 are indicated by horizontal arrows. The A/T-rich sequence region and DRs are indicated by a shaded box. The smallest region that allows for replication in the streptococcal host is marked by two vertical arrows. The relative locations of the recombinant and ExoIII derivatives are listed below. The position of the spe in the derivatives is indicated by vertical arrows. (B) Stability of pFW213 derivatives in S. parasanguinis under nonselective conditions.
Stability of pFW213 replicons and detection of ssDNA intermediates.
Recombinant plasmids pCTLr2, pCTLr3, pCTLr4, and pCTLr5 were generated by PCR using pT16R as a template to characterize the effect of relBE on plasmid stability (Fig. 3A). As expected, the full-length pCTL14 was stable in S. parasanguinis, and no plasmid loss was observed after 117 generations of culturing (Fig. 3B). Both pT16R and pCTLr5 were also stably maintained over 117 generations (Fig. 3B), indicating that ORFs 1, 2, 3, and 4 are not essential for replication and stability. When the deletion was extended to nt 6897, 150 bp upstream of the translation start codon of relB, plasmid pCTLr4 (Fig. 3A) became unstable (Fig. 3B). After 65 generations of culturing, 18% of the colonies had lost the plasmid; at the 117th generation, the percentage of the cells that did not contain pCTLr4 was increased to 60%, indicating that the intergenic region between relBE and ORF1 critically affects the stability of the plasmid. The instability of the plasmid was relatively unchanged when the deletion was extended to nt 6180; plasmid pCTLr3 (Fig. 3A) was lost from 25% and 65% of the population after 65 and 117 generations of culturing, respectively (Fig. 3B). On the other hand, the stability of the replicon decreased significantly after the deletion was extended to nt 5129; plasmid pCTLr2 (Fig. 3A) was absent from 62% of the population after 65 generations of culturing and completely lost after 104 generations of culturing (Fig. 3B), demonstrating that the region upstream of rep, from nt 5129 to 6180, was crucial for the plasmid's stability. It is tempting to suggest that ORF6 regulates the stability of the pFW213 replicon through either the mRNA or protein generated from ORF6. Taken together, these results indicated that ORF6, the relBE, and the intergenic region between relBE and ORF1 all were important to the stability of pFW213. The toxin-antitoxin (TA) system is one of the mechanisms used by low-copy-number plasmids to maintain segregational stability (33). The inherent instability of the antitoxin (RelB) leads to activation of the toxin (RelE) in plasmid-free cells and, thus, ensures plasmid maintenance. Gotfredsen et al. (16) demonstrated that the E. coli relBEF operon could stabilize a mini-R1 replicon, whereas the relBE system of plasmid p307 is essential for the segregational stability of the plasmid (17). The RelB antitoxin is sensitive to the Lon protease in E. coli, and reduced segregational stability of the plasmid was observed in the Lon-deficient E. coli host (17). Thus, the results of the deletion analysis suggest that the relBE of pFW213 encodes a functional TA system that is essential for plasmid maintenance. Of note, among plasmids from Gram-positive bacteria, the RelBE system has been described in several Lactobacillus spp. plasmids (12, 19, 41). However, no homology of primary amino acid sequences was observed between the RelBE of pFW213 and those of Lactobacillus spp. plasmids.
The sequence homology between pFW213 Rep and other known RepB proteins suggests that pFW213 replicates via theta replication. To test whether pFW213 replicates via RCR, experiments were designed to determine whether ssDNA was present in streptococcal transformants containing the pFW213 derivatives pCTLr2 and pCTLr5. Plasmid pCTLr2 is the smallest replicon that replicates in both S. parasanguinis and S. gordonii, while plasmid pCTLr5 contains rep, relBE, and a partial ORF1. If pFW213 replicated via RCR, then pCTLr2 contains only dso, since pCTLr2 was not stably maintained in S. parasanguinis in the absence of selection (Fig. 3B) and, thus, accumulation of ssDNA intermediates would be apparent. However, we did not observe any S1 nuclease-sensitive targets in either strain, regardless of prior denaturation, supporting the concept that pCTLr2 does not replicate via RCR (Fig. 4). In addition, wild-type FW213 was also subject to the same analysis, and ssDNA was not detected (data not shown).
Fig. 4.
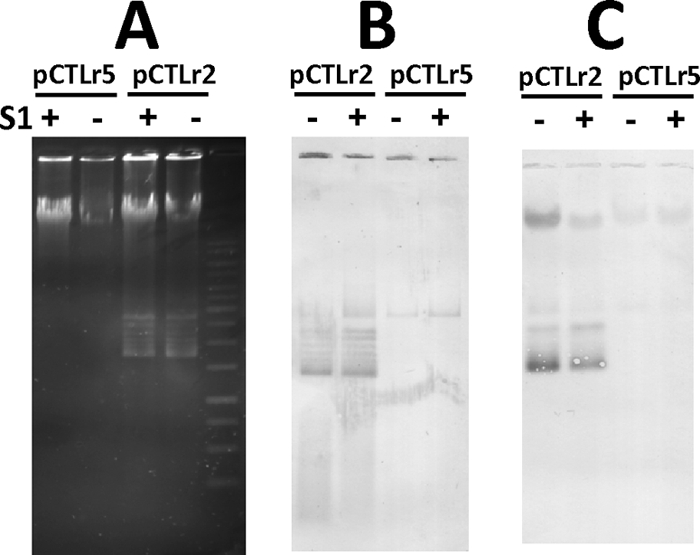
Absence of detectable pFW213-specific ssDNA in total cellular DNA from FW213 strains containing pCTLr2 and pCTLr5. (A) Agarose gel electrophoresis of plasmids isolated from recombinant S. parasanguinis FW213. (B) Southern transfer of samples shown in panel A without denaturation of DNA. (C) Southern transfer of samples shown in panel A with denaturation of DNA. Both blots were hybridized with DIG-labeled spe. +, S1 nuclease treated; −, no S1 nuclease treatment prior to electrophoresis.
Incompatibility.
The presence of pCTL14 and pDL276 in S. parasanguinis in the absence of selection was monitored to determine the compatibility of pFW213 and the pVA380-1 replicon. Both plasmids were stably maintained over 80 generations, indicating that these two replicons belong to different Inc groups.
Properties of the shuttle cloning vector pCG1.
The smallest pFW213 derivative, pCTLr2, was chosen as the basis for the construction of pCG1 (Fig. 5). The construction is detailed in Materials and Methods. Plasmid pCG1 was maintained stably in E. coli DH10B over 100 generations, as determined by the lack of alterations in the restriction enzyme patterns of isolated plasmids. Attempts to construct fusions with a pUC origin of replication failed, indicating that pCTLr2 cannot be stably maintained at high copy numbers in E. coli.
Fig. 5.
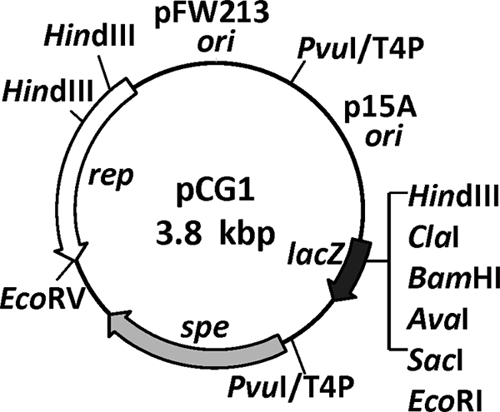
Restriction endonuclease map of the Streptococcus-E. coli shuttle vector pCG1. The locations of the fusion sites (PvuI/T4P, T4 polymerase-treated PvuI sites) are indicated.
The transformation efficiency of pCG1 in naturally competent, plasmid-free S. gordonii CH1 and S. mutans GS5 is approximately 400 colonies per μg of DNA isolated from a streptococcal host. When plasmid DNA was isolated from an E. coli host, a significant reduction (P ≤ 0.05, Student's t test) in transformation efficiency was observed (Table 4). The transformation efficiency in S. sanguinis SK36 was lower than that in S. gordonii and S. mutans, especially when plasmid DNA was isolated from recombinant S. gordonii. Restriction analysis of plasmids isolated from all streptococcal hosts confirmed that pCG1 was introduced and maintained successfully without any structural rearrangement. Furthermore, various streptococcal chromosomal fragments of 1 to 4 kb containing sequences covering an intact ORF and the promoter region were cloned into pCG1 in E. coli and then successfully transferred to streptococcal hosts by natural transformation or electroporation. Again, the recombinant plasmids were stably maintained in both hosts.
Table 4.
Transformation of streptococcal hosts with pCG1
| Species and strain | Transformation frequency (CFU μg DNA−1)a |
|
|---|---|---|
| pCG1/S. gordonii | pCG1/E. coli | |
| S. gordonii CH1 | 446 ± 114 | 193 ± 46 |
| S. mutans GS5 | 416 ± 55 | 129 ± 26 |
| S. sanguinis SK36 | 8 ± 2 | 53 ± 14 |
Plasmid pCG1 isolated from recombinant S. gordonii (pCG1/S. gordonii) or E. coli (pCG1/E. coli) was used. The numbers are the means and standard deviations of the results of three independent experiments.
To further confirm the compatibility of pCG1 and pVA380-1 replicon-based vectors in streptococcal hosts, we compared the transformation efficiency of pCG1 in native S. gordonii CH1, S. mutans GS5, and S. sanguinis SK36 with that of the same streptococcal hosts harboring pDL276. In agreement with the observation that pCTL14 and pDL276 can coexist in S. parasanguinis, a comparable transformation efficiency was detected with pCG1 in the same streptococcal host regardless of the presence of pDL276, confirming that the pFW213 and pVA380-1 replicons are compatible with each other.
Summary.
A theta-replicating Streptococcus-E. coli shuttle vector, pCG1, that allows for blue-white (X-Gal) selection in the E. coli host and is compatible with pVA380-1-based streptococcal vectors was constructed. Plasmid pCG1 contains the E. coli p15A origin of replication, the replication region of pFW213, a spectinomycin resistance gene expressed constitutively in both streptococcal and E. coli host strains, and the multiple cloning region of plasmid pSU21. This shuttle vector will facilitate experiments that require two compatible streptococcal plasmids in the same host.
ACKNOWLEDGEMENTS
This work was supported in part by the Chang Gung Memorial Hospital (grants CMRPD170173 and CMRPD170263) and the National Science Council (grant NMRPD 180973).
We thank P. Fives-Taylor, L. Lee, S. T. Liu, and S. Silver for careful review of the manuscript.
Footnotes
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Anderson D. G., McKay L. L. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ballester S., Lopez P., Espinosa M., Alonso J. C., Lacks S. A. 1989. Plasmid structural instability associated with pC194 replication functions. J. Bacteriol. 171:2271–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartolome B., Jubete Y., Martinez E., de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78 [DOI] [PubMed] [Google Scholar]
- 4. Buckley N. D., Lee L. N., LeBlanc D. J. 1995. Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J. Bacteriol. 177:5028–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burnette-Curley D., et al. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 63:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y. Y., Weaver C. A., Mendelsohn D. R., Burne R. A. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. 1974. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J. Bacteriol. 117:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole R. M., Calandra G. B., Huff E., Nugent K. M. 1976. Attributes of potential utility in differentiating among “group H” streptococci or Streptococcus sanguis. J. Dent. Res. 55:A142–A153 [DOI] [PubMed] [Google Scholar]
- 9. del Solar G., Giraldo R., Ruiz-Echevarria M. J., Espinosa M., Diaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Franco C., Pisaneschi G., Beccari E. 2000. Molecular analysis of two rolling-circle replicating cryptic plasmids, pBMYdx and pBMY1, from the soil gram-positive Bacillus mycoides. Plasmid 44:280–284 [DOI] [PubMed] [Google Scholar]
- 11. Dunny G. M., Lee L. N., LeBlanc D. J. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang F., et al. 2008. Characterization of endogenous plasmids from Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 74:3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Francia M. V., et al. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79–100 [DOI] [PubMed] [Google Scholar]
- 14. Garcillan-Barcia M. P., Francia M. V., de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657–687 [DOI] [PubMed] [Google Scholar]
- 15. Gfeller K. Y., Roth M., Meile L., Teuber M. 2003. Sequence and genetic organization of the 19.3-kb erythromycin- and dalfopristin-resistance plasmid pLME300 from Lactobacillus fermentum ROT1. Plasmid 50:190–201 [DOI] [PubMed] [Google Scholar]
- 16. Gotfredsen M., Gerdes K. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065–1076 [DOI] [PubMed] [Google Scholar]
- 17. Gronlund H., Gerdes K. 1999. Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 285:1401–1415 [DOI] [PubMed] [Google Scholar]
- 18. Gruss A., Ehrlich S. D. 1988. Insertion of foreign DNA into plasmids from gram-positive bacteria induces formation of high-molecular-weight plasmid multimers. J. Bacteriol. 170:1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagen K. E., Tramp C. A., Altermann E., Welker D. L., Tompkins T. A. 2010. Sequence analysis of plasmid pIR52-1 from Lactobacillus helveticus R0052 and investigation of its origin of replication. Plasmid 63:108–117 [DOI] [PubMed] [Google Scholar]
- 20. Henikoff S. 1984. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351–359 [DOI] [PubMed] [Google Scholar]
- 21. Horn N., Wegmann U., Narbad A., Gasson M. J. 2005. Characterisation of a novel plasmid p9785S from Lactobacillus johnsonii FI9785. Plasmid 54:176–183 [DOI] [PubMed] [Google Scholar]
- 22. LeBlanc D. J., Chen Y. Y., Lee L. N. 1993. Identification and characterization of a mobilization gene in the streptococcal plasmid, pVA380-1. Plasmid 30:296–302 [DOI] [PubMed] [Google Scholar]
- 23. LeBlanc D. J., Hassell F. P. 1976. Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J. Bacteriol. 128:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeBlanc D. J., Lee L. N. 1984. Physical and genetic analyses of streptococcal plasmid pAM beta 1 and cloning of its replication region. J. Bacteriol. 157:445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. LeBlanc D. J., Lee L. N., Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145 [DOI] [PubMed] [Google Scholar]
- 26. LeBlanc D. J., Lee L. N., Inamine J. M. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macrina F. L., et al. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145–150 [DOI] [PubMed] [Google Scholar]
- 28. Macrina F. L., Tobian J. A., Jones K. R., Evans R. P., Clewell D. B. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19:345–353 [DOI] [PubMed] [Google Scholar]
- 29. Macrina F. L., Wood P. H., Jones K. R. 1980. Genetic transformation of Streptococcus sanguis (Challis) with cryptic plasmids from Streptococcus ferus. Infect. Immun. 28:692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michel B., Ehrlich S. D. 1986. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 5:3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miki T., Easton A. M., Rownd R. H. 1980. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J. Bacteriol. 141:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morelli L., Sarra P. G., Bottazzi V. 1988. In vivo transfer of pAM beta 1 from Lactobacillus reuteri to Enterococcus faecalis. J. Appl. Bacteriol. 65:371–375 [DOI] [PubMed] [Google Scholar]
- 33. Nordstrom K., Austin S. J. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37–69 [DOI] [PubMed] [Google Scholar]
- 34. Pavlova S. I., et al. 2002. Characterization of a cryptic plasmid from Lactobacillus fermentum KC5b and its use for constructing a stable Lactobacillus cloning vector. Plasmid 47:182–192 [DOI] [PubMed] [Google Scholar]
- 35. Priebe S. D., Lacks S. A. 1989. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J. Bacteriol. 171:4778–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pucci M. J., Monteschio M. E., Kemker C. L. 1988. Intergeneric and intrageneric conjugal transfer of plasmid-encoded antibiotic resistance determinants in Leuconostoc spp. Appl. Environ. Microbiol. 54:281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Southern E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 38. Tannock G. W. 1987. Conjugal transfer of plasmid pAM beta 1 in Lactobacillus reuteri and between lactobacilli and Enterococcus faecalis. Appl. Environ. Microbiol. 53:2693–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu H., Zeng M., Fives-Taylor P. 2007. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect. Immun. 75:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wyckoff H. A., Barnes M., Gillies K. O., Sandine W. E. 1996. Characterization and sequence analysis of a stable cryptic plasmid from Enterococcus faecium 226 and development of a stable cloning vector. Appl. Environ. Microbiol. 62:1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang W., et al. 2008. Complete nucleotide sequence of plasmid plca36 isolated from Lactobacillus casei Zhang. Plasmid 60:131–135 [DOI] [PubMed] [Google Scholar]