Abstract
We previously isolated a mutant hypersensitive to l-alanyl-l-alanine from a non-l-alanine-metabolizing Escherichia coli strain and found that it lacked an inducible l-alanine export system. Consequently, this mutant showed a significant accumulation of intracellular l-alanine and a reduction in the l-alanine export rate compared to the parent strain. When the mutant was used as a host to clone a gene(s) that complements the dipeptide-hypersensitive phenotype, two uncharacterized genes, ygaW and ytfF, and two characterized genes, yddG and yeaS, were identified. Overexpression of each gene in the mutant resulted in a decrease in the intracellular l-alanine level and enhancement of the l-alanine export rate in the presence of the dipeptide, suggesting that their products function as exporters of l-alanine. Since ygaW exhibited the most striking impact on both the intra- and the extracellular l-alanine levels among the four genes identified, we disrupted the ygaW gene in the non-l-alanine-metabolizing strain. The resulting isogenic mutant showed the same intra- and extracellular l-alanine levels as observed in the dipeptide-hypersensitive mutant obtained by chemical mutagenesis. When each gene was overexpressed in the wild-type strain, which does not intrinsically excrete alanine, only the ygaW gene conferred on the cells the ability to excrete alanine. In addition, expression of the ygaW gene was induced in the presence of the dipeptide. On the basis of these results, we concluded that YgaW is likely to be the physiologically most relevant exporter for l-alanine in E. coli and proposed that the gene be redesignated alaE for alanine export.
INTRODUCTION
Bacteria are known to export xenobiotic substances, such as heavy metals (34), antibiotics (35), or organic solvents (49), to survive under harsh circumstances. In the last 15 years, it has been shown that in addition to harmful substances, normal metabolites, such as amino acids (10), purine ribonucleosides (13), and sugars (28), are exported by specific exporters. However, a physiological function of the exporters remains obscure. In regard to amino acids, after the identification of LysE as the exporter for lysine in Corynebacterium glutamicum (47), more than 10 transporters have been shown to export amino acids and their analogues. In C. glutamicum, BrnFE (20), NCgl1221 (32), and ThrE (43) were found to mediate the efflux of l-isoleucine, l-glutamic acid, and l-threonine, respectively. In Escherichia coli, exporters for l-cysteine (YdeD, YfiK, CydDC, and Bcr) (7, 12, 37, 50), l-aromatic amino acids (YddG) (9), l-leucine (YeaS) (26), l-threonine (RhtA and RhtC) (29, 51), l-arginine (YggA) (33), l-valine (YgaZH) (36), and l-homoserine (RhtB) (51) were identified.
The specific exporter of alanine has not been identified so far in E. coli and other bacteria except for a peculiar case in Tetragenococcus halophilus, where AspT has been found to function as an l-asparate:l-alanine exchanger (1). On the one hand, a wide range of wild-type and metabolically engineered bacterial strains (14, 17, 19, 21) have been observed to excrete alanine. In addition, E. coli, the wild-type strain of which does not intrinsically excrete alanine (21), has been engineered to produce l-alanine in the culture medium (53), indicating that E. coli possesses an l-alanine export system(s). To clarify the alanine export system in this bacterium, we previously isolated mutants hypersensitive to extracellularly added l-alanyl-l-alanine (Ala-Ala) (18) based on the knowledge that an amino acid exporterless mutant exhibits growth arrest in the presence of a peptide containing the amino acid of concern (20, 43, 47). Consequently, we found that the mutants lacked an inducible l-alanine export system (18).
In this study, we describe the identification of genes that complemented the hypersensitivity of the mutant to the dipeptide and show lines of evidence demonstrating that one of the genes identified probably plays a physiologically important role in l-alanine efflux in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains and plasmids used in this study are listed in Table 1. The primers used are listed in Table 2. Cells were grown aerobically at 37°C in L broth medium containing 1% tryptone, 0.5% yeast extract, and 0.5% NaCl (pH 7.2) or minimal medium (11) containing 22 mM glucose, 7.5 mM (NH4)2SO4, 1.7 mM MgSO4, 7 mM K2SO4, 22 mM NaCl, and 100 mM sodium phosphate (pH 7.1). When necessary, d-alanine (50 μg ml−1), l-alanine (50 μg ml−1), gentamicin (GM; 6.25 μg ml−1), kanamycin (KM; 6.25 μg ml−1), chloramphenicol (CP; 12.5 μg ml−1), tetracycline (TC; 12.5 μg ml−1), and ampicillin (AP; 100 μg ml−1) were added to the medium. Growth was monitored by measuring the optical density at 660 nm (OD660).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant property(ies)a | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| MG1655 | Wild type | Laboratory strain |
| W3110 | Wild type | Laboratory strain |
| JM109 | recA1 endA1 gyrA96 thi hsdR17(rK− mK+) e14− (mcrA) supE44 relA1 Δ(lac-proAB)/F[traD36 proAB+lacIqlacZΔM15] | Laboratory strain |
| BL21(DE3) | F−dcm ompT hsdS (rB− mB−) gal λ(DE3) | Novagen |
| MLA301 | MG1655 alr::FRT dadX::FRT avtA::GMryfbQ::KMryfdZ::FRT | 18 |
| LAX12 | MLA301 hypersensitive to Ala-Ala | 18 |
| MLA301ΔytfF | MLA301 with a deletion in the ytfF gene | This study |
| MLA301ΔygaW | MLA301 with a deletion in the ygaW gene | This study |
| Bacillus sphaericus ATCC 10208 | Wild type | Laboratory strain |
| Streptococcus bovis JCM5802 | Wild type | Laboratory strain |
| Plasmids | ||
| pBR322 | CPr TCr; pMB1 ori | Takara |
| pSTV29 | CPrlacZ′; p15A ori | Takara |
| pET-15b | APrlacI; promoter T7; pMB1 ori | Novagen |
| pTH18cs1 | cat1 lacZ′ repAts1; derivative of pSC101 | 14 |
| pYeaS | pSTV29 harboring a 1.1-kb PCR fragment of the yeaS gene | This study |
| pYddG | pSTV29 harboring a 1.5-kb PCR fragment of the yddG gene | This study |
| pYtfF | pSTV29 harboring a 1.4-kb PCR fragment of the ytfF gene | This study |
| pYgaW | pSTV29 harboring a 1.0-kb PCR fragment of the ygaW gene | This study |
| pET-YgaW | pET-15b harboring a 0.5-kb PCR fragment of the ygaW gene | This study |
| pΔYtfF | pYtfF with a deletion of a 0.3-kb fragment in the ytfF gene | This study |
| p18cΔYtfF | pTH18cs1 harboring a BamHI fragment of pΔYtfF | This study |
| pΔYgaW | pYgaW with a deletion of a 0.4-kb fragment in the ygaW gene | This study |
| p18cΔYgaW | pTH18cs1 harboring a BamHI fragment of pΔYgaW | This study |
| pAlaD | pSTV29 harboring a 1.8-kb PCR fragment of the Bacillus sphaericusalaD gene preceded by the Streptococcus bovis ldh promoter | This study |
| pAldD-YgaW | pAlaD harboring a 1.0-kb PCR fragment of the ygaW gene | This study |
Abbreviations for antibiotics: GM, gentamicin; KM, kanamycin; CP, chloramphenicol; TC, tetracycline; AP, ampicillin.
Table 2.
Primers used in this study
| Primer | Nucleotide sequencea | Restriction site |
|---|---|---|
| yeaS-F | 5′-CGGGATCCCGACGTCAGCGAAAGTGC-3′ | BamHI |
| yeaS-R | 5′-CGGGATCCCAGAAAGGCGTTGAGCG-3′ | BamHI |
| yddG-F | 5′-CGGGATCCAGTGCTTGCTTCGGGG-3′ | BamHI |
| yddG-R | 5′-CGGGATCCATAGACTTCGGCAGCGTG-3′ | BamHI |
| ytfF-F | 5′-CGGGATCCCGGCACTGAAAAGCGTCG-3′ | BamHI |
| ytfF-R | 5′-CGGGATCCAGAGAGCGCCAGTTCACC-3′ | BamHI |
| ygaW-F | 5′-CGGGATCCCGTTACCTCACCCCCAAAC-3′ | BamHI |
| ygaW-R | 5′-CGGGATCCCGCGAATGGGACGTACCG-3′ | BamHI |
| ygaW15b-F | 5′-CGGGATCCATATGTTCTCACCGCAGTCACG-3′ | NdeI |
| ygaW15b-R | 5′-CGGGATCCTCAGGCTTTTACCTGCTGGT-3′ | BamHI |
| ΔytfF-F | 5′-CGGATATCGATGATGTGGGCAACAGC-3′ | EcoRV |
| ΔytfF-R | 5′-CGGATATCTCATCGTGGAAACAGGCG-3′ | EcoRV |
| BSalaD-F | 5′-GATGTTTAGATAAATGAAGATTGGTATTCC-3′ | |
| BSalaD-R | 5′-AAACTGCAGCTAATCCACCATAAATG-3′ | PstI |
| SBPldh-F | 5′-GCTGGATCCATAAATTGATGAATCAC-3′ | BamHI |
| SBPldh-R | 5′-GGAATACCAATCTTCATTTATCTAAACATC-3′ | |
| yeaSRT-F | 5′-CGGTTATCTTGCGGCCTG-3′ | |
| yeaSRT-R | 5′-TTGCAGCGTCGCCAGTCG-3′ | |
| yddGRT-F | 5′-AGGCGGTGACAATGGGTTAC-3′ | |
| yddGRT-R | 5′-TTTAATCATGACGGGCGTGC-3′ | |
| ytfFRT-F | 5′-GCAGGGTTGATGTGGGGG-3′ | |
| ytfFRT-R | 5′-GAATGACCACCGGCAGGG-3′ | |
| ygaWRT-F | 5′-TTCTCACCGCAGTCACGC-3′ | |
| ygaWRT-R | 5′-CTGCTGGTAACGGCTGAC-3′ | |
| gapART-F | 5′-TGAATGGCAAACTGACTGGTATGGC-3′ | |
| gapART-R | 5′-AACCGGTTTCGTTGTCGTACCAGGA-3′ |
Underlines represent restriction sites in the tag sequences added.
Shotgun cloning.
DNA manipulation was performed according to the standard protocol described previously (40). Chromosomal DNA of E. coli W3110 was isolated according to a previously described method (39). The DNA library of the strain W3110 was constructed by ligating Sau3AI-digested DNA fragments (2 to 10 kb) with pBR322 (Takara, Japan), which had been treated with BamHI and bacterial alkaline phosphatase (Toyobo, Japan), using E. coli JM109 as host cells. The genome library purified from the transformants was introduced into the dipeptide-hypersensitive mutant LAX12 by electroporation with the Gene Pulser II (Bio-Rad) according to the manufacturer's instructions. Selection was performed on minimal medium containing 50 μg ml−1 d-alanine and 100 μg ml−1 AP in the presence of 3 mM Ala-Ala at 37°C for 3 days.
Construction of recombinant plasmids expressing the yeaS, yddG, ytfF, and ygaW genes.
To construct the plasmids harboring yeaS, yddG, ytfF, and ygaW, the gene fragments were amplified by PCR using the primer sets (forward/reverse) of yeaS-F/yeaS-R, yddG-F/yddG-R, ytfF-F/ytfF-R, and ygaW-F/ygaW-R, respectively, and the chromosome of MG1655 as a template. Each amplified DNA was treated with BamHI and cloned into the BamHI site of pSTV29 (Takara, Japan). We then carried out restriction analyses of the resulting plasmids and obtained the recombinant plasmids, pYeaS, pYddG, pYtfF, and pYgaW, carrying the yeaS, yddG, ytfF, and ygaW genes, respectively, genes that were placed in the opposite orientation relative to the lac promoter. Thus, their expression is under the control of their own promoters.
Cellular localization of YgaW.
To assess cellular localization of YgaW, we constructed the histidine (His)-tagged ygaW gene. The open reading frame (ORF) of the ygaW gene was amplified by PCR using the primer set ygaW15b-F/ygaW15b-R. The resulting fragment was digested with NdeI and BamHI and cloned into pET-15b (Novagen) restricted with the same enzymes, leading to pET-YgaW. We then introduced pET-YgaW into E. coli BL21(DE3) cells, and the resulting transformant was grown in L broth containing 100 μg ml−1 AP at 37°C to a mid-log phase (OD660 = 0.6). The expression of His-tagged YgaW was initiated by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside and continued at 37°C for 2 h. Cells were then harvested, washed once with 50 mM Tris-HCl buffer (pH 8.0), and suspended in a breaking buffer containing 50 mM Tris-HCl (pH 8.0) and 1 mM phenylmethanesulfonyl fluoride. After disruption of the cells by sonicating them five times for 15 s with 45-s intermittent cooling at the maximum level using the Bioruptor (model UCD-200T; CosmoBio Co., Japan), the supernatant obtained by centrifugation (15,000 × g, 4°C, 20 min) was ultracentrifugated at 100,000 × g at 4°C for 1 h to obtain the membrane fraction. To purify His-tagged YgaW, the membrane fraction was resuspended in a solubilization buffer containing 50 mM Tris-HCl (pH 8.0), 0.5% sodium dodecyl sulfate (SDS), 10 mM imidazole, and 300 mM NaCl, followed by the addition of Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Germany). After incubation with gentle agitation at 4°C for 1 h, the resin was washed with the solubilization buffer containing 20 mM imidazole and the protein was eluted with an elution buffer containing 50 mM Tris-HCl (pH 8.0), 0.5% SDS, and 250 mM imidazole. Each fraction was mixed with an equal volume of a sample buffer containing 125 mM Tris-HCl (pH 6.8), 4% SDS, 10% sucrose, 0.004% bromophenol blue, and 10% 2-mercaptoethanol at 23°C for 5 min and then subjected to SDS-polyacrylamide gel (15%) electrophoresis. The proteins were visualized by staining with Coomassie brilliant blue R250.
Construction of gene deletion mutants.
To construct ygaW and ytfF deletion mutants, we made the chromosomal deletion plasmids p18cΔYgaW and p18cΔYtfF, respectively. For the ygaW deletion plasmid, the NruI fragment containing the final 401 bp of the ygaW gene and 12 bp downstream was removed by NruI digestion and religation to generate pΔYgaW. pΔYgaW retained a putative stem-and-loop structure of the ygaW gene, thus it is most unlikely to exert any influence on expression of the downstream gene. The BamHI fragment of pΔYgaW was excised and cloned into the BamHI site of pTH18cs1 (15), leading to p18cΔYgaW. For the ytfF deletion plasmid, inverse PCR was performed using the primer set ΔytfF-F/ΔytfF-R and pYtfF as a template. The resulting fragment was digested with EcoRV and religated to yield pΔYtfF. The BamHI fragment of the resulting plasmid was excised and cloned into the BamHI site of pTH18cs1, leading to p18cΔYtfF. After transformation of p18cΔYgaW and p18cΔYtfF into E. coli MLA301, the resulting transformants were grown in L broth containing 50 μg ml−1 d-alanine, 6.25 μg ml−1 GM, 6.25 μg/ml KM, and 12.5 μg ml−1 CP at 42°C overnight, and integrants were selected on L agar containing 50 μg ml−1 d-alanine, 6.25 μg ml−1 GM, 6.25 μg ml−1 KM, and 12.5 μg ml−1 CP. Subsequently, the ygaW and ytfF deletion mutants MLA301ΔygaW and MLA301ΔytfF, respectively, were obtained by selecting CP susceptible clones. Deletion of each gene in the chromosome was verified by PCR analysis using the primer sets (forward/reverse) of ygaW-F/ygaW-R and ytfF-F/ytfF-R, respectively.
Amino acid accumulation assay.
To determine intracellular and extracellular l-alanine concentrations, cells grown in minimal medium containing 50 μg ml−1 d- and l-alanine were inoculated into minimal medium containing 50 μg ml−1 d-alanine and 1% tryptone. Cells cultivated to a mid-log phase were washed twice with ice-cold minimal medium and suspended in prewarmed minimal medium (37°C) to give an OD660 of 3.0, which corresponds to 1.14 mg of cells (dry weight) ml−1. After a 10-min preincubation at 37°C, the reaction was initiated by the addition of 6 mM Ala-Ala. Separation of the intracellular and extracellular fractions was performed by the silicone oil method (22), in which the cells were placed onto the upper layer of a 3:2 mixture of silicone oil AR20 and AR200 (Wacker Chemie, Germany), with the lower layer consisting of 20% (wt/wt) perchloric acid, followed by centrifugation (20,000 × g, 23°C, 1 min). The cell suspension medium remaining above the silicone layer was recovered as the extracellular fraction. The cell pellets were sonicated briefly, and the resulting cell suspension was centrifuged (20,000 × g, 23°C, 5 min). The supernatant was neutralized with 2 M Na2CO3 to obtain the intracellular fraction. Amino acids in each fraction were quantified in terms of their o-phthalaldehyde derivatives by a cation exchange column (Shim-pack AMINO NA; Shimadzu, Japan) with a high-performance liquid chromatography system (LC-10A; Shimadzu, Japan). To calculate the intracellular amino acid concentration, the intracellular volume was assumed to be 2.03 μl/mg (dry weight) cells (42).
Total RNA isolation and RT-PCR analysis.
Cells of the mid-log phase, which had been prepared as described for the amino acid accumulation assay, were incubated with 6 mM Ala-Ala at 37°C for 5 min and then mixed with RNAprotect bacterial reagent (Qiagen, Germany). After centrifugation (5,000 × g, 23°C, 10 min), total RNA was isolated using the RNeasy minikit with a spin column (Qiagen, Germany) according to the manufacturer's instructions. To remove contaminating DNA, the isolated RNA was treated twice with RNase-free DNase. Reverse transcription (RT) was performed with 300 ng of total RNA as a template by 2.5 units of AMV reverse transcriptase XL (Takara, Japan) in a total volume of 10 μl using the following reaction conditions: 30°C for 10 min, 42°C for 30 min, 95°C for 5 min, and 4°C for 5 min. PCRs were then performed using 0.5 μl of cDNA products (corresponding to 15 ng total RNA), Quick Taq HS DyeMix (Toyobo, Japan), and the primer sets yeaSRT-F/yeaSRT-R, yddGRT-F/yddGRT-R, ytfFRT-F/ytfFRT-R, ygaWRT-F/ygaWRT-R, and gapART-F/gapART-R in a total volume of 5 μl. The amplifying condition was as follows: 94°C for 2 min, 30 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 30 s, and final extension at 68°C for 5 min. The PCR products were analyzed by electrophoresis on a 2% agarose gel.
Alanine fermentation.
The modified overlap extension method (16) was used to construct an expression plasmid for alanine dehydrogenase (AlaDH) of Bacillus sphaericus (25). The gene fragment including the ORF and the downstream region of the alaD gene from B. sphaericus ATCC 10208 was amplified by PCR using the primer set BSalaD-F/BSalaD-R. The promoter region of the ldh gene from Streptococcus bovis JCM5802, which is efficiently transcribed in E. coli, was amplified by PCR using the primer set SBPldh-F/SBPldh-R. The amplified fragments were then fused by PCR using the primer set SBPldh-F/BSalaD-R, followed by cloning into pSTV29 restricted with BamHI and PstI, leading to pAlaD, in which the fused alaD gene was placed in the opposite orientation relative to the lac promoter. Next, the ygaW gene fragment amplified by PCR using the primer set ygaW-F/ygaW-R was digested with BamHI and cloned into the BamHI site of pAlaD, followed by restriction analysis to determine the direction of the ygaW gene. The resulting plasmid, pAlaD-YgaW, appeared to have the ygaW gene in the opposite orientation relative to alaD. MG1655 cells harboring pAlaD-YgaW, pAlaD, or pSTV29 grown aerobically in L broth containing 12.5 μg ml−1 CP were inoculated into a fermentation medium containing 1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.5% glucose, 1% NH4Cl, 100 mM morpholinepropanesulfonic acid (MOPS; pH 7.2), and 12.5 μg ml−1 CP and cultivated at 37°C with gentle shaking under anaerobic conditions where nitrogen gas constituted the gas phase of a test tube. The concentrations of alanine produced and glucose remaining in the culture medium were determined by a high-performance liquid chromatography system and the Glucose CII test (Wako Pure Chemical Industries, Japan), respectively.
Other methods.
Protein concentration was determined by the method of Lowry et al., with bovine serum albumin as the standard (30). The nucleotide sequences of the yeaS, yddG, ytfF, and ygaW genes were determined by the dideoxy chain termination method (41).
RESULTS
Identification of a gene(s) complementing the Ala-Ala-hypersensitive mutant.
Since the sensitivity of Ala-Ala-hypersensitive mutants has been found to be associated with extensive accumulation of l-alanine derived from externally added Ala-Ala (18), we predicted that the growth inhibition could be relieved if the mutants gained a gene encoding an l-alanine exporter. To identify the gene, we performed a shotgun cloning experiment using the dipeptide-hypersensitive mutant LAX12 as a host and obtained several transformants after a 3-day incubation in the presence of the dipeptide, during which time the host cells did not form colonies. When we isolated plasmids from 11 independent clones and retransformed them into LAX12, all recombinant plasmids conferred on the mutant the ability to grow on minimal medium containing Ala-Ala, indicating that the growth restoration was plasmid borne. Restriction analysis, subcloning, and sequencing of the DNA fragments cloned in each plasmid resulted in identification of four open reading frames (ORFs) responsible for the growth recovery: two characterized genes, yddG and yeaS, and two uncharacterized genes, ytfF and ygaW. Since these genes rendered LAX12 cells free of the growth inhibition caused by Ala-Ala, their products, YddG, YeaS, YtfF, and YgaW, might function as exporters of l-alanine. The characterized ORFs, yddG (9) and yeaS (26), have indeed been found to encode exporters for l-aromatic amino acids and l-leucine, respectively, suggesting that l-alanine can be recognized as a minor substrate. The uncharacterized ORFs, ytfF and ygaW, are predicted to encode inner membrane proteins (http://gib.genes.nig.ac.jp/single/index.php?spid=Ecol_K12_MG1655). If YtfF and YgaW export l-alanine, they should be localized in the membrane. YtfF belongs to the RhtA family, most members of which possess 10 predicted transmembrane segments (TMSs) and a similar size of about 300 amino acid residues (29). The E. coli proteins of this family, YdeD (7), YddG (9, 38), and RhtA (29), appear to be integrated in the membrane and export amino acids. These findings suggest that YtfF also is a membrane protein with export activity for l-alanine. In contrast, YgaW was peculiar in that it is a small polypeptide of 149 amino acid residues with four predicted TMSs (Fig. 1A). Accordingly, YgaW was assumed to be a membrane protein and to function as the exporter for l-alanine. Therefore, we evaluated whether or not YgaW is a membrane protein. A protein band with a size of about 17.5 kDa was observed in the membrane fraction of cells harboring a recombinant plasmid with a His-tagged ygaW gene (Fig. 1B, lane 7) but not in that of host cells harboring an empty vector (Fig. 1B, lane 6), which corresponded to that of the affinity-purified His-tagged YgaW protein (Fig. 1B, lane 8). These findings taken together clearly indicate that YgaW is localized in the membrane. It is noted that the apparent size of the recombinant YgaW protein was different from the predicted size of 19 kDa, which is possibly due to the high hydrophobicity of YgaW.
Fig. 1.
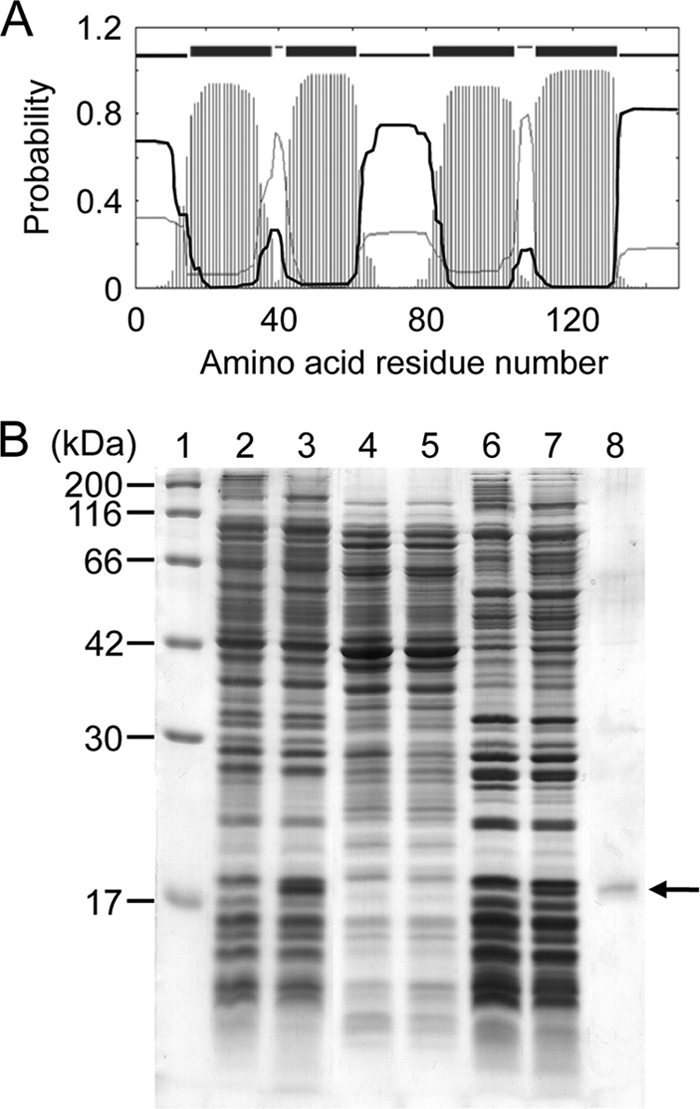
(A) Transmembrane topology and probability profile for YgaW predicted using the TMHMM program, version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (23). The top line shows the predicted topology with the four predicted transmembrane helices. The thin black and gray curves show the posterior probabilities for the inside and outside loops, respectively. The striped profile shows the probability for the transmembrane helix. (B) Cellular localization of YgaW. Whole-cell lysate (lanes 2 and 3), soluble fraction (lanes 4 and 5), and membrane fraction (lanes 6 and 7) were subjected to 15% SDS-polyacrylamide gel electrophoresis (40 μg protein each) and visualized by staining with Coomassie brilliant blue. Lane 1, molecular marker; lanes 2, 4, and 6, BL21(DE3)/pET-15b; lanes 3, 5, and 7, BL21(DE3)/pET-YgaW; lane 8, purified His-tagged YgaW. The arrow indicates His-tagged YgaW.
l-Alanine export mediated by the identified genes.
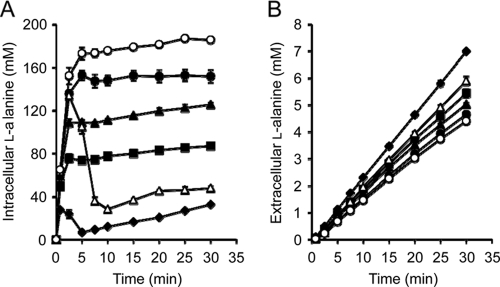
To investigate whether the products of the cloned genes are associated with l-alanine export activity, we determined the intra- and extracellular levels of l-alanine in the mutant LAX12, harboring a recombinant plasmid with each gene, and in the parent strain, MLA301 (Fig. 2). When the cells were incubated with 6 mM Ala-Ala, LAX12 harboring an empty vector accumulated l-alanine to a high level in the cells. In contrast, the intracellular l-alanine level in LAX12 bearing each gene showed lower steady-state levels (Fig. 2A). It should be noted that the intracellular l-alanine level of LAX12 possessing the ygaW gene was lower than that in the parent, MLA301. Correspondingly, the extracellular level of l-alanine in each transformant was higher than that in the mutant with the empty vector (Fig. 2B). Notably, the ygaW-bearing strain had the highest extracellular l-alanine level. Based on the l-alanine export profile, we calculated the l-alanine export rates to be 141, 153, 163, and 226 nmol/mg of cells (dry weight)/min for strains containing YeaS, YddG, YtfF, and YgaW, respectively, which were higher than the rate of export for LAX12 with the empty vector, 135 nmol/mg of cells (dry weight)/min. In particular, only the export rate of the mutant bearing the cloned ygaW gene was higher than that of the parent MLA301, 181 nmol/mg of cells (dry weight)/min. This experiment shows that the l-alanine accumulation resulting from the defect in LAX12 is completely complemented by introduction of pYgaW and partly reduced by pYtfF, pYddG, or pYeaS. Figure 2B shows that pYgaW nearly doubled the rate of extracellular l-alanine accumulation compared to the level for LAX12, to rates slightly higher than that for MLA301, while pYtfF, pYddG, or pYeaS had smaller effects. The results indicated that the product of each gene identified possesses an export activity for l-alanine. It is interesting to note that ygaW-bearing LAX12 showed a profile of transient increase in the intracellular l-alanine level similar to that of the parent, although the concentration was significantly lower (Fig. 2A). This result suggested that the plasmid-borne ygaW gene might be induced in the presence of Ala-Ala as observed in the non-l-alanine-metabolizing strain MLA301 (Fig. 2A).
Fig. 2.
Time course of intracellular (A) and extracellular (B) concentrations of l-alanine. Cells of the Ala-Ala-hypersensitive mutant LAX12 harboring pSTV29 (open circles), pYeaS (solid circles), pYddG (solid triangles), pYtfF (solid squares), or pYgaW (solid diamonds) and MLA301 harboring pSTV29 (open triangles) were incubated in minimal medium containing 6 mM Ala-Ala. The intra- and extracellular fractions were recovered by the silicone oil method at the time intervals indicated. l-Alanine in both fractions was determined by high-performance liquid chromatography. Data are means of results from two (LAX12 harboring pSTV29, pYddG, or pYgaW and MLA301 harboring pSTV29) or three (LAX12 harboring pYeaS or pYtfF) independent experiments. Vertical bars indicate the standard errors of the means.
Effect of ygaW gene inactivation on l-alanine export.
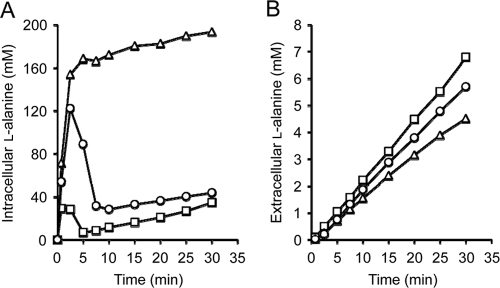
Since the ygaW gene had the most striking impact on the intracellular and extracellular l-alanine levels in the dipeptide-sensitive mutant (Fig. 2), it was assumed that YgaW would be a major l-alanine exporter in E. coli. If this is the case, introduction of a loss-of-function mutation in the ygaW gene in the strain MLA301 should have a strong influence on the intra- and extracellular l-alanine levels. To address this possibility, we constructed an MLA301-derived mutant lacking ygaW, MLA301ΔygaW. This mutant showed the same sensitivity to Ala-Ala (MIC, 39 μg ml−1) as that of LAX12, whereas the parent MLA301 showed an MIC of >10 mg ml−1. We then determined the intra- and extracellular l-alanine levels of MLA301ΔygaW in the presence of 6 mM Ala-Ala (Fig. 3). The intracellular l-alanine level rapidly increased as in MLA301 and leveled off at around 180 mM (Fig. 3A). It should be noted that the plateau level of l-alanine in the mutant was much higher than the basal level observed in MLA301. Correspondingly, the extracellular l-alanine level in the mutant was lower than that in MLA301 (Fig. 3B). The ygaW-borne plasmid counteracted the effect of gene inactivation on both the intra- and the extracellular l-alanine levels. These results with the ygaW mutant were almost the same as those of LAX12 obtained by random chemical mutagenesis (Fig. 2) and suggest that YgaW would significantly contribute to l-alanine export in E. coli.
Fig. 3.
Effect of ygaW gene inactivation on intracellular (A) and extracellular (B) l-alanine concentrations. l-Alanine in the intracellular and extracellular fractions obtained was determined as described in the legend to Fig. 2. Symbols: circles, MLA301; triangles, MLA301ΔygaW; squares, MLA301ΔygaW/pYgaW. Typical data from three independent experiments are shown.
Identification of a mutation in the ygaW gene.
Since the ygaW knockout mutant MLA301ΔygaW showed almost the same l-alanine export deficiency as that in LAX12 (Fig. 2 and 3), it is highly probable that LAX12 may have a loss-of-function mutation in the ygaW gene. To clarify this, we determined the nucleotide sequence of the chromosomal ygaW gene region in LAX12. Consequently, we found that LAX12 had a nucleotide change from guanine to adenine at the 374th nucleotide from the initiation codon of ygaW, leading to the substitution of glutamic acid for glycine at codon 125. Interestingly, this amino acid change (G125E) resided in the fourth predicted TMS (Fig. 1A). It is well known that an unpaired charged amino acid residue in the transmembrane α-helixes destabilizes the overall structure, thereby leading to dysfunction of integral membrane proteins (46). Thus, the mutation identified in the ygaW gene of LAX12 is most likely to cause dysfunction of its product. Indeed, MLA301ΔygaW possessing the ygaW gene cloned from LAX12 showed a level of Ala-Ala sensitivity similar to that shown by MLA301ΔygaW (data not shown), indicating clearly that the mutation had led to dysfunction of YgaW. It should be noted that there was no mutation in the chromosome regions of the other three genes, yeaS, yddG, and ytfF, in LAX12.
YgaW-mediated alanine excretion in the wild-type background.
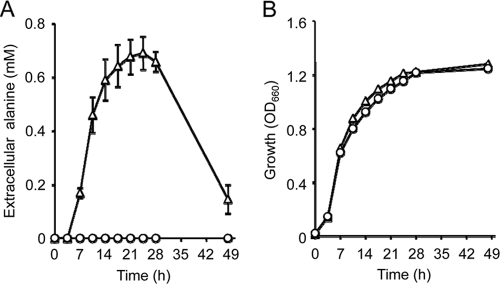
The l-alanine export activities of the four exporters identified might not precisely reflect the physiological functionality, since intracellular l-alanine reached a level beyond the physiological concentration (31) in the presence of Ala-Ala. Therefore, we assessed l-alanine export in the wild-type background with respect to the internal amino acid pools. For this purpose, wild-type MG1655 harboring the recombinant plasmid with each gene was grown in minimal medium, and amino acids in the culture supernatant were measured over time until glucose had been exhausted. The plasmid-borne ygaW gene provided MG1655 with an ability to excrete alanine into the medium, reaching the highest level of about 0.7 mM, whereas MG1655 with the empty vector did not excrete alanine (Fig. 4A). Both cells showed almost the same growth (Fig. 4B). The decrease in extracellular alanine after reaching the highest level was probably due to reabsorption of the excreted alanine since the glucose was exhausted at this time (data not shown). In contrast, MG1655 harboring the recombinant plasmid with yeaS, yddG, or ytfF did not excrete alanine into the medium (data not shown). These results clearly indicated that YgaW, but not the other transporters, contributes to the excretion of alanine under physiological conditions. Since alanine was excreted by the wild-type strain, alanine exported by YgaW could be the l form and/or the d form. To clarify whether YgaW could export d-alanine, we determined the MICs of l-alanine and d-alanine for MLA301ΔygaW and MLA301. MLA301ΔygaW showed a decreased MIC of l-alanine (1.25 mg ml−1) compared to the level for the parent MLA301 (5 mg ml−1), whereas both strains showed the same MIC of d-alanine (20 mg ml−1), suggesting that YgaW exhibits stereospecificity for l-alanine.
Fig. 4.
YgaW-mediated alanine excretion in wild-type E. coli. (A) Alanine concentrations in the culture supernatant. (B) Growth of MG1655 transformants. MG1655 harboring pSTV29 (circles) or pYgaW (triangles) was grown in minimal medium, and alanine in the culture supernatants was determined by high-performance liquid chromatography. Data are means of results from three independent experiments. Vertical bars in panel A indicate the standard errors of the means.
Expression of genes involved in l-alanine export.
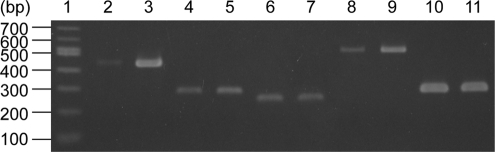
We previously found that LAX12 lacks an inducible l-alanine export system (18). In addition, we identified that LAX12 has a loss-of-function mutation in the ygaW gene as described above; accordingly, YgaW is strongly expected to be an inducible l-alanine exporter. Thus, we investigated the transcription level of ygaW as well as those of ytfF, yddG, and yeaS in the presence or absence of Ala-Ala in MLA301 using the RT-PCR method. As expected, the transcription level of ygaW was increased in the presence of Ala-Ala (Fig. 5, lanes 2 and 3), which was in good agreement with the results indicating that the l-alanine export activity of YgaW was induced (Fig. 2A). Similarly, an elevated transcription level of yeaS was observed in the presence of Ala-Ala (Fig. 5, lanes 8 and 9), which is consistent with a result reported previously (26). Expression of ytfF was slightly increased in the presence of Ala-Ala (Fig. 5, lanes 4 and 5).
Fig. 5.
Expression analysis of the ygaW, ytfF, yddG, and yeaS genes. The non-l-alanine-metabolizing strain MLA301 was incubated in minimal medium with (lanes 3, 5, 7, 9, and 11) or without (lanes 2, 4, 6, 8, and 10) 6 mM Ala-Ala. After a 5-min incubation, total RNA isolation and RT-PCR were performed as described in Materials and Methods. Lane 1, molecular size marker in base pairs; lanes 2 and 3, ygaW; lanes 4 and 5, ytfF; lanes 6 and 7, yddG; lanes 8 and 9, yeaS; lanes 10 and 11, gapA.
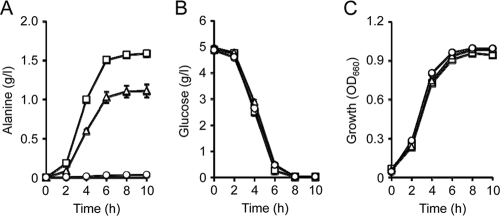
Influence of YgaW overexpression on alanine production in an alanine-producing strain.
The next question to arise is whether or not enhancement of alanine export capacity can promote alanine production in an alanine-producing strain. To address this issue, we employed an alanine dehydrogenase gene, alaD, from Bacillus sphaericus, overexpression of which renders E. coli cells capable of producing alanine (27). MG1655 cells coexpressing the alaD and ygaW genes from a multicopy plasmid produced more alanine than the strain possessing the alaD gene alone (Fig. 6A). As a result, alanine yield after a 10-h cultivation was increased from 22.5% (MG1655/pAlaD) to 32.7% (MG1655/pAlaD-YgaW) on a weight basis (g/g glucose). It is noted that all strains tested exhibited almost the same glucose consumption and growth (Fig. 6B and C). The results indicated that enhancement of alanine export by YgaW improved alanine production.
Fig. 6.
Effect of ygaW gene amplification on alanine production. MG1655 harboring pSTV29 (circles), pAlaD (triangles), or pAlaD-YgaW (squares) was grown anaerobically in fermentation medium. Alanine production (A), glucose consumption (B), and growth (C) were determined at the time intervals indicated. Data are means of results from three independent experiments. Vertical bars in panels A and B indicate the standard errors of the means.
DISCUSSION
We identified in this study two uncharacterized genes, ygaW and ytfF, and two characterized genes, yddG and yeaS, which complemented the Ala-Ala hypersensitivity of a mutant lacking an inducible l-alanine export system. Among these, the ygaW gene product, YgaW, seemed to be the most physiologically relevant exporter of l-alanine on the basis of the following lines of evidence: (i) the l-alanine exporterless-mutant, LAX12, overexpressing YgaW showed the lowest intracellular level and the highest export rate of l-alanine (Fig. 2), (ii) disruption of the ygaW gene in the non-l-alanine-metabolizing mutant MLA301 resulted in intracellular accumulation and a reduced export rate of l-alanine in the presence of extracellular Ala-Ala (Fig. 3), (iii) yddG (9) and yeaS (26) have been characterized to encode exporters for l-aromatic amino acids and l-leucine, respectively, but not for l-alanine, (iv) disruption of the ytfF gene in MLA301 did not lead to a change in the levels of intra- and extracellular l-alanine in the presence of Ala-Ala (data not shown), (v) wild-type MG1655 harboring the ygaW-bearing plasmid, but not harboring the control plasmid pSTV29 and the recombinant plasmid with yeaS, yddG, or ytfF, excreted alanine when grown in minimal medium (Fig. 4), and (vi) expression of the ygaW gene was significantly induced under conditions where the intracellular l-alanine level was increased (Fig. 5).
Amino acid exporters characterized to date and putative exporters as well have been classified into several transporter families. YeaS belongs to the RhtB/LysE family, whose members are distributed widely in prokaryotes (3). YddG and YtfF are classified into the RhtA family, the members of which are found in prokaryotes and eukaryotes (29). However, a BLAST search (4) showed that YgaW had a limited number of orthologues with high homology (data not shown). Indeed, YgaW homologues found by the homology search through the MBGD (Microbial Genome Database; http://mbgd.genome.ad.jp/) (45) under default conditions were located only in gammaproteobacteria and alphaproteobacteria, suggesting that the first ancestral gene of ygaW might have originated in proteobacteria some time after divergence from other bacteria. Most of the YgaW homologues possessed four predicted TMSs and a small size of 140 to 160 amino acid residues. Although the structural characteristics were somewhat similar to that of a small multidrug resistance protein family (4 TMSs, approximately 100 to 140 amino acid residues) (5), no significant similarity was observed in the primary sequences between a homologue of E. coli YgaW and a protein in the small multidrug resistance family. All of these characteristic features of the YgaW homologues suggested that YgaW is a member of a novel protein family.
Exporters for amino acids and their analogues are assumed to play important roles in bacterial physiology under natural conditions: these include excretion of intercellular signal molecules such as homoserine lactone (52), maintenance of a balanced intracellular pool of amino acids (7, 33), and the prevention of a buildup of the intracellular amino acid concentration to a toxic level under certain circumstances, such as peptide-rich conditions (6, 33, 47). Although there is no direct evidence that shows the physiological roles of the four exporters identified in this study, the physiological function of YgaW can be predicted in relation to the distribution of its homologues in the bacterial world. Orthologues with high homology to E. coli YgaW exist in several enteric bacterial groups, such as Salmonella enterica serovar Typhimurium (identity 87.2%) and Citrobacter koseri (identity 87.2%), a natural habitat of which is the mammalian intestine, and pathogenic bacterial groups, such as Vibrio cholera (identity 62.8%), which also infect the mammalian intestine. The environment in the intestine is assumed to be peptide rich. For example, the human intestine has a basal level of small peptides, and the level of amino acids in peptide form increases to over 100 mM upon consumption of protein-rich meals (2). In addition, l-alanine is the second most abundant amino acid in proteins in nature (8). It is, therefore, noteworthy that 1 mM Ala-Ala caused growth inhibition of the ygaW-deficient derivative of wild-type MG1655 on minimal medium (data not shown). Accordingly, it is suggested that YgaW and its homologues may act as a safety valve for l-alanine to prevent excessive accumulation of peptide-derived l-alanine in the cells.
Several amino acid exporters have been demonstrated to enhance the production of their amino acid substrates when they were overexpressed in amino acid-producing strains (9, 12, 24, 26, 29, 36, 50). This beneficial effect is also applicable to YgaW, since overexpression of YgaW in an alanine-producing strain enhanced alanine production (Fig. 6). However, the yield of alanine produced was lower than that reported previously for E. coli (44, 48, 53). The reason for this relatively low yield is partly due to the use of the wild-type strain for constructing the alanine producer strain.
Based on the findings on the l-alanine export activity of the ygaW gene product, we propose that the gene be redesignated alaE for alanine export.
Footnotes
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Abe K., et al. 2002. Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction. J. Bacteriol. 184:2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adibi S. A., Mercer D. W. 1973. Protein digestion in human intestine as reflected in luminal, mucosal, and plasma amino acid concentrations after meals. J. Clin. Invest. 52:1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aleshin V. V., Zakataeva N. P., Livshits V. A. 1999. A new family of amino-acid-efflux proteins. Trends Biochem. Sci. 24:133–135 [DOI] [PubMed] [Google Scholar]
- 4. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bay D. C., Rommens K. L., Turner R. J. 2008. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778:1814–1838 [DOI] [PubMed] [Google Scholar]
- 6. Bellmann A., et al. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774 [DOI] [PubMed] [Google Scholar]
- 7. Dassler T., Maier T., Winterhalter C., Böck A. 2000. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 36:1101–1112 [DOI] [PubMed] [Google Scholar]
- 8. Doolittle R. F. 1989. Redundancies in protein sequences, p. 599–623 In Fasman G. D. (ed.), Predictions of protein structure and the principles of protein conformation . Plenum Press, New York, NY [Google Scholar]
- 9. Doroshenko V., et al. 2007. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 275:312–318 [DOI] [PubMed] [Google Scholar]
- 10. Eggeling L., Sahm H. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 180:155–160 [DOI] [PubMed] [Google Scholar]
- 11. Fisher R., Tuli R., Haselkorn R. 1981. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylylated. Proc. Natl. Acad. Sci. U. S. A. 78:3393–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franke I., Resch A., Dassler T., Maier T., Böck A. 2003. YfiK from Escherichia coli promotes export of O-acetylserine and cysteine. J. Bacteriol. 185:1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gronskiy S. V., et al. 2005. The yicM (nepI) gene of Escherichia coli encodes a major facilitator superfamily protein involved in efflux of purine ribonucleosides. FEMS Microbiol. Lett. 250:39–47 [DOI] [PubMed] [Google Scholar]
- 14. Hashimoto S., Katsumata R. 1998. l-Alanine fermentation by an alanine racemase-deficient mutant of the dl-alanine hyperproducing bacterium Arthrobacter oxydans HAP-l. J. Ferment. Bioeng. 86:385–390 [Google Scholar]
- 15. Hashimoto-Gotoh T., et al. 2000. A set of temperature sensitive-replication/-segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185–191 [DOI] [PubMed] [Google Scholar]
- 16. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 17. Hols P., et al. 1999. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17:588–592 [DOI] [PubMed] [Google Scholar]
- 18. Hori H., Ando T., Isogai E., Yoneyama H., Katsumata R. 2011. Identification of an L-alanine export system in Escherichia coli and isolation and characterization of export deficient mutants. FEMS Microbiol. Lett. 316:83–89 [DOI] [PubMed] [Google Scholar]
- 19. Katsumata R., Hashimoto S. September 1996. Process for producing alanine. U.S. patent 5559016.
- 20. Kennerknecht N., et al. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J. Bacteriol. 184:3947–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kinoshita S., Udaka S., Shimono M. 1957. Studies on the amino acid fermentation partI. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193–205 [PubMed] [Google Scholar]
- 22. Klingenberg M., Pfaff E. 1967. Means of terminating reactions. Methods Enzymol. 10:680–684 [Google Scholar]
- 23. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 24. Kruse D., et al. 2002. Influence of threonine exporters on threonine production in Escherichia coli. Appl. Microbiol. Biotechnol. 59:205–210 [DOI] [PubMed] [Google Scholar]
- 25. Kuroda S., Tanizawa K., Sakamoto Y., Tanaka H., Soda K. 1990. Alanine dehydrogenases from two Bacillus species with distinct thermostabilities: molecular cloning, DNA and protein sequence determination, and structural comparison with other NAD(P)+-dependent dehydrogenases. Biochemistry 29:1009–1015 [DOI] [PubMed] [Google Scholar]
- 26. Kutukova E. A., et al. 2005. The yeaS (leuE) gene of Escherichia coli encodes an exporter of leucine, and the Lrp protein regulates its expression. FEBS Lett. 579:4629–4634 [DOI] [PubMed] [Google Scholar]
- 27. Lee M., Smith G. M., Eiteman M. A., Altman E. 2004. Aerobic production of alanine by Escherichia coli aceF ldhA mutants expressing the Bacillus sphaericus alaD gene. Appl. Microbiol. Biotechnol. 65:56–60 [DOI] [PubMed] [Google Scholar]
- 28. Liu J. Y., Miller P. F., Willard J., Olson E. R. 1999. Functional and biochemical characterization of Escherichia coli sugar efflux transporters. J. Biol. Chem. 274:22977–22984 [DOI] [PubMed] [Google Scholar]
- 29. Livshits V. A., Zakataeva N. P., Aleshin V. V., Vitushkina M. V. 2003. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 154:123–135 [DOI] [PubMed] [Google Scholar]
- 30. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 31. Mengin-Lecreulx D., Flouret B., van Heijenoort J. 1982. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 151:1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura J., Hirano S., Ito H., Wachi M. 2007. Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl. Environ. Microbiol. 73:4491–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nandineni M. R., Gowrishankar J. 2004. Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J. Bacteriol. 186:3539–3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nies D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730–750 [DOI] [PubMed] [Google Scholar]
- 35. Nikaido H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park J. H., Lee K. H., Kim T. Y., Lee S. Y. 2007. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. U. S. A. 104:7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pittman M. S., et al. 2002. Cysteine is exported from the Escherichia coli cytoplasm by CydDC, an ATP-binding cassette-type transporter required for cytochrome assembly. J. Biol. Chem. 277:49841–49849 [DOI] [PubMed] [Google Scholar]
- 38. Rapp M., et al. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 13:937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saito H., Miura K. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619–629 [PubMed] [Google Scholar]
- 40. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Sanger F., Nicklen S., Coulson R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schneider F., Krämer R., Burkovski A. 2004. Identification and characterization of the main β-alanine uptake system in Escherichia coli. Appl. Microbiol. Biotechnol. 65:576–582 [DOI] [PubMed] [Google Scholar]
- 43. Simic P., Sahm H., Eggeling L. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith G. M., Lee S. A., Reilly K. C., Eiteman M. A., Altman E. 2006. Fed-batch two-phase production of alanine by a metabolically engineered Escherichia coli. Biotechnol. Lett. 28:1695–1700 [DOI] [PubMed] [Google Scholar]
- 45. Uchiyama I., Higuchi T., Kawai M. 2010. MBGD update 2010: toward a comprehensive resource for exploring microbial genome diversity. Nucleic Acids Res. 38:D360–D365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venkatesan P., Kaback H. R. 1998. The substrate-binding site in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:9802–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vrljic M., Sahm H., Eggeling L. 1996. A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22:815–826 [DOI] [PubMed] [Google Scholar]
- 48. Wada M., Narita K., Yokota A. 2007. Alanine production in an H+-ATPase- and lactate dehydrogenase-defective mutant of Escherichia coli expressing alanine dehydrogenase. Appl. Microbiol. Biotechnol. 76:819–825 [DOI] [PubMed] [Google Scholar]
- 49. White D. G., Goldman J., Demple B., Levy S. B. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamada S., et al. 2006. Effect of drug transporter genes on cysteine export and overproduction in Escherichia coli. Appl. Environ. Microbiol. 72:4735–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zakataeva N. P., Aleshin V. V., Tokmakova I. L., Troshin P. V., Livshits V. A. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228–232 [DOI] [PubMed] [Google Scholar]
- 52. Zakataeva N. P., et al. 2006. Export of metabolites by the proteins of the DMT and RhtB families and its possible role in intercellular communication. Mikrobiologiia 75:509–520 [PubMed] [Google Scholar]
- 53. Zhang X., Jantama K., Moore J. C., Shanmugam K. T., Ingram L. O. 2007. Production of L-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:355–366 [DOI] [PubMed] [Google Scholar]