Abstract
The distribution of membrane lipids of 17 different strains representing 13 species of subdivisions 1 and 3 of the phylum Acidobacteria, a highly diverse phylum of the Bacteria, were examined by hydrolysis and gas chromatography-mass spectrometry (MS) and by high-performance liquid chromatography-MS of intact polar lipids. Upon both acid and base hydrolyses of total cell material, the uncommon membrane-spanning lipid 13,16-dimethyl octacosanedioic acid (iso-diabolic acid) was released in substantial amounts (22 to 43% of the total fatty acids) from all of the acidobacteria studied. This lipid has previously been encountered only in thermophilic Thermoanaerobacter species but bears a structural resemblance to the alkyl chains of bacterial glycerol dialkyl glycerol tetraethers (GDGTs) that occur ubiquitously in peat and soil and are suspected to be produced by acidobacteria. As reported previously, most species also contained iso-C15 and C16:1ω7C as major fatty acids but the presence of iso-diabolic acid was unnoticed in previous studies, most probably because the complex lipid that contained this moiety was not extractable from the cells; it could only be released by hydrolysis. Direct analysis of intact polar lipids in the Bligh-Dyer extract of three acidobacterial strains, indeed, did not reveal any membrane-spanning lipids containing iso-diabolic acid. In 3 of the 17 strains, ether-bound iso-diabolic acid was detected after hydrolysis of the cells, including one branched GDGT containing iso-diabolic acid-derived alkyl chains. Since the GDGT distribution in soils is much more complex, branched GDGTs in soil likely also originate from other (acido)bacteria capable of biosynthesizing these components.
INTRODUCTION
Acidobacteria form one of the most diverse phyla of the domain Bacteria. Twenty-six subdivisions have been reported, mainly based on environmental sequences (4), but only a few of these include taxonomically characterized representatives. Subdivision 1 is best characterized by Acidobacterium capsulatum (25), Terriglobus roseus (12), Edaphobacter aggregans, Edaphobacter modestus (26), and four Granulicella species (40). Bryobacter aggregatus (30) is the only species described for subdivision 3, and subdivision 8 contains three described species: Holophaga foetida (33), Geothrix fermentans (7), and Acanthopleuribacter pedis (16). A whole-genome study has been performed for A. capsulatum and two isolates, “Candidatus Koribacter versatilis” and “Candidatus Solibacter usitatus,” from subdivisions 1 and 3, respectively (48). These studies indicate that acidobacteria are generally versatile heterotrophs that are common in peats and acidic soils. Molecular ecology studies have indicated that acidobacteria form a substantial part of the microbial community of wetlands (10, 29, 34, 39) and soils (19, 21). In soils, the relative contribution of these bacteria to the overall bacterial community is negatively correlated with pH (21); especially species from subdivision 1 are more abundant at lower pH (12).
The abundance of acidobacteria in soils and peat has led to the suggestion that they may be the biological source of a group of structurally uncommon tetraether membrane lipids (49). These components (e.g., structure 1 in Fig. 1) were first identified in peat (45) and subsequently turned out to occur ubiquitously in soils (51). Determination of the stereochemistry of the glycerol moieties in these molecules revealed that they must derive from Bacteria (50), possibly Acidobacteria (49). The natural 13C abundance of these bacterial tetraethers in soil is consistent with a heterotrophic lifestyle of its source organism(s) (53). Examination of a suite of bacterial cultures, including a few acidobacterial strains, for the bacterial tetraethers did not reveal their origin (49). However, in recent years many more members of the phylum Acidobacteria have been characterized.
Fig. 1.
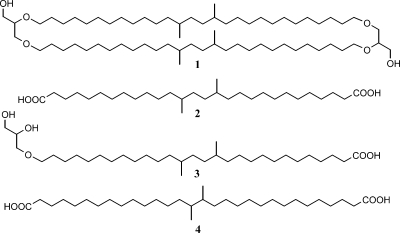
Structures of lipids discussed in the text. Shown are the branched GDGT with four methyl substituents (structure 1), iso-diabolic acid (structure 2), the glycerol ether derivative of iso-diabolic acid (structure 3), and diabolic acid (structure 4).
In the present paper, we describe in detail the lipid compositions of 17 different strains representing 13 species of subdivisions 1 and 3 of the phylum Acidobacteria and discuss their distributions.
MATERIALS AND METHODS
Cultures.
The acidobacteria used in this study are listed in Table 1. Four members of the genus Granulicella, i.e., Granulicella paludicola OB1010T, G. paludicola LCBR1, G. aggregans TPB6028T, and G. pectinivorans TPB6011T, were grown at the Winogradsky Institute in liquid medium MM1 with 0.5 g liter−1 fructose as described by Pankratov and Dedysh (40). Two strains of B. aggregatus, Bryocella elongata SN10, Terriglobus sp. KMR, A. capsulatum 161T, and Acidobacteriaceae bacterium KA1 were grown at the Winogradsky Institute in liquid medium MM with 0.5 g liter−1 glucose (30). B. aggregatus MPL3T and Acidobacteriaceae bacterium KA1 were grown at both pHs 4.2 and 7.0 in medium MM buffered with 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) or HEPES, respectively. For most strains, the cell biomass used for analysis was collected after 1 week of incubation at 24°C. The biomass of the slow-growing species B. aggregatus was collected after 2 weeks of incubation.
Table 1.
Acidobacteria used in this study
| Species | Subdivision | Origin | Substrates used | pH range | pH optimum | Reference(s) |
|---|---|---|---|---|---|---|
| Acidobacteriaceae strain 277 | 1 | Sandy soil from river bank, Okavango, Namibia | Most sugars, some sugar alcohols, starch, some amino acids, acetate, succinate, some heteropolysaccharides | 3.5–7.5 | 5.0–6.5 | DSMZa |
| Acidobacteriaceae strain 307 | 1 | Sandy soil from river bank, Okavango, Namibia | Most sugars, some sugar alcohols, starch, some amino acids, fumarate, succinate, some heteropolysaccharides | 3.5–8.5 | 5.0–6.5 | DSMZa |
| Acidobacteriaceae strain A2-1c | 1 | Savanna soil, Erichsfelde, Namibia | Most sugars, some sugar alcohols, starch, some amino acids, gluconate, pyruvate, succinate, some heteropolysaccharides | NDb | ND | DSMZa |
| Acidobacteriaceae strain KA1 (= DSM 23886) | 1 | Sphagnum peat, Tver region, European north Russia | Most sugars, glucuronate, galacturonate, some heteropolysaccharides | 3.5–7.3 | 5.0–5.5 | WIMc |
| Acidobacteriaceae strain A2-4c | 1 | Savanna soil, Erichsfelde, Namibia | ND | ND | ND | DSMZa |
| A. capsulatum 161T (= DSM 11244T) | 1 | Acidic mine drainage, Japan | Most sugars, glucuronate, starch, some heteropolysaccharides | 3.0–6.0 | ND | 12, 25 |
| B. elongata SN10T (= DSM 22489T) | 1 | Sphagnum peat, Archangelsk region, European north Russia | Most sugars, lactate, ethanol, some heteropolysaccharides | 3.2–6.6 | 4.7–5.2 | 9 |
| G. pectinivorans TPB6011T (– DSM 21001T) | 1 | Sphagnum peat, Tomsk region, western Siberia | Several sugars, acetate, lactate, galacturonate, gluconate, starch, some heteropolysaccharides and polyalcohols | 3.0–7.5 | 3.8–4.5 | 40 |
| G. aggregans TPB6028T (= LMG 25274T) | 1 | Sphagnum peat, Tomsk region, western Siberia | Most sugars, malate, pyruvate, galacturonate, gluconate, starch, some heteropolysaccharides and sugar alcohols | 3.0–7.5 | 3.8–4.5 | 40 |
| G. paludicola OB1010T (= DSM 22464T) | 1 | Sphagnum peat, Yaroslavl region, European north Russia | Most sugars, starch, some heteropolysaccharides, galacturonate | 3.0–7.5 | 3.8–4.5 | 40 |
| G. paludicola LCBR1 | 1 | Cladonia sp. collected from peat bog, Tomsk region, western Siberia | Most sugars, starch, some heteropolysaccharides, galacturonate, gluconate, glucuronate | 3.0–7.5 | 3.8–4.5 | 40 |
| T. roseus KBS 63T (= DSM 18391T) | 1 | Agricultural soil, Michigan | Several sugars, succinate, glucuronate, gluconate | 5.0–7.0 | 6.0 | 12 |
| Terriglobus sp. KMR (= ATCC BAA-1395) | 1 | Sphagnum peat, Yaroslavl region, European north Russia | Most sugars, starch, some heteropolysaccharides | 4.5–7.2 | 5.5–6.5 | 41 |
| E. modestus Jbg-1T (= DSM 18101T) | 1 | Soil deciduous forest, Würzburg, Germany | Most sugars, some sugar alcohols, glutamate, glutamine | 4.0–7.0 | 5.5 | 26 |
| E. aggregans Wbg-1T (= DSM 19364T) | 1 | Alpine soil, Kochel, Germany | Several sugars, glutamate, glutamine, aspartate, ornithine, glucuronate | 4.5–7.0 | 5.5 | 26 |
| B. aggregatus MPL3T (= DSM 18758T) | 3 | Sphagnum peat, Tomsk region, western Siberia | Most sugars, pyruvate, starch, some heteropolysaccharides, galacturonate, glucuronate | 4.5–7.2 | 5.5–6.5 | 30 |
| B. aggregatus MPL1011 | 3 | Sphagnum peat, Tomsk region, western Siberia | Most sugars, pyruvate, lactate, starch, some heteropolysaccharides, galacturonate, glucuronate | 4.5–7.2 | 5.5–6.5 | 30 |
Isolated at DSMZ.
ND, not determined.
WIM, isolated at the Winogradsky Institute of Microbiology.
Cells of all other acidobacterial strains and A. capsulatum 161T were grown at the DSMZ at room temperature with vigorous shaking for 1 to 5 weeks, depending on the strain. Strains 277, 307, A2-1c, and A2-4c were grown at pH 5.5 in a medium consisting of soil solution equivalent (1) with the iron content changed from 5 to 50 μM and buffered with 10 mM MES as the basal medium and supplemented with 10-fold-diluted HD medium (0.5 g liter−1 of peptone, 0.25 g liter−1 of yeast extract, and 0.1 g liter−1 of glucose), Balch's vitamin mixture (2) 10-fold diluted, and trace element solution SL-10 (47). The two Edaphobacter strains (Jbg-1T and Wbg-1T) were grown at pH 5.5 in a 5-fold-diluted HD medium containing 1.0 g liter−1 of peptone, 0.5 g liter−1 of yeast extract, 0.2 g liter−1 of glucose, and 10 mM MES. T. roseus KBS 63T was grown at pH 7.2 in DSMZ medium 830 (see http://www.dsmz.de/microorganisms/media_list.php). A. capsulatum 161T was grown at pH 3.5 in DSMZ medium 269 with 0.1 g liter−1 instead of 0.3 g liter−1 yeast extract. Biomass was harvested by ultracentrifugation (9,000 × g, 30 min; Avanti-J26 XPI, Beckman Coulter), frozen (−20°C, overnight), and lyophilized (5 Pa at −30°C).
Lipid analysis.
For all of the strains studied, lyophilized cells were hydrolyzed with 1 N 5% HCl in methanol by reflux for 3 h. The hydrolysate was adjusted to pH 4 with 2 N KOH-methanol (MeOH) (1:1, vol/vol) and, after addition of water to a final 1:1 ratio of H2O-MeOH, extracted three times with dichloromethane (DCM). The DCM fractions were collected and dried over sodium sulfate. The extract obtained was methylated with diazomethane and separated over an activated Al2O3 column into an apolar and a polar fraction using DCM and DCM-MeOH (1:1, vol/vol) as the eluent, respectively. The apolar fraction (containing the fatty acid methyl esters [FAMEs]) was analyzed by gas chromatography (GC) and GC-mass spectrometry (MS). Double-bound positions of the monounsaturated FAMEs were determined on the basis of the mass spectra of their dimethyl disulfide derivatives as described by Nichols et al. (35). The polar fraction was dissolved in hexane–2-propanol (99:1, vol/vol), filtered over a 0.45-μm polytetrafluoroethylene filter, and analyzed by high-performance liquid chromatography (HPLC)–atmospheric pressure chemical ionization (APCI) mass spectrometry (MS) for branched GDGTs.
For selected strains, intact polar lipids were extracted from the lyophilized cells using a modified Bligh-Dyer technique (5) as described by Pitcher et al. (42). An aliquot of the extract obtained was dissolved in hexane–2-propanol–water (72:27:1), filtered through a 0.45-μm regenerated cellulose filter, and analyzed by HPLC-electrospray ionization (ESI) MS.
For G. aggregans TPB6028T, a more extensive lipid characterization was performed. The lyophilized cells were also hydrolyzed with 1 N KOH-MeOH (96%) by reflux for 1 h. The hydrolysate was neutralized with 2 N HCl-MeOH (1:1, vol/vol) to pH 4 and, after addition of water, extracted with DCM three times. The extract was methylated with diazomethane, silylated with N,O-bis(trimethylsilyl)-trifluoroacetamide in pyridine at 60°C for 20 min and analyzed by GC and GC-MS. Lyophilized cells were also directly extracted using a modified Bligh-Dyer technique (see above) without prior acid or base hydrolysis, and the extract was methylated and silylated as described above and analyzed by GC and GC-MS. Aliquots of the Bligh-Dyer extract were also hydrolyzed with HCl-MeOH and with KOH-MeOH, derivatized, and analyzed by GC and GC-MS as described above. The cell residue after Bligh-Dyer extraction was hydrolyzed with HCl-MeOH and KOH-MeOH, derivatized, and analyzed by GC and GC-MS as described above.
GC and GC-MS.
GC was performed using a Hewlett-Packard (HP6890) instrument equipped with an on-column injector and a flame ionization detector. A fused silica capillary column (25 m by 0.32 mm) coated with CP Sil-5 CB (film thickness, 0.12 μm) was used with helium as the carrier gas. The samples were injected at 70°C, and the oven temperature was programmed to 130°C at 20°C/min and then at 4°C/min to 320°C, at which it was held for 10 min. GC-MS was performed on a Finnigan Trace ultra gas chromatograph interfaced with a Finnigan Trace DSQ mass spectrometer operated at 70 eV with a mass range of m/z 40 to 800 and a cycle time of 1.7 s (resolution, 1,000). The gas chromatograph was equipped with a fused silica capillary column as described for GC. The carrier gas was helium. The same temperature program as for GC was used.
HPLC-APCI-MS.
GDGT lipids were analyzed by HPLC-APCI-MS according to Hopmans et al. (17), with minor modifications. Analyses were performed on an Agilent 1100 series HPLC-MS instrument equipped with an autoinjector and HP Chemstation software. Separation was achieved on an Alltech Prevail Cyano column (150 mm by 2.1 mm; 3 μm). The flow rate of the hexane–2-propanol (99:1, vol/vol) eluent was 0.2 ml min−1, isocratically for the first 5 min and thereafter with a linear gradient to 1.8% propanol in 45 min. The column was subsequently rinsed by eluting 10% propanol in hexane for 10 min and back to 1% propanol for the last 10 min. The injection volume of the samples was usually 10 μl. Branched GDGTs were analyzed mostly by APCI-MS in selected ion monitoring mode (SIM) (44).
IPL analysis by HPLC-ESI-MS2.
Intact polar lipids (IPLs; i.e., glycerol ester membrane lipids with attached polar head groups) were analyzed according to a previously published method (46), with some modifications. The Agilent 1200 series liquid chromatograph used was equipped with a thermostat-controlled autoinjector and a column oven and coupled to a Thermo LTQ XL linear ion trap with an Ion Max source with an ESI probe (Thermo Scientific, Waltham, MA). Separation was achieved on a Lichrosphere diol column (250 mm by 2.1 mm, 5-μm particles; Alltech Associates Inc.) maintained at 30°C using the mobile-phase system described by Sturt et al. (46) but with a modified elution program of 100% A for 1 min, followed by a linear gradient to 66% A and 34% B in 17 min, maintenance for 12 min, followed by a linear gradient to 35% A and 65% B in 15 min, where A is 79:20:0.12:0.04 hexane–2-propanol–formic acid–14.8 M aqueous NH3 and B is 88:10:0.12:0.04 2-propanol—water–formic acid–14.8 M aqueous NH3. The flow rate was 0.2 ml min−1, and the total run time was 60 min, with a reequilibration period of 20 min. Positive-ion ESI settings were as follows: capillary temperature, 275°C; sheath gas (N2) pressure, 25 arbitrary units (AU); auxiliary gas (N2) pressure, 15 AU; sweep gas (N2) pressure, 20 AU; spray voltage, 4.5 kV. The lipid extract was analyzed by positive-ion scanning (m/z 400 to 2,000), followed by data-dependent, dual-stage tandem MS (MS2), where the four most abundant masses in the mass spectrum were fragmented successively (normalized collision energy, 25; isolation width, 5.0; activation Q, 0.175). Each MS2 was followed by data-dependent, triple-stage tandem MS (MS3), where the base peak of the MS2 spectrum was fragmented under identical fragmentation conditions.
Preparation of an authentic standard.
13,16-Dimethyloctacosanedioic acid (ca. 3 mg; 96% pure by GC) was isolated by thin-layer chromatography from a large-size culture of Thermoanaerobacter thermohydrosulfuricus strain TLOT (3). Its structure was confirmed by two-dimensional nuclear magnetic resonance techniques, which gave spectra identical to those reported previously (24).
RESULTS
Seventeen strains belonging to the phylum Acidobacteria were analyzed for their lipid composition; 15 of them are species in subdivision 1 (A. capsulatum, two Terriglobus strains, two Edaphobacter species, four Granulicella species, B. elongata SN10, and five additional representatives of the family Acidobacteriaceae), and two are in subdivision 3 (two B. aggregatus strains) (Table 1; see Fig. S1 in the supplemental material).
Composition of total lipid fractions.
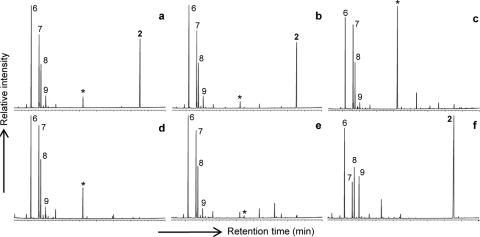
Fig. 2 a shows a typical gas chromatogram of a total lipid fraction obtained after acid hydrolysis of cells of G. aggregans TPB6028T. The dominant regular fatty acids are iso-C15, n-C16:1ω7c, and n-C16, a distribution commonly observed in the acidobacterial strains studied, although other dominant regular fatty acids are iso-C17:1ω8c, iso-C17, and n-C18:1ω9, depending on the phylogenetic positions of the strains studied (Table 2). In addition to these regular fatty acids, a more unusual, much later-eluting (Fig. 2a) lipid was detected in relatively large amounts (22 to 43% of the total fatty acids; Table 2). This lipid was identified as a long-chain dicarboxylic acid, 13,16-dimethyloctacosanedioic acid (or iso-diabolic acid; structure 2 in Fig. 1), based on mass spectral data reported previously (24) and coelution experiments with an authentic standard isolated from cell material of T. thermohydrosulfuricus (3).
Fig. 2.
Gas chromatograms of various lipid extracts of G. aggregans TPB6028T. Shown are the fatty acids released after acid (a) and alkaline (b) hydrolyses of whole-cell material and the fatty acids present in the Bligh-Dyer extract (c), the acid-hydrolyzed Bligh-Dyer extract (d), the alkali-hydrolyzed Bligh-Dyer extract (e), and the alkali-hydrolyzed residue after Bligh-Dyer extraction (f). Fatty acids were derivatized to the corresponding methyl esters prior to GC analysis. Key: 2, iso-diabolic acid; 6, iso-C15 fatty acid; 7, C16:1ω7c fatty acid; 8, C16:0 fatty acid; 9, iso-C17 fatty acid. An asterisk denotes a plasticizer contaminant (phthalate). Note the abundance of iso-diabolic acid in the hydrolysates from the cell material and residue after extraction and its absence from the solvent extract.
Table 2.
Fatty acid composition after acid hydrolysis of cell material and presence of branched GDGTs in the acidobacterial strains studied
| Component | % of total fatty acidsa in strainb: |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1c | 2c | 3c | 4d | 5c | 6c | 6d | 7d | 8d | 9d | 10d | 11d | 12c | 13d | 14c | 15c | 16d | 17d | |
| iso-C13 | 0.2 | 0.2 | 5.4 | 5.2 | 2.3 | |||||||||||||
| iso-C14 | 2.0 | 1.6 | ||||||||||||||||
| C14:1ω5 | 0.1 | 1.7 | 0.1 | 0.4e | 0.2 | 0.4 | 0.1 | 0.5 | 0.6 | 0.1 | 0.3 | |||||||
| C14:0 | 0.5 | 0.5 | 0.5 | 1.8 | 0.8 | 0.9 | 0.5 | 1.5 | 1.9 | 1.1 | 1.0 | 1.2 | 1.7 | |||||
| iso-C15:1ω6 | 0.4 | 3.8f | 0.2 | 0.1 | 0.2 | 0.1 | ||||||||||||
| iso-C15 | 38.7 | 34.2 | 29.2 | 46.0 | 32.1 | 36.1 | 38.8 | 35.0 | 37.0 | 30.0 | 33.0 | 32.5 | 35.1 | 27.0 | 22.4 | 26.0 | 16.0 | 12.0 |
| anteiso-C15 | 7.3 | 0.2 | 0.1 | 1.1 | 0.8 | 0.5 | 0.8 | |||||||||||
| C15:1ω6 | 0.1 | 0.8 | 0.3 | 4.4g | 0.4 | 0.1 | 0.3 | 0.5 | ||||||||||
| C15:0 | 0.2 | 1.4 | 0.2 | 0.2 | 0.6 | 0.4 | ||||||||||||
| iso-C16 | 0.8 | 2.6 | 0.6 | |||||||||||||||
| C16:1ω7c | 0.2 | 0.3 | 9.9 | 2.0 | 3.2 | 26.0 | 21.0 | 22.0 | 8.4 | 18.5 | 20.3 | 18.0 | 21.5 | 23.7 | 23.0 | 32.0 | ||
| C16:1ω7t | 0.4 | 0.3 | 1.2 | 0.4 | 0.2 | 0.3 | 2.1 | 0.8 | 0.4 | |||||||||
| C16:1ω6+ω5 | 0.3 | 1.6 | 0.1 | |||||||||||||||
| C16:0 | 3.0 | 0.6 | 4.8 | 5.8 | 3.2 | 7.0 | 5.0 | 11.0 | 9.8 | 6.0 | 6.8 | 7.3 | 7.6 | 5.6 | 15.0 | 18.0 | ||
| iso-C17:1ω8c | 19.5 | 14.6 | 17.4 | 18.4 | 1.0 | 0.5 | 0.8 | 0.8 | 0.2 | 0.6 | 0.1 | 0.2 | 0.4 | 0.3 | 1.1 | 0.3 | ||
| iso-C17:1ω8t | 0.9 | 1.3 | 0.5 | 1.5 | ||||||||||||||
| iso-C17 | 5.8 | 6.1 | 8.5 | 6.8 | 1.0 | 1.7 | 1.5 | 1.7 | 1.6 | 3.0 | 2.2 | 1.7 | 0.5 | 0.7 | 0.7 | 2.0 | 2.7 | 1.0 |
| anteiso-C17 | 0.4 | 0.9 | 4.1 | 1.8 | 0.2 | 0.6 | 0.6 | 1.0 | 1.0 | 0.7 | 2.4 | 0.5 | ||||||
| C17:1ω8 | 0.3 | 1.4 | 0.9 | 0.2h | 0.1h | 0.1 | ||||||||||||
| C17:0 | 0.8 | 0.2 | 1.3 | 0.3 | 0.1 | 0.5 | 0.4 | 0.6 | 0.7 | 0.3 | 0.5 | 0.4 | ||||||
| C18:1ω9 | 3.4 | 11.1 | 15.4 | 0.4 | 2.0 | |||||||||||||
| C18:1ω7 | 2.9 | 0.7 | 0.1 | 0.1 | 3.8 | 0.2 | 0.9 | 0.4 | ||||||||||
| C18:0 | 2.6 | 0.6 | 4.0 | 1.8 | 0.4 | 0.4 | 1.0 | 0.5 | 0.5 | 1.4 | 0.8 | 1.5 | 1.2 | |||||
| iso-C19 | 2.7 | |||||||||||||||||
| iso-Diabolic acidi | 34.7 | 29.9 | 30.3 | 21.5 | 39.4 | 35.6 | 33.4 | 26.0 | 28.0 | 29.0 | 29.0 | 31.5 | 33.1 | 36.0 | 42.8 | 36.1 | 33.0 | 29.0 |
| iso-C15-monoglycerol ether | 0.2 | 0.5 | ||||||||||||||||
| C30 glycerol etherj | 3.2 | 3.5 | ||||||||||||||||
| GDGTk | − | ± | − | − | √ | − | − | − | − | − | − | − | − | − | − | √ | − | − |
Values for major components (i.e., >3% of the total fatty acids) are underlined.
Strains: 1, Acidobacteriaceae strain 277; 2, Acidobacteriaceae strain 307; 3, Acidobacteriaceae strain A2-1c; 4, Acidobacteriaceae strain KA1; 5, Acidobacteriaceae strain A2-4c; 6, A. capsulatum 161T; 7, B. elongata SN10T; 8, G. pectinivorans TPB6011T; 9, G. aggregans TPB6028T; 10, G. paludicola LCBR1; 11, G. paludicola OB1010T; 12, T. roseus KBS63T; 13, Terriglobus sp. strain KMR; 14, E. modestus Jbg-1T; 15, E. aggregans Wbg-1T; 16, B. aggregatus MPL3T; 17, B. aggregatus MPL1011T.
Grown at DSMZ.
Grown at the Winogradsky Institute of Microbiology.
ω7 instead of ω5.
ω8 instead of ω6.
ω7 instead of ω6.
ω6 instead of ω8.
Structure 2 in Fig. 1.
Structure 3 in Fig. 1.
Structure 1 in Fig. 1. −, not detected; ±, detected by single-ion monitoring MS but not confirmed by full mass spectrum; √, present, confirmed by full mass spectrum.
iso-Diabolic acid could be identified only after hydrolysis of the cell material. We used G. aggregans TPB6028T to examine the mode of occurrence of iso-diabolic acid in the phylum Acidobacteria. The lipids released after alkaline hydrolysis of the cell material were similar to those obtained after acid hydrolysis (Fig. 2b). However, iso-diabolic acid was absent from the Bligh-Dyer extract of G. aggregans TPB6028T (Fig. 2c) and also did not appear after base or acid hydrolysis of the Bligh-Dyer extract (Fig. 2d and e). Acid and base hydrolyses of the residue of the cell material after Bligh-Dyer extraction did release iso-diabolic acid; the distribution of the lipids released revealed an enrichment of iso-diabolic acid relative to the regular fatty acids (e.g., Fig. 2f). Bligh-Dyer extraction of G. aggregans TPB6028T at an elevated temperature (i.e., 40°C) did not result in release of iso-diabolic acid in any form in the extract, as revealed by acid hydrolysis of the obtained extract and subsequent GC-MS analysis.
Acid hydrolysis of cell material of E. aggregans Wbg-1T and Acidobacteriaceae bacterium A2-4c released not only iso-diabolic acid but also its glycerol ether derivative (i.e., structure 3 in Fig. 1). This component was tentatively identified by its mass spectrum, which was virtually identical to that of 13,14-dimethyl-28-glyceryloxydodecanoic acid (8), which was identified on the basis of the full identification of its homologue (11). Although the mass spectrum was almost identical, there was a difference in the retention time, consistent with the different methyl substitution pattern of the glycerol ether derivative of iso-diabolic acid.
Distribution of intact polar lipids in Acidobacteria.
To characterize the IPLs of the species belonging to the phylum Acidobacteria, the Bligh-Dyer solvent extract of G. aggregans TPB6028T, T. roseus KBS 63T, and E. aggregans Wbg-1T were analyzed by HPLC-ESI-MS2. The IPLs were dominated by phosphatidylethanolamine (PE) lipids with iso-C15, n-C16:1, and n-C16:0 diglyceride moieties as determined by the molecular weight and characteristic fragmentation. However, no membrane-spanning IPLs were detected in any of these Bligh-Dyer extracts.
Presence of branched GDGTs in Acidobacteria.
The acid-hydrolyzed biomass of the acidobacterial cultures was also analyzed for the presence of GDGTs by HPLC-APCI-MS using SIM. In the cases of E. aggregans Wbg-1T and Acidobacteriaceae bacteria A2-4c and 307, positive identification of a branched GDGT (i.e., structure 1 in Fig. 1) was obtained, while the other cultures were negative (Table 2). The concentration in strain 307 was low, but in the cases of E. aggregans Wbg-1T and strain A2-4c, its identification by SIM was confirmed by full-scan HPLC-APCI-MS; the APCI mass spectrum was characterized by peaks at m/z 1,022 [M+H]+, m/z 1,004 [M+H]+-18 (loss of water), and m/z 948 [M+H]+-74 (loss of glycerol), the characteristic fragmentation pattern for isoprenoid (17) and nonisoprenoidal GDGTs (45). In both cases, no other branched GDGTs with additional methyl groups, which normally co-occur with branched GDGT in peats and soils (50, 52), were detected.
DISCUSSION
Fatty acid distribution.
All of the fatty acid distributions of the acidobacteria studied show a quite consistent pattern; they all contain iso-C15:0 as an abundant fatty acid (12 to 39%; Table 2) in combination with C16:1ω7C and C16:0, C18:1ω9, or iso-C17 fatty acids (Table 2). This is generally consistent with data reported in the literature for these species (9, 12, 26, 30, 40), except that most of these studies report “summed feature 1” (i.e., 16:1ω7c and/or 15:0 iso-2-OH). Our GC-MS data indicate that in all of these species, C16:1ω7C is a dominant fatty acid and that 2-hydroxy iso-C15 is not. Another apparent mismatch with the literature data is observed for E. aggregans Wbg-1T: Koch et al. (26) reported C17:1ω8C (49%), iso-C16:0 (19%), and C17:0 (8%) to be the dominant fatty acids, in strong contrast to the fatty composition reported here, i.e., dominated by iso-C15:0 (26%), C16:1ω7C (24%), and C16:0 (6%) (Table 2). The fatty acid distribution reported here matches those of phylogenetically related species (e.g., E. modestus Jbg-1T; Table 2) quite well, and we therefore cannot explain the results reported by Koch et al. (26).
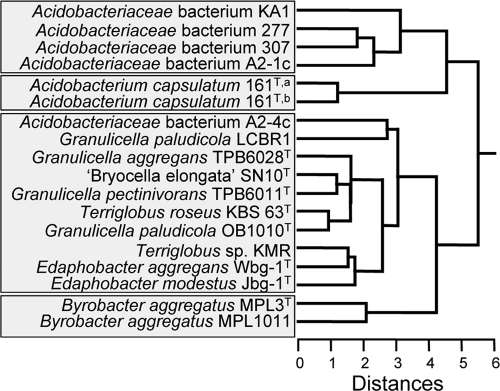
Multivariate statistical analysis of the distribution of the lipids released by acid hydrolysis of cell material revealed four distinct clusters (Fig. 3). The first cluster is composed of Acidobacteriaceae bacterial strains 277, 307, A2-1c, and KA1; they are all characterized by relatively large amounts of (un)saturated iso-C17 fatty acids (23 to 30%; Table 2). The second cluster is formed by A. capsulatum 161T, containing C18:1ω9 as an important constituent (ca. 13%), which is uncommon in the other acidobacteria studied (Table 2). The third cluster, composed of many of the other species, is characterized by the presence of C16:0 and C16:1 fatty acids (Table 2). The Bryobacter species, characterized by a relatively high abundance of the C16:0 fatty acid, form a fourth distinct cluster. This classification is generally in line with the 16S rRNA gene phylogeny (see Fig. S1 in the supplemental material), which also shows Acidobacteriaceae bacterial strains 277, 307, A2-1c, and KA1 and Bryobacter species as distinct clusters. In the 16S rRNA gene-based tree, A. capsulatum is most closely related to Acidobacteriaceae strain A2-4c, whereas this is not apparent from its lipid profile, which most closely resembles that of the Granulicella-Terriglobus-Edaphobacter group (Fig. 3). The apparent ordering of the different Granulicella, Terriglobus, and Edaphobacter species in the 16S rRNA gene tree is not so strictly followed in the clustering on the basis of the lipid profiles, which is likely caused by the rather similar lipid distributions of these species (Table 2).
Fig. 3.
Multivariate statistical analysis of the distribution of the lipids released by acid hydrolysis of cell material of the acidobacteria studied. The input of the cluster analysis was the Bray-Curtis similarity matrix of lipid profiles (% of total lipids; Table 2). A hierarchical clustering was performed in SYSTAT 13 using Euclidian distance and the average-linking method. A. capsulatum 161 was grown at both the DSMZ (a) and the Winogradsky Institute (b).
iso-Diabolic acid in acidobacteria.
In contrast to all previous studies, iso-diabolic acid was detected in all examined species of subdivisions 1 and 3 of the phylum Acidobacteria in relatively large amounts (22 to 43% of all fatty acids). This is most likely due to the fact that it could be released only by hydrolysis of total cell material, a procedure that is not typically applied in most microbiological studies. iso-Diabolic acid has previously been encountered only in thermophilic Thermoanaerobacter species (3, 24, 32), where it fulfills a role as a membrane-spanning lipid. In our study of T. thermohydrosulfuricus (3), iso-diabolic acid was also detected only after hydrolysis of total cell material. However, Lee et al. (32) reported IPLs containing iso-diabolic acid in an esterified form in an extract obtained by extracting T. ethanolicus at an elevated temperature (i.e., 40°C). When G. aggregans TPB6028T cells were subjected to hot extraction, no material could be released that, upon hydrolysis, generated iso-diabolic acid. This indicates that these acidobacteria probably contain complex lipids that are hard to extract and contain iso-diabolic acid in a bound form in substantial amounts. Since iso-diabolic acid is released by both acid and base hydrolyses, it is likely that it is predominantly bound via ester linkages and not by glycosidic or amide bonds, which are not hydrolyzed by treatment with a base.
Membrane-spanning lipids are far less common in the bacterial than in the archaeal domain but do occur there. Clarke et al. (6) provided evidence that diabolic acids (e.g., structure 4 in Fig. 1) may act as linkers between two glycerol moieties in polar membrane lipids of Butyrivibrio spp. Diabolic acids and their ether derivatives have also been identified in members of the order Thermotogales (11, 18, 20, 54) and in Sarcina ventriculi (22). In all of these cases, the membrane-spanning lipids could be extracted from the cells, in contrast to what is reported here for the acidobacteria. Sinninghe Damsté et al. (8) examined nine different species of the order Thermotogales by HPLC-MS and demonstrated the presence of membrane-spanning diglycerol lipids comprised of diabolic acid-derived moieties. In Thermotoga spp., the core membrane lipids were characterized by the presence of both ester and ether bonds, whereas in the phylogenetically more distinct Thermosipho and Fervidobacterium spp., only ester bonds occurred.
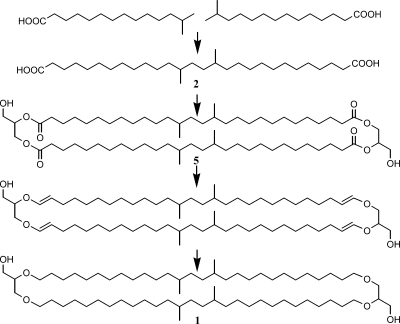
It is interesting that in the acidobacteria E. aggregans Wbg-1T and Acidobacteriaceae strain A2-4c, we detected the glycerol ether derivative of iso-diabolic acid and a branched GDGT (Table 2; structures 1 and 3 in Fig. 1). Both components bear a strong structural resemblance to iso-diabolic acid since the carbon skeleton of the diether-bound alkyl moieties is the same as that of iso-diabolic acid (Fig. 1). In E. aggregans Wbg-1T and strain A2-4c, we detected only a branched GDGT with four methyl groups and not any further methylated branched GDGTs, which normally co-occur with branched GDGT in peats and soils (50, 52). We also detected only one monoether (i.e., structure 3 in Fig. 1). At the same time, iso-diabolic acid was the only dicarboxylic acid encountered in this species (and all other species). This suggests a biosynthetic relationship between the branched GDGT, the glycerol ether derivative of iso-diabolic acid, and iso-diabolic acid. Ring et al. (43) provided evidence that ether linkages in IPLs in the aerobic bacterium Myxococcus xanthus were formed by the reduction of ester linkages via vinyl ether intermediates. This pathway could perhaps also explain the presence of both ester and ether bonds in the membrane-spanning lipids of Thermotoga species and the presence of only tetraesters in Thermosipho and Fervidobacterium spp. (8), of which the latter two would thus apparently lack the enzymes to perform this reaction. Similarly, this may explain the presence of branched GDGT and the glycerol ether derivative of iso-diabolic acid in E. aggregans Wbg-1T and their absence in the other acidobacteria biosynthesizing iso-diabolic acid. This would indicate that a glycerol dialkyl glycerol tetraester in which iso-diabolic acid forms the backbone would be an intermediate (structure 5 in Fig. 4) which in the other acidobacteria (which would lack the ability to transform an ester into an ether linkage) could form the core of the difficult-to-extract IPLs. The presence of mixed esters/ethers with iso-diabolic acid as the core lipid in E. aggregans Wbg-1T and strain A2-4c would also explain the presence of the glycerol ether derivative of iso-diabolic acid after hydrolysis and its absence in the other acidobacteria studied.
Fig. 4.
Hypothetical scheme outlining a potential biosynthesis route from the C15 iso fatty acid to branched GDGT 1. Intermediates are iso-diabolic acid formed by the coupling of two C15 iso fatty acids as previously proposed by Jung et al. (23), a glycerol dialkyl glycerol tetraester in which iso-diabolic acid forms the backbone, and an intermediate in its conversion into branched GDGT. The latter step is based on a biosynthetic route of branched dialkyl glycerol diethers in M. xanthus proposed by Ring et al. (43).
It has been demonstrated that diabolic acids are produced from condensation reactions of fatty acids at the ω-1 positions (15). This proposed biosynthetic pathway is in good agreement with the carbon number distributions of lipids in various species of Thermotogales (8). Jung et al. (23) suggested that the iso-diabolic acids would be biosynthesized by ω,ω′ coupling of two iso fatty acids. In Archaea, isoprenoid GDGTs are also thought to be produced from (partial) condensation of two glycerol diethers by ω,ω′ coupling of isoprenoid alkyl chains (27, 28, 36); note that the tail (where the condensation takes place) of the isoprenoid chain is identical to that of iso fatty acids. The proposed ω,ω′ coupling of two iso fatty acids to obtain iso-diabolic acids is in full agreement with the results obtained here: iso-C15 fatty acid is the dominant iso fatty acid present, and the C30 iso-diabolic acid is the only dicarboxylic acid present. This would suggest the hypothetical overall biosynthetic pathway indicated in Fig. 4. Incorporation of isovaleryl coenzyme A in the pathway of fatty acid biosynthesis results in the formation of iso-C15 fatty acid. Subsequent ω,ω′ coupling of two iso-C15 fatty acids results in the formation of iso-diabolic acid, which can be incorporated into tetraester (structure 5) and tetraether (structure 1) membrane-spanning lipids that subsequently are attached to polar head groups to produce membrane-spanning IPLs. Some of these IPLs have previously been identified in T. ethanolicus (32). It should be noted that the membrane-spanning IPLs do not necessarily contain only iso-diabolic acid but can also contain, in addition, other fatty acid moieties, as observed for the extractable PE IPLs. However, the results of the hydrolysis of the residue after Bligh-Dyer extraction indicate that iso-diabolic acid represents the majority (Fig. 2f). The identity of the head groups in these membrane-spanning IPLs remains unknown, as it was not possible to extract them, even at elevated temperature.
Most bacterial species that contain membrane-spanning lipids are moderate or extreme thermophiles (various Thermotoga, Thermosipho, Fervidobacterium, and Thermoanaerobacter species and S. ventriculi), although Butyrivibrio sp. is a mesophile. All of the analyzed acidobacteria studied here are also mesophilic. Therefore, the occurrence of membrane-spanning IPLs in bacteria seems not only to be an adaptation to temperature. In their study of the presence of diabolic acids in S. ventriculi, Jung et al. (23) showed that this bacterium produced these lipids only when grown at pH 3 and not when grown at pH 7, suggesting a response to pH. When, at neutral pH, the growth temperature was increased from 37 to 45 to 55°C, diabolic acids were also produced. Some of the bacteria studied that produce membrane-spanning IPLs are (slightly) acidophilic, and the acidobacteria studied here are also acidophilic. However, when Acidobacteriaceae strain KA1 and B. aggregatus MPL3T, which are representatives of subdivisions 1 and 3, respectively, were grown at pH 7.0 instead of pH 4.2, iso-diabolic acid was still produced in similar relative quantities (Table 3). Therefore, it remains to be seen why some bacteria produce membrane-spanning IPLs.
Table 3.
Major fatty acids (>5% of the total fatty acids) present in the acid hydrolysates of two acidobacteria grown at different pHs
| Fatty acid | % of total fatty acids |
|||
|---|---|---|---|---|
|
Acidobacteriaceae strain KA1 |
B. aggregatus MPL3T |
|||
| pH 4.2 | pH 7.0 | pH 4.2 | pH 7.0 | |
| iso-C15 | 44.5 | 49.1 | 14.4 | 16.9 |
| C16:1ω7c | 28.1 | 34.1 | ||
| C16:0 | 8.7 | 8.8 | ||
| iso-C17:1ω8c | 15.1 | 9.5 | ||
| iso-C17:0 | 8.4 | 9.5 | ||
| iso-Diabolic acida | 21.3 | 22.1 | 39.8 | 33.7 |
Structure 2 in Fig. 1.
Geobiological implications.
In sedimentary records, soil-derived branched GDGTs are widely used to reconstruct past pHs and temperatures. This is based on empirical relationships developed on the basis of differences in branched GDGT distributions observed in soils from a wide variety of geographical locations and their relationship with environmental variables such as temperature and pH (52). This was explained by the presence of ubiquitously occurring bacteria in soil that adjust their membrane composition in response to temperature and pH. Acidobacteria were proposed as likely candidates (49) because they occur in substantial cell numbers in soil (13, 14, 19) and peat (10, 34) and because branched GDGT concentrations in soil are higher at lower pH (51) and acidobacteria, especially those in subdivision 1, are often relatively more abundant in soil at lower pH (21, 31).
The identification of a branched GDGT in E. aggregans Wbg-1T and Acidobacteriaceae strain A2-4c is the first evidence that the bacterial branched GDGTs may indeed be produced by acidobacteria, as suggested earlier (49). The presence of iso-diabolic acid in all examined species of Acidobacteria in subdivisions 1 and 3 is a further confirmation of this, since the carbon skeleton of iso-diabolic acid is the most common alkyl moiety in the branched GDGTs in soil (51). A potential acidobacterial origin of branched GDGTs is consistent with their heterotrophic lifestyle (i.e., they use a wide variety of organic components; Table 1); the natural 13C abundance of branched GDGTs in soil also suggests that they must derive from heterotrophic microbes (37, 38, 53). Nevertheless, there is still an important mismatch with the GDGTs occurring in E. aggregans Wbg-1T and Acidobacteriaceae strain A2-4c and those occurring in soil: in E. aggregans Wbg-1T and Acidobacteriaceae strain A2-4c, only a branched GDGT with four methyl groups was detected, whereas soils contain branched GDGTs with additional methyl substituents (51, 52). Another issue is that the acidobacteria analyzed here are all aerobes, while in peatlands the highest concentrations of branched GDGTs are found below the water table, in the zone where oxygen quickly becomes limiting (49, 50). Therefore, branched GDGTs in soil likely also originate from other (acido)bacteria capable of biosynthesizing these components, and until we have discovered what they are, biological experiments using cultures to validate the relationships between the degree of methylation and cyclization of branched GDGTs and environmental parameters such as temperature and pH (52) have to wait.
Supplementary Material
ACKNOWLEDGMENTS
J.S.S.D. and E.C.H. received funding from the ERC project PACEMAKER. J.W.H.W. was funded by the Netherlands Organization for Scientific Research (NWO) through a VENI grant. J.O. received support by the BMBF programs BIOLOG (01LC0621C) and The Future Okavango (01LL0912M). S.N.D. was supported by the Program MCB RAS.
We thank Timofey Pankratov, Irina Kulichevskaya, and Lilia Kirsanova (Winogradsky Institute) and Martina Mayer, Pia Wüst, and Alicia Geppert (DSMZ) for help with cultivating acidobacteria for this study.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Angle J. S., Mcgrath S. P., Chaney R. L. 1991. New culture-medium containing ionic concentrations of nutrients similar to concentrations found in the soil solution. Appl. Environ. Microbiol. 57:3674–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. 1979. Methanogens—re-evaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balk M., et al. 2009. Isolation and characterization of a new CO-utilizing strain, Thermoanaerobacter thermohydrosulfuricus subsp. carboxydovorans, isolated from a geothermal spring in Turkey. Extremophiles 13:885–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barns S. M., Cain E. C., Sommerville L., Kuske C. R. 2007. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 73:3113–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 6. Clarke N. G., Hazlewood G. P., Dawson R. M. C. 1980. Structure of diabolic acid-containing phospholipids isolated from Butyrivibrio sp. Biochem. J. 191:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coates J. D., Ellis D. J., Gaw C. V., Lovley D. R. 1999. Geothrix fermentans gen. nov., sp nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49:1615–1622 [DOI] [PubMed] [Google Scholar]
- 8. Damsté J. S. S., et al. 2007. Structural characterization of diabolic acid-based tetraester, tetraether and mixed ether/ester, membrane-spanning lipids of bacteria from the order Thermotogales. Arch. Microbiol. 188:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dedysh S. N., et al. Bryocella elongata gen. nov., sp. nov., a novel member of subdivision 1 of the Acidobacteria isolated from a methanotrophic enrichment culture. Int. J. Syst. Evol. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 10. Dedysh S. N., Pankratov T. A., Belova S. E., Kulichevskaya I. S., Liesack W. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeRosa M., et al. 1988. A new 15,16-dimethyl-30-glyceryloxytriacontanoic acid from lipids of Thermotoga maritima. J. Chem. Soc. Chem. Commun. 1988:1300–1301 [Google Scholar]
- 12. Eichorst S. A., Breznak J. A., Schmidt T. M. 2007. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faoro H., et al. 2010. Influence of soil characteristics on the diversity of bacteria in the Southern Brazilian Atlantic Forest. Appl. Environ. Microbiol. 76:4744–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fierer N., Bradford M. A., Jackson R. B. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 15. Fitz W., Arigoni D. 1992. Biosynthesis of 15,16-dimethyltriacontanedioic acid (diabolic acid) from [16-(H-2)3]- and [14-(H-2)2]-palmitic acids. J. Chem. Soc. Chem. Commun. 1992:1533–1534 [Google Scholar]
- 16. Fukunaga Y., Kurahashi M., Yanagi K., Yokota A., Harayama S. 2008. Acanthopleuribacter pedis gen. nov., sp. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagaceae fam. nov., Holophagales ord. nov. and Holophagae classis nov. in the phylum ‘Acidobacteria’. Int. J. Syst. Evol. Microbiol. 58:2597–2601 [DOI] [PubMed] [Google Scholar]
- 17. Hopmans E. C., Schouten S., Pancost R. D., Van der Meer M. T. J., Sinninghe Damsté J. S. 2000. Analysis of intact tetraether lipids in archaeal cell material and sediments by high performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 14:585–589 [DOI] [PubMed] [Google Scholar]
- 18. Huber R., et al. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 oC. Arch. Microbiol. 144:324–333 [Google Scholar]
- 19. Janssen P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeanthon C., et al. 1995. Thermotoga subterranea sp. nov., a new thermophilic bacterium isolated from a continental oil reservoir. Arch. Microbiol. 164:91–97 [PubMed] [Google Scholar]
- 21. Jones R. T., et al. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jung S., Hollingsworth R. I. 1994. Structures and stereochemistry of the very long α,ω-bifunctional alkyl species in the membrane of Sarcina ventriculi indicate that they are formed by tail-to-tail coupling of normal fatty acids. J. Lipid Res. 35:1932–1945 [PubMed] [Google Scholar]
- 23. Jung S., Lowe S. E., Hollingsworth R. I., Zeikus J. G. 1993. Sarcina ventriculi synthesizes very long chain dicarboxylic acids in response to different forms of environmental stress. J. Biol. Chem. 268:2828–2835 [PubMed] [Google Scholar]
- 24. Jung S., Zeikus J. G., Hollingsworth R. I. 1994. A new family of very long chain α,ω-dicarboxylic acids is a major structural fatty acyl component of the membrane lipids of Thermoanaerobacter ethanolicus 39E. J. Lipid Res. 35:1057–1065 [PubMed] [Google Scholar]
- 25. Kishimoto N., Kosako Y., Tano T. 1991. Acidobacterium capsulatum gen. nov., sp. nov., an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1–7 [DOI] [PubMed] [Google Scholar]
- 26. Koch I. H., Gich F., Dunfield P. F., Overmann J. 2008. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int. J. Syst. Evol. Microbiol. 58:1114–1122 [DOI] [PubMed] [Google Scholar]
- 27. Koga Y., Nishihara M., Morii H., Akagawa-Matsushita M. 1993. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol. Rev. 57:164–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kon T., Nemoto N., Oshima T., Yamagishi A. 2002. Effects of a squalene epoxidase inhibitor, terbinafine, on ether lipid biosyntheses in a thermoacidophilic archaeon, Thermoplasma acidophilum. J. Bacteriol. 184:1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kraigher B., et al. 2006. Microbial activity and community structure in two drained fen soils in the Ljubljana Marsh. Soil Biol. Biochem. 38:2762–2771 [Google Scholar]
- 30. Kulichevskaya I. S., Suzina N. E., Liesack W., Dedysh S. N. 2010. Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemo-organotroph from subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 60:301–306 [DOI] [PubMed] [Google Scholar]
- 31. Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S., Kang S., Kim J. N., Jung S. 2002. Structural analyses of the novel phosphoglycolipids containing the unusual very long bifunctional acyl chain, α,ω-13,16-dimethyloctacosanedioate in Thermoanaerobacter ethanolicus. Bull. Korean Chem. Soc. 23:1778–1784 [Google Scholar]
- 33. Liesack W., Bak F., Kreft J.-U., Stackebrandt E. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85–90 [DOI] [PubMed] [Google Scholar]
- 34. Morales S. E., et al. 2006. Comparison of bacterial communities in New England Sphagnum bogs using terminal restriction fragment length polymorphism (T-RFLP). Microb. Ecol. 52:34–44 [DOI] [PubMed] [Google Scholar]
- 35. Nichols P. D., Guckert J. B., White D. C. 1986. Determination of monounsaturated fatty acid double-bond position and geometry for microbial monocultures and complex consortia by capillary GC-MS of their dimethyl disulphide adducts. J. Microbiol. Methods 5:49–55 [Google Scholar]
- 36. Nishihara M., Morii H., Koga Y. 1989. Heptads of polar ether lipids of an archaebacterium, Methanobacterium thermoautotrophicum: structure and biosynthetic relationship. Biochemistry 28:95–102 [Google Scholar]
- 37. Oppermann B. I., et al. 2010. Soil microbial community changes as a result of long-term exposure to a natural CO2 vent. Geochim. Cosmochim. Acta 74:2697–2716 [Google Scholar]
- 38. Pancost R. D., Sinninghe Damsté J. S. 2003. Carbon isotopic compositions of prokaryotic lipids as tracers of carbon cycling in diverse settings. Chem. Geol. 195:29–58 [Google Scholar]
- 39. Pankratov T. A., Belova S. E., Dedysh S. N. 2005. Evaluation of the phylogenetic diversity of prokaryotic microorganisms in Sphagnum peat bogs by means of fluorescence in situ hybridization (FISH). Microbiology 74:722–728 [PubMed] [Google Scholar]
- 40. Pankratov T. A., Dedysh S. N. 2010. Granulicella paudicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymers-degrading Acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 60:2951–2959 [DOI] [PubMed] [Google Scholar]
- 41. Pankratov T. A., Serkebaeva Y. M., Kulichevskaya I. S., Liesack W., Dedysh S. N. 2008. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2:551–560 [DOI] [PubMed] [Google Scholar]
- 42. Pitcher A., Hopmans E. C., Schouten S., Sinninghe Damsté J. S. 2009. Separation of core and intact polar archaeal tetraether lipids using silica columns: insights into living and fossil biomass contributions. Org. Geochem. 40:12–19 [Google Scholar]
- 43. Ring M. W., et al. 2006. Novel iso-branched ether lipids as specific markers of developmental sporulation in the myxobacterium Myxococcus xanthus. J. Biol. Chem. 281:36691–36700 [DOI] [PubMed] [Google Scholar]
- 44. Schouten S., Huguet C., Hopmans E. C., Kienhuis M. V. M., Sinninghe Damsté J. S. 2007. Analytical methodology for TEX86 paleothermometry by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal. Chem. 79:2940–2944 [DOI] [PubMed] [Google Scholar]
- 45. Sinninghe Damsté J. S., Hopmans E. C., Pancost R. D., Schouten S., Geenevasen J. A. J. 2000. Newly discovered non-isoprenoid glycerol dialkyl glycerol tetraether lipids in sediments. J. Chem. Soc. Chem. Commun. 2000:1683–1684 [Google Scholar]
- 46. Sturt H. F., Summons R. E., Smith K., Elvert M., Hinrichs K.-U. 2004. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18:617–628 [DOI] [PubMed] [Google Scholar]
- 47. Tschech A., Pfennig N. 1984. Growth-yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163–167 [Google Scholar]
- 48. Ward N. L., et al. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weijers J. W. H., et al. 2009. Constraints on the biological source(s) of the orphan branched tetraether membrane lipids. Geomicrobiol. J. 26:402–414 [Google Scholar]
- 50. Weijers J. W. H., et al. 2006. Membrane lipids of mesophilic anaerobic bacteria thriving in peats have typical archaeal traits. Environ. Microbiol. 8:648–657 [DOI] [PubMed] [Google Scholar]
- 51. Weijers J. W. H., Schouten S., Spaargaren O. C., Sinninghe Damsté J. S. 2006. Occurrence and distribution of tetraether membrane lipids in soils: implications for the use of the TEX86 proxy and the BIT index. Org. Geochem. 37:1680–1693 [Google Scholar]
- 52. Weijers J. W. H., Schouten S., van den Donker J. C., Hopmans E. C., Sinninghe Damsté J. S. 2007. Environmental controls on bacterial tetraether membrane lipid distribution in soils. Geochim. Cosmochim. Acta 71:703–713 [Google Scholar]
- 53. Weijers J. W. H., Wiesenberg G. L. B., Bol R., Hopmans E. C., Pancost R. D. 2010. Carbon isotopic composition of branched tetraether membrane lipids in soils suggest a rapid turnover and a heterotrophic life style of their source organism(s). Biogeosciences 7:2959–2973 [Google Scholar]
- 54. Windberger E., Huber R., Trincone A., Fricke H., Stetter K. O. 1989. Thermotoga thermarum sp. nov. and Thermotoga neapolitana occurring in African continental solfataric springs. Arch. Microbiol. 151:506–512 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.