Abstract
Aim
To evaluate Y-chromosomal diversity of the Moravian Valachs of the Czech Republic and compare them with a Czech population sample and other samples from Central and South-Eastern Europe, and to evaluate the effects of genetic isolation and sampling.
Methods
The first sample set of the Valachs consisted of 94 unrelated male donors from the Valach region in northeastern Czech Republic border-area. The second sample set of the Valachs consisted of 79 men who originated from 7 paternal lineages defined by surname. No close relatives were sampled. The third sample set consisted of 273 unrelated men from the whole of the Czech Republic and was used for comparison, as well as published data for other 27 populations. The total number of samples was 3244. Y-short tandem repeat (STR) markers were typed by standard methods using PowerPlex® Y System (Promega) and Yfiler® Amplification Kit (Applied Biosystems) kits. Y-chromosomal haplogroups were estimated from the haplotype information. Haplotype diversity and other intra- and inter-population statistics were computed.
Results
The Moravian Valachs showed a lower genetic variability of Y-STR markers than other Central European populations, resembling more to the isolated Balkan populations (Aromuns, Csango, Bulgarian, and Macedonian Roma) than the surrounding populations (Czechs, Slovaks, Poles, Saxons). We illustrated the effect of sampling on Valach paternal lineages, which includes reduction of discrimination capacity and variability inside Y-chromosomal haplogroups. Valach modal haplotype belongs to R1a haplogroup and it was not detected in the Czech population.
Conclusion
The Moravian Valachs display strong substructure and isolation in their Y chromosomal markers. They represent a unique Central European population model for population genetics.
Y-chromosomal variation of Central European populations and the possible appearance of genetic isolates in these populations are of increasing interest to forensic and human population geneticists.
Y-chromosomal data for the population of the Czech Republic is still fractional. Kráčmarová et al published a short report on paleolithic and neolithic Y chromosomal haplogroups in the Czech population (1) and Luca et al performed a refined study of the same data (2). Zastera et al published a major study on Czech Y-chromosomal data (3). Other authors have also reported on Czech Y-chromosomal variation, usually with other population data from Europe (4-7). A recent study compared Czechs with other West Slavic populations (8). In this range of reports regarding, genetic variation of possible or confirmed genetic isolates within Central European populations is virtually absent. Here we present the intra-population diversity of such an isolated population, the Moravian Valachs.
So far, a limited number of studies that illustrate the variety of Y-chromosomal polymorphisms in the countries and populations supposedly connected or similar to the Moravian Valachs – the supposed isolate – have been published. Rebala et al (9) focused on the Slavic population from Eastern and Central Europe. As historical sources suggest, immigration from Slavic populations was one of the major sources for the emergence of the Valach population of the Czech Republic, therefore the study of Rebala et al (9) is certainly of great interest to us, as well as other studies on southern European Slavic populations (10). Bosch et al (11) analyzed paternal (and maternal) lineages of the Aromuns and other surrounding Balkan populations, thus offering excellent material for their comparison with the Valachs. They clearly documented the differences between Aromuns (ie, isolated populations) and the major populations that surround them, not only in haplogroup and haplotype lineages, but also in intra-population genetic variability.
The Valachs (or Wallachs/Vlachs as they are sometimes called) are one of the most distinct ethnographic and cultural subpopulations of Central Europe. Today, they can be found not only in the Czech Republic – in its eastern border mountain ranges and highlands (Beskydy in Moravia) – but also in south-southeast Poland and several parts of Slovakia (far western, northern, and central region). Originally, this group spread from the Maramures region of Romania, roughly following the Carpathian Mountain range. The arrival of the Valachs to the area of today’s Czech Republic took place at the very end of the 15th or beginning of the 16th century (12). The migration was not spontaneous, but rather encouraged and subsidized by the local nobility, and it lasted at least until the end of the 18th century, with immigrants supposedly coming not only from Romania, but also from Ukraine, Poland, and Slovakia (13).
Until the beginning of the 20th century, the Moravian Valachs’ way of life was similar to other Romanian ethnic groups in the Balkans, especially the Aromuns (seasonal mountain sheep herding, production of cheese, wool, and leather products). An admixture of the newly-arrived Valachs with autochthonous (Slavic and German) Moravian population also began soon after the arrival of the first immigrants – so we can assume a steady genetic and cultural flow between these two populations. Nonetheless, the core of the Valach settlement was located in a previously uninhabited high altitude region, neighboring with the indigenous population from lowlands. The result of the admixture process was a complete merging of both populations, and the disappearance of any distinction between “new” Valachs and indigenous Moravians during the 18th century, and the creation of one ethnogeographic region with all its properties and people – the Moravian Valachs.
Demographic data (13,14) show only a small increase in the Valach population during the 17-18th century. In combination with population depression during and after the Thirty Years' War (1618-1648), the conditions in the Valach population favored inbreeding, an effect reinforced by isolation-by-distance from the surrounding populations.
To investigate how severe this isolation effect was on Y chromosomal polymorphisms in the Moravian Valachs and whether it is still detectable in modern Valach population is the main aim of our study. Another topic of interest was how the intra-population variability and the sampling bias can affect forensic and population analyses performed on these data.
Material and methods
Hundred and seventy-three DNA samples of male Valachs from the Czech Republic were analyzed. These samples were divided into two groups because of the important differences in sampling procedure and are consistently referred to separately throughout this article.
The first group consisted of 94 samples of unrelated donors (code: VALACH, Moravian Valachs). All donors identified themselves as belonging to the Valach ethnic group in a short interview that was held immediately before DNA sampling in the form of mouth swabs. Only donors whose paternal lineage was present in the region of the Valach country for at 3 generations were included into the study. Informed consent was provided by the donors and no other data (including name, address, etc.) were gathered. The data were rendered fully anonymous.
The second Valach sample set consisted of 79 samples (code: VLIN, Moravian Valachs lineages). The sampling process in this case differed significantly from the VALACH sample set. VLIN sample set came from 7 Valach paternal lineages. These were defined primarily by surname, as well as by geographic localization in the Valach region and self-identification of the donors. Although the samples came from broad families, no first, second, third, and fourth degree relatives were included in the study, virtually making this Valach sample set composed of unrelated, non-randomly selected Valachs, carrying 7 different surnames.
The control sample set consisted of 273 unrelated male donors from the whole Czech Republic. Donors did not identify themselves as having the Valach origin, however, there was no other information gathered about their ethnicity or origin. Data are available on request and will be submitted to the Y-chromosome haplotype reference database (http://www.yhrd.org/) database.
We gathered published samples for Y-short tandem repeat (STR) loci from other populations, concentrating on Eastern European and Balkan populations. Our total set, Moravian Valachs included, consisted of 30 populations encompassing 3244 individuals (Table 1). Due to the limitation of the published data, only minimal haplotype loci (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and DYS385a/b) were used for the analysis of intra-population statistics computing and the comparison between populations. For detailed analysis, 12 loci haplotypes were utilized, which also included all extended haplotype loci (minimal haplotype loci + DYS437, DYS438, DYS439).
Table 1.
Population set used in this study
| Population | Code | Data source | Number of samples |
| Albanians (Tirana) | ALB | (11) | 34 |
| Aromuns (Romania) | ARO | (11) | 48 |
| Aromuns (Andon Poci, Albania) | AAA | (11) | 19 |
| Aromuns (Dukasi, Albania) | AAD | (11) | 39 |
| Aromuns (Krusevo, Macedonia) | AMK | (11) | 43 |
| Aromuns (Stip, Macedonia) | AMS | (11) | 58 |
| Bulgarian Gypsies | BUG | (35) | 81 |
| Bulgarian Turks | BUT | (35) | 61 |
| Bulgars | BUL | (35) | 122 |
| Croatians (Rijeka area) | CRO | (36) | 101 |
| Csango (Lunca de Sus) | CSA | (37) | 84 |
| Czechs (Czech Republic) | CZE | this study | 273 |
| Greeks (Thrace) | GRE | (11) | 39 |
| Hungarians (Budapest) | HUN | (38) | 116 |
| Albanians (Kosovo) | ALK | (29) | 117 |
| Luzice Sorbs | SORB | (32) | 29 |
| Macedonians | MACE | (39) | 84 |
| Macedonian Romani | RMA | (39) | 68 |
| Macedonians (Skopje) | MAC | (11) | 51 |
| Moravian Valachs | VALACH | this study | 94 |
| Moravian Valachs (lineages) | VLIN | this study | 79 |
| Poles (Central Poland) | POLC | (40) | 254 |
| Poles (SE. Poland) | POSE | YA003352* | 161 |
| Romanians (Constanta) | ROMC | (11) | 31 |
| Romanians (Ploesti) | ROMP | (11) | 36 |
| Russians (European part) | RUS | (41) | 541 |
| Saxons (Dresden) | SAX | (32) | 89 |
| Serbia and Monte Negro | SEMN | (42) | 237 |
| Slovaks (Bratislava) | SLO | (9) | 164 |
| Szekely (Miercurea Ciuc) | SZE | (37) | 91 |
| Total | 3244 |
*POSE population comes from http://www.yhrd.org/ database, accession number provided.
The Y-chromosomal STRs of VALACH samples were assessed using PowerPlex® Y System (Promega, Madison, WI, USA). It contained 12 microsatellite polymorphic sites (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS438, and DYS439), including all recommended minimal haplotype loci (minHT), and all extended haplotype loci (extHT; SWGDAM recommended loci).
CZE samples and VLIN samples were typed using AmpFiSTR Yfiler® PCR Amplification Kit (Applied Biosystems, Carslbad, CA, USA), that included all of the above mentioned loci plus DYS456, DYS458, DYS635, Y GATA H4, DYS448. All 17 markers were used only in evaluating discrimination capacity of Y-STR haplotypes in CZE and VLIN sample sets.
Using the Y-STR information, we estimated also the Y chromosomal haplogroups in our samples by the free internet software tool ‘Haplogroup Predictor’ by Whit Athey (http://www.hprg.com/hapest5/) (15,16). We were aware of the issues present in estimating Y-chromosomal haplogroups from Y-STR frequencies (17), thus for the subsequent analyses (median networks) we used only the samples with Hg estimate probability higher than 90%.
Genetic intra-population indices were computed in Arlequin v3.1 software (18). For construction of median networks, we used Neworks 4.60 (available at http://www.fluxus-engineering.com) (19,20). We used 12 Y-STR loci (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS437, DYS438, and DYS439) for the network analysis. A reduced median algorithm (r = 2) was followed by a median joining procedure (epsilon = 0) to reduce the reticulation of the networks (21). The results from the median joining procedure were post-processed by maximum parsimony calculation to further simplify the final network.
The multidimensional scaling analysis was performed in Statistica 9.0 software (StatSoft Inc., Tulsa, OK, USA).
Results
The Moravian Valachs of the Czech Republic showed remarkably low values of intra-population genetic diversity. This low differentiation, as compared with other populations in our study, is shown by the haplotype (gene) diversity values ± standard deviation (0.9792 ± 0.0075), the average gene diversity per locus (0.476607 ± 0.268081), and the mean number of pairwise differences (3.812857 ± 1.936064) (Table 2). This is especially true if we compare the Valachs to population samples from adjacent populations, ie, Czechs, Slovaks, Saxons, and both Polish samples. In this comparison, the Valachs’ Y-chromosomal variability was lower, and their haplotypes were more similar to each other. Our second Valach data set of Valach lineages (VLIN) showed even more extreme values of haplotype diversity – the 5th lowest value from our population set (0.9335 ± 00192) – which can be expected given that they come from paternal lineages. The average diversity per locus of the VLIN data set (0.573799 ± 0.315593) and the mean number of pairwise differences (4.590393 ± 2.278308) were still lower than the cross-population average, but not as extreme as their haplotype diversity.
Table 2.
Intrapopulation genetic diversity parameters of our population sample set
| Population | Code | Gene (haplotype) diversity* |
Average gene diversity over loci* |
Mean number of pairwise differences* |
||||
|---|---|---|---|---|---|---|---|---|
| SD† | Rank‡ | SD† | SD† | |||||
| Albanians (Kosovo) |
ALK |
0.9621 |
0.0079 |
7 |
0.546309 |
0.301059 |
4.370469 |
2.175093 |
| Albanians (Tirana) |
ALB |
0.9911 |
0.0093 |
14 |
0.608734 |
0.338317 |
4.869875 |
2.434188 |
| Aromuns (Andon Poci, Albania) |
AAA |
0.7076 |
0.0690 |
1 |
0.462719 |
0.273324 |
3.701754 |
1.956638 |
| Aromuns (Dukasi, Albania) |
AAD |
0.7976 |
0.0615 |
2 |
0.498988 |
0.283267 |
3.991903 |
2.039691 |
| Aromuns (Krusevo, Macedonia) |
AMK |
0.9911 |
0.0077 |
15 |
0.612957 |
0.338176 |
4.903654 |
2.436244 |
| Aromuns (Romania) |
ARO |
0.9645 |
0.0141 |
8 |
0.644393 |
0.352587 |
5.155142 |
2.541297 |
| Aromuns (Stip, Macedonia) |
AMS |
0.9062 |
0.0204 |
4 |
0.566773 |
0.313685 |
4.53418 |
2.262519 |
| Bulgarian Gypsies |
BUG |
0.9556 |
0.0149 |
6 |
0.53561 |
0.297067 |
4.284877 |
2.144699 |
| Bulgarian Turks |
BUT |
0.9978 |
0.0035 |
28 |
0.66127 |
0.359114 |
5.290164 |
2.590619 |
| Bulgars |
BUL |
0.9977 |
0.0015 |
27 |
0.617328 |
0.33504 |
4.938626 |
2.420761 |
| Croatians (Rijeka area) |
CRO |
0.9949 |
0.0030 |
21 |
0.618688 |
0.336276 |
4.949505 |
2.42891 |
| Csango (Lunca de Sus) |
CSA |
0.9791 |
0.0057 |
9 |
0.626757 |
0.340851 |
5.014056 |
2.46101 |
|
Czechs (Czech Republic) |
CZE |
0.9964 |
0.0010 |
26 |
0.625512 |
0.337410 |
5.004094 |
2.439948 |
| Greeks (Thrace) |
GRE |
0.9906 |
0.0100 |
13 |
0.618084 |
0.341532 |
4.944669 |
2.459228 |
| Hungarians (Budapest) |
HUN |
0.9987 |
0.0013 |
30 |
0.659389 |
0.355352 |
5.275112 |
2.567318 |
| Luzice Sorbs |
SORB |
0.9852 |
0.0136 |
11 |
0.5508 |
0.311564 |
4.406404 |
2.239319 |
| Macedonian Romani |
RMA |
0.9008 |
0.0242 |
3 |
0.477996 |
0.269877 |
3.823968 |
1.947508 |
| Macedonians |
MACE |
0.9885 |
0.0058 |
12 |
0.583333 |
0.319942 |
4.666667 |
2.310046 |
| Macedonians (Skopje) |
MAC |
0.9929 |
0.0058 |
17 |
0.647843 |
0.353808 |
5.182745 |
2.550724 |
|
Moravian Valachs |
VALACH |
0.9792 |
0.0075 |
10 |
0.476607 |
0.268081 |
3.812857 |
1.936064 |
|
Moravian Valachs (lineages) |
VLIN |
0.9335 |
0.0192 |
5 |
0.573799 |
0.315593 |
4.590393 |
2.278308 |
| Poles (Central Poland) |
POLC |
0.9957 |
0.0012 |
23 |
0.570041 |
0.311005 |
4.560331 |
2.248893 |
| Poles (SE. Poland) |
POSE |
0.9929 |
0.0024 |
18 |
0.547438 |
0.300886 |
4.379503 |
2.174803 |
| Romanians (Constanta) |
ROMC |
0.9978 |
0.0089 |
29 |
0.636828 |
0.353164 |
5.094624 |
2.539503 |
| Romanians (Ploesti) |
ROMP |
0.9952 |
0.0078 |
22 |
0.604563 |
0.335687 |
4.836508 |
2.416084 |
| Russians (European part) |
RUS |
0.9943 |
0.0009 |
20 |
0.60807 |
0.328484 |
4.864558 |
2.376197 |
| Saxons (Dresden) |
SAX |
0.9959 |
0.0030 |
25 |
0.632629 |
0.343441 |
5.061032 |
2.480027 |
| Serbia and Monte Negro |
SEMN |
0.9917 |
0.0024 |
16 |
0.571837 |
0.311953 |
4.574698 |
2.255625 |
| Slovaks (Bratislava) |
SLO |
0.9932 |
0.0021 |
19 |
0.590762 |
0.32161 |
4.726096 |
2.324641 |
| Szekely (Miercurea Ciuc) | SZE | 0.9958 | 0.0029 | 24 | 0.679151 | 0.365707 | 5.433211 | 2.640936 |
*The descriptive statistics were computed from the Y-short tandem repeat haplotype frequencies based on minimal haplotype loci (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and DYS385a/b).
†SD – standard deviation.
‡The ranking sequence according to the gene diversity values (from the lowest to the highest).
To assess the power of discrimination of the used Y-STR markers, we computed the discrimination capacity for both of the Valach sample sets (VALACH, VLIN) and Czech sample set (CZE) for 9, 12, and 17 Y-STR marker haplotypes (Table 3). Discrimination values of Y-STR haplotypes were markedly low in VLIN, which is due to the effect of the sampling procedure.
Table 3.
Comparison of discrimination capacity of 9, 12, and 17 Y-short tandem repeat (STR) loci haplotypes in two Moravian Valach sample sets and Czech population sample set
|
Code |
n |
9 Y-STR (minHT) loci* |
12 Y-STR loci (PowerPlex Y)† |
17 Y-STR loci (Yfiler)‡ |
||||||
| No. of haplotypes |
discrimination capacity (%) |
gene diversity |
No. of haplotypes |
discrimination capacity (%) |
gene diversity |
No. of haplotypes |
discrimination capacity (%) |
gene diversity |
||
|
VALACH |
94 |
66 |
70.21 |
0.9792 |
72 |
76.60 |
0.9840 |
N/A |
N/A |
N/A |
|
VLIN |
79 |
35 |
44.30 |
0.9335 |
39 |
49.37 |
0.9507 |
49 |
62.03 |
0.9786 |
| CZE | 273 | 214 | 78.39 | 0.9964 | 247 | 90.48 | 0.9987 | 266 | 97.44 | 0.9998 |
*DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b.
†* + DYS437, DYS438, DYS439.
‡* + † + DYS456, DYS458, DYS635, Y GATA H4, DYS448.
Table 4. Genetic distances (FST – below diagonal; RST – above diagonal*) between Valach populations (VALACH, VLIN) and the geographically surrounding populations
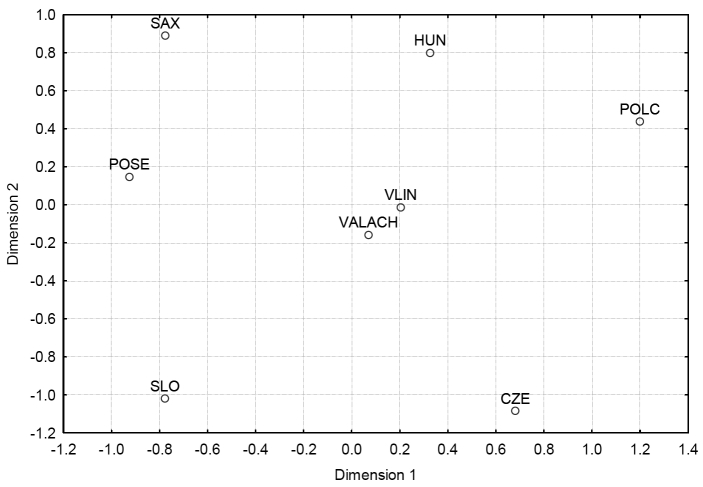
The values of intra-population statistics showed a decrease in Y-chromosomal diversity as the result of isolation. Further evidence of the isolation are the pairwise genetic distances between the Valachs (VALACH, VLIN) and other surrounding populations from our sample set: Czechs (CZE), Slovaks (SLO), Saxons (SAX), Hungarians (HUN), and Poles from Central and Southeast Poland (POLC, POSE). The pairwise FST distances between the Valachs and other populations were all significant at 0.10% level (10 000 permutations). The RST distances displayed a more random pattern, which could have been caused by small allele length differences between the populations. We computed the complete FST and RST genetic distances between the selected populations (Table 4). The FST genetic distances were visualized by multidimensional scaling analysis (MDS). Its results for first and second MDS dimensions are plotted in Figure 1. The Valachs showed marked genetic isolation from the neighboring populations.
Figure 1.
Plot of the first two dimensions of the multidimensional scaling analysis (stress value = 0.163) from the FST genetic distance matrix. VALACH – Moravian Valachs, VLIN – Moravian Valachs (lineages), CZE – Czechs, SLO – Slovaks, SAX – Saxons, HUN – Hungarians, POLC – Poles from Central Poland, POSE – Poles from Southeast Poland.
Distribution of Y chromosomal haplogroups in the VALACH, VLIN, and CZE populations was not uniform (Figure 2). While our Czech population sample well reflected the Central European Y haplogroup pool, the Valach sample set showed some deviation from the expected frequencies of the Y-haplogroups. This was especially noticeable in the VLIN sample set, with the overrepresentation of haplogroup I2a and N, each of them being a dominant haplogroup in one of the 7 paternal lineages sampled.
Figure 2.
Distribution of Y chromosomal haplogroups in Moravian Valach (closed bars), Moravian Valach (lineages) (gray bars), and Czech (open bars) populations.
Variation within selected haplogroups is displayed in Figure 3. Haplogroups R1a, N, I2a, and E1b1 were chosen because of their major representation in different paternal lineages of VLIN sample set. Marked isolation of Valach haplotypes within the haplogroups can be seen in R1a, I2a and E1b1b networks. This reflects the substructure of the examined populations. The effect of sampling can be seen when we compare the distribution of the VLIN and VALACH haplotypes (Figure 3). Closely related paternal lineages of the VLIN sample set demonstrate as clustered, low diversity, branches within the networks. Unrelated VALACH samples, while they still form almost exclusive Valach branches of the network, are separated by more mutation steps and exhibit higher diversity. VLIN in N and I2a come each from a different surname-defined paternal lineage. VLIN samples belonging to E1b1b originated in two paternal lineages. The correspondent clusters are clearly seen in Figure 3D. VLIN samples in R1a are from 3 paternal lineages with an intermingled substructure.
Figure 3.
Median networks diagrams of internal Y chromosomal haplogroups variability for Czech (black), Moravian Valach (white), and Moravian Valach (lineages) (gray) sample sets. (A) Haplogroup R1a; (B) Haplogroup N; (C) Haplogroup I2a; (D) Haplogroup E1b1b.
R1a is the most prevalent and the most diverse haplogroup in the Valach and Czech population. Therefore, we selected this haplogroup for further analysis and identification of its modal haplotype.
Valach-specific modal haplotype
The Y-chromosome haplotype that is common in the Euro-Atlantic populations is called “Atlantic Modal Haplotype” (22). We propose that the common haplotype found in the population sample of 94 unrelated Moravian Valachs is called “Valach Modal Haplotype” (VMH). This modal haplotype belongs to the R1a haplogroup and is defined by 8 Y-STR polymorphisms (DYS390, DYS391, DYS392, DYS393, DYS385a, DYS385b, DYS437, DYS439) (Table 5). The comparison of the VMH with the control population samples CZE (n = 273) used in this study or the other published Czech population sample (3) (n = 1148) resulted in no matches. Further comparison with the European metapopopulation of Y-chromosome haplotype reference database (YHRD) (23,24) release 35 (built on December 30, 2010, 12:56 GMT, consisting of 91 601 haplotypes within 710 populations) resulted in 2 exact matches and a set of matches with neighbor haplotypes (difference of 1 repeat in 1 DYS included in the VMH) (Table 6).
Table 5.
Valach Modal Haplotype within the R1a haplogroup in population sample of 94 unrelated Moravian Valachs (VALACH).
| Haplotype | n | DYS19 | DYS389I | DYS389II | DYS390 | DYS391 | DYS392 | DYS393 | DYS385a | DYS385b | DYS437 | DYS439 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VALACH8 |
4 |
14 |
13 |
29 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH15 |
1 |
14 |
13 |
30 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH19 |
5 |
14 |
13 |
31 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH38 |
2 |
15 |
13 |
29 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH52 |
2 |
16 |
13 |
30 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH55 |
1 |
16 |
13 |
31 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH72 |
1 |
18 |
13 |
30 |
24 |
10 |
11 |
13 |
11 |
14 |
15 |
12 |
| VALACH MODAL HAPLOTYPE | 24 | 10 | 11 | 13 | 11 | 14 | 15 | 12 | ||||
Table 6.
Comparison of Valach Modal Haplotype* with the European metapopulation
| Locus difference† | Population‡, number of matches, (YHRD code) |
|---|---|
|
exact match |
Slovakia, 1x, (YA003557); Argentina, 1x, (YA003561) |
| DYS392 = 12 |
Bihor, Romania, 2x, (YA003605); Majorca, Spain, 1x, (YA003489) |
| DYS392 = 10 |
Taktakoz Romani, Hungary, 1x, (YA003659) |
| DYS391 = 11 |
Novi Sad, Serbia, 3x, (YA003632) |
| DYS437 = 14 |
Ivanava, Belorus, 3x, (YA003507); Santa Ninfa, Italy, 2x, (YA003110); Novi Sad, Serbia, 1x, (YA003632); Rjasan, Russia, 1x, (YA003178); Klimaviči, Belarus, 1x, (YA003508); Stuttgart, Germany, 1x, (YA003031); Cologne, Germany, 1x, (YA002963) |
| DYS390 = 25 |
Sverdlovsk, Russia, 1x, (YA003678) |
| DYS393 = 12 |
Eastern Slovakia, 1x, (YA003546) |
| DYS439 = 13 | Eastern Slovakia, 1x, (YA003546) |
*Valach Modal Haplotype: 24-10-11-13-11-14-15-12; DYS390-391-392-393-385a-385b-437-439.
†Exact matches and 1 allele difference matches are shown.
‡European metapopulation from YHRD database, release 35 (http://www.yhrd.org/).
Discussion
We found the traces of isolation and substructure in the Moravian Valachs’ Y-chromosomal genetic variation. Studies with well defined Y-chromosomal data for Central Europe are scarce. Previously mentioned studies of Czech population Y-STR variability reported no inner differentiation of the population (1,3). A substructure of Y-chromosomal lineages was reported in the Brabant region of Belgium and the Netherlands (25). Also, a strong Y-chromosomal breakpoint in Romanian population, based on ethnic origin, was demonstrated (26). Petrejčíková et al (27) have analyzed men from Eastern Slovakia and found non-significant separation from the surrounding Slavic populations. In Belarus population, a limited population substructure was observed (28), although only detectable when using 7-12 Y-STR haplotypes. With 17 Y-STR haplotypes (Yfiler loci), the substructure was no longer detectable. The population of Moravian Valachs analyzed in our study displayed signs of isolation and substructure, which are noticeable in 9, 12, and 17 Y-STR haplotypes. The isolation of the Valach population, low effective population size and, thus, the faster operation of genetic drift are expressed in low haplotype diversity in the Valachs. These effects of isolation are also evident in the average diversity values over loci and the mean number of pairwise differences. Regarding these parameters, the Moravian Valachs more resemble rather isolated Balkan populations (Aromuns, Csango, Bulgarian Roma, and Macedonian Roma) (11,29), population from eastern Adriatic coast islands (30,31), or isolated Slavic populations like the Lužice Sorbs (32), than the surrounding Central European populations of the Czech Republic, Slovak Republic, or Poland. Zalán et al (33) have also reported such low values of Y-chromosomal diversity in Hungarian Vlax Roma (not related to the Valachs, despite the similar name). A common feature of all these populations is geographical and/or cultural isolation, which in the case of the Moravian Valachs lasted at least until the beginning of the 20th century. The isolation of the Valachs from the surrounding populations was already documented by FST pairwise distances. Non-significant pairwise FST genetic distances based on 12 Y-STRs were shown between the regions surrounding the Valach country (Moravia-Silesia and Zlin regions, data by Zastera et al) and CZE, but highly significant between the Valach samples and both CZE and Zastera’s data (P < 0.010; 10000 permutations). Thus the Valachs are isolated not only from the general Czech population, but also from the non-Valach Czech population from the regions neighboring the Valach region.
Y-STR analysis of the VALACH population sample revealed a previously unnoted modal haplotype within the haplogroup R1a (VMH). The reason for comparing the VMH with the European metapopopulation of Y-chromosome haplotype reference database was to test whether the VMH was specific just for the typical Moravian Valach region or can be found in populations where we expect the occurrence of Valach individuals. The majority of close and neighbor matches were from the populations and geographic locations where the migration of the Romanian or Moravian Valachs was recorded. The matches in non-Slavic regions (Argentina, Spain, Italy, Germany) are in concordance with the emigration waves of the Valachs in the 19th and 20th century.
Forensic analysis of Y-chromosome loci requires the highest possible haplotype diversity. Some studies have shown (34) that commercially available kits like PowerPlex Y or Yfiler cannot provide a sufficient discrimination power to discern the haplotypes inside broad families, and a definition of a region-specific set of Y-STR loci is a must. The results of haplotype diversity of the population samples tested within this study (VALACH, VLIN, and CZE) revealed that the sampling in one close geographic region or inside a group of people that recognize themselves as members of a certain “clan” brings lower diversity. However, this phenomenon enables us to find a founding haplotype for that group. The modal haplotype within VALACH population sample belonging to the haplogroup R1a can be of high forensic importance as it defines a relatively large group of individuals that can be identified through Y-chromosome STR analysis of modality. Defining such modal haplotype is only possible in the case of detailed knowledge of the genetic (sub)structure of the population in question.
Besides the genetic or forensic aspects, the ethical aspects of data gathering and presentation should be predominant, especially if we want to investigate not-so-distant paternal lineages. Moreover, genealogical, linguistic, and historical information is also of foremost interest to the researcher. The optimal number of Y-chromosomal STR polymorphic loci to be used in such a study differs vastly according to the objectives. We confirmed that 17 loci of Yfiler kit could be insufficient for forensic application if we were to analyze a cluster of related paternal lineages. On the other hand, for population genetic applications, a set of a few precisely defined core Y-STR could very well describe the population and could be used for interpopulation comparison. Properties of the sample set under examination are also strongly influenced by the sampling procedure, which is not always known or properly administered. We demonstrated this sampling effect on the differences between our Moravian Valach sample sets (VALACH vs VLIN).
Our analyses confirmed that the Moravian Valachs represent a unique population data set from the Czech Republic and the whole region of Central Europe due to their ethnographic coherence and isolation that is clearly detectable in their Y-chromosomal diversity. The VMH can be used for further studies on Valach migration or in the evaluation of forensic analysis results.
| SLO | SAX | POLC | POSE | HUN | VALACH | VLIN | CZE | |
|---|---|---|---|---|---|---|---|---|
|
SLO |
- |
0.03393 |
0.01043 |
-0.00571 |
0.05227 |
0.01966 |
0.04191 |
-0.00406 |
|
SAX |
0.02052 |
- |
0.05743 |
0.02967 |
-0.00808 |
0.00198 |
0.00897 |
0.03808 |
|
POLC |
0.00188 |
0.03079 |
- |
-0.00474 |
0.07888 |
0.03122 |
0.10654 |
0.00163 |
|
POSE |
0.00532 |
0.03252 |
-0.00043 |
- |
0.04802 |
0.01960 |
-0.00224 |
-0.00355 |
|
HUN |
0.02450 |
0.00532 |
0.03532 |
0.04190 |
- |
0.00270 |
0.02325 |
0.06064 |
|
VALACH |
0.08979 |
0.05805 |
0.10999 |
0.10114 |
0.08175 |
- |
0.00623 |
0.03159 |
|
VLIN |
0.02830 |
0.06106 |
0.04693 |
0.05861 |
0.06207 |
0.14301 |
- |
-0.00510 |
| CZE | 0.01108 | -0.00114 | 0.01783 | 0.02046 | 0.00482 | 0.06421 | 0.04759 | - |
*Statistically significant pairwise distances are printed in bold (P < 0.01; 10 000 permutations).
Acknowledgments
Authors thank the following colleagues for their input and consultations of presented study: H. Šimková (KUP, Czech Republic), J. M. Rocha (IPATIMUP, Portugal), R. Brdička (ÚHKT, Czech Republic), MUDr Jaroslav Ostřížek (general practitioner in Hutisko-Solanec, Valach region, Czech Republic), Lenka Sasková, Marcela Šilerová, and Vladislava Urbanová (Forensic DNA Service, Czech Republic).
Funding EE was supported by grants MSM 0021620843 (Ministry of Education, Youth, and Sports of the Czech Republic). EE and VV were supported by grant UK 218/2005/B-BIO/PedF (Charles University in Prague, Czech Republic). VS was supported by the grant VD20072010B14 (Ministry of the Interior of the Czech Republic).
Ethical approval No ethical approval was demanded by our institutions (Faculty of Science, Charles University in Prague; Institute of Criminalistics, Prague) or by the financing bodies (Charles University in Prague; Ministry of Education, Youth, and Sports of the Czech Republic; Ministry of the Interior of the Czech Republic). Nevertheless, informed consent was gathered from the donors and we otherwise followed our institutional guidelines regarding working with the human samples.
Declaration of authorship EE provided a part of the data, performed the analysis, and wrote the majority of the manuscript. DV performed the research and wrote the manuscript. VS provided part of the data and participated in the analysis and proof reading of the manuscript. VV was a member of the research team – common grant project with the first author, and participated in the proof-reading of the manuscript.
Competing interests All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work.
References
- 1.Kráčmarová A, Bruchová H, Černý V, Brdička R.The ratio of palolithic and neolithic Y chromosomal haplogroups in Czech population [in Czech]Archeologické rozhledy 200658237–49. [Google Scholar]
- 2.Luca F, Di Giacomo F, Benincasa T, Popa LO, Banyko J, Kracmarova A, et al. Y-Chromosomal variation in the Czech Republic. Am J Phys Anthropol. 2007;132:132–9. doi: 10.1002/ajpa.20500. [DOI] [PubMed] [Google Scholar]
- 3.Zastera J, Roewer L, Willuweit S, Sekera P, Benesova L, Minarik M. Assembly of a large Y-STR haplotype database for the Czech population and investigation of its substructure. Forensic Sci Int Genet. 2010;4:e75–8. doi: 10.1016/j.fsigen.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, et al. The genetic legacy of paleolithic Homo sapiens sapiens in extant Europeans: A Y chromosome perspective. Science. 2000;290:1155–9. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 5.Cruciani F, La Fratta R, Trombetta B, Santolamazza P, Sellitto D, Colomb EB, et al. Tracing past human male movements in Northern/Eastern Africa and Western Eurasia: New clues from Y-chromosomal haplogroups E-M78 nad J-M12. Mol Biol Evol. 2007;24:1300–11. doi: 10.1093/molbev/msm049. [DOI] [PubMed] [Google Scholar]
- 6.Rootsi S, Magri C, Kivisild T, Benuzzi G, Help H, Bermisheva M, et al. Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am J Hum Genet. 2004;75:128–37. doi: 10.1086/422196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, et al. The Eurasian Heartland: A continental perspective on Y-chromosome diversity. Proc Natl Acad Sci U S A. 2001;98:10244–9. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wozniak M, Malyarchuk B, Derenko M, Vanecek T, Lazur J, Gomolcak P, et al. Similarities and distinctions in Y chromosome gene pool of Western Slavs. Am J Phys Anthropol. 2010;142:540–8. doi: 10.1002/ajpa.21253. [DOI] [PubMed] [Google Scholar]
- 9.Rebala K, Mikulich AI, Tsybovsky IS, Sivakova D, Dzupinkova Z, Szczerkowska-Dobosz A, et al. Y-STR variation among Slavs: evidence for the Slavic homeland in the middle Dnieper basin. J Hum Genet. 2007;52:406–14. doi: 10.1007/s10038-007-0125-6. [DOI] [PubMed] [Google Scholar]
- 10.Pericic M, Lauc LB, Klaric IM, Rootsi S, Janicijevic B, Rudan I, et al. High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations. Mol Biol Evol. 2005;22:1964–75. doi: 10.1093/molbev/msi185. [DOI] [PubMed] [Google Scholar]
- 11.Bosch E, Calafell F, Gonzalez-Neira A, Flaiz C, Mateu E, Schneil HG, et al. Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–87. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 12.Kadlec K. Valachs and Valach law in Slavic and Hungarian lands [in Czech]. Prague: Česká akademie císaře Františka Josefa pro vědy, slovesnost a umění; 1916. [Google Scholar]
- 13.Štika J. Ethnographic region of Moravian Valachia, its origin and development [in Czech]. Ostrava (the Czech Republic): Profil; 1973. [Google Scholar]
- 14.Holík J. In the name of Valachs, the Valach revolt (1620-1644), last glory of Lukov castle [in Czech]. Zlín (the Czech Republic): Spolek přátel na hradu Lukova; 2006. [Google Scholar]
- 15.Athey TW. Haplogroup prediction from Y-STR values using a Bayesian-allele frequency approach. Journal of Genetic Genealogy. 2006;2:34–9. [Google Scholar]
- 16.Athey TW. Haplogroup prediction from Y-STR values using an allele-frequency approach. Journal of Genetic Genealogy. 2005;1:1–7. [Google Scholar]
- 17.Muzzio M, Ramallo V, Motti JM, Santos MR, Camelo JS, Bailliet G. Software for Y-haplogroup predictions: a word of caution. Int J Legal Med. 2011;125:143–7. doi: 10.1007/s00414-009-0404-1. [DOI] [PubMed] [Google Scholar]
- 18.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 19.Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–53. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 21.Forster P, Rohl A, Lunnermann P, Brinkmann C, Zerjal T, Tyler-Smith C, et al. A short tandem repeat-based phylogeny for the human Y chromosome. Am J Hum Genet. 2000;67:182–96. doi: 10.1086/302953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson JF, Weiss DA, Richards M, Thomas MG, Bradman N, Goldstein DB. Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc Natl Acad Sci U S A. 2001;98:5078–83. doi: 10.1073/pnas.071036898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roewer L, Krawczak M, Willuweit S, Nagy M, Alves C, Amorim A, et al. Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci Int. 2001;118:106–13. doi: 10.1016/S0379-0738(00)00478-3. [DOI] [PubMed] [Google Scholar]
- 24.Willuweit S, Roewer L. Y chromosome haplotype reference database (YHRD): Update. Forensic Sci Int Genet. 2007;1:83–7. doi: 10.1016/j.fsigen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Larmuseau MH, Vanderheyden N, Jacobs M, Coomans M, Larno L, Decorte R. Micro-geographic distribution of Y-chromosomal variation in the central-western European region Brabant. Forensic Sci Int Genet. 2011;5:95–9. doi: 10.1016/j.fsigen.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 26.Bembea M, Patocs A, Kozma K, Jurca C, Skrypnyk C. Y-chromosome STR haplotype diversity in three ethnically isolated population from North-Western Romania. Forensic Sci Int Genet. 2011;5:e99–100. doi: 10.1016/j.fsigen.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Petrejcikova E, Sotak M, Bernasovska J, Bernasovsky I, Rebala K, Sovicova A, et al. Allele frequencies and population data for 11 Y-chromosome STRs in samples from Eastern Slovakia. Forensic Sci Int Genet. 2011;5:e53–62. doi: 10.1016/j.fsigen.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Rebala K, Tsybovsky IS, Bogacheva AV, Kotova SA, Mikulich AI, Szczerkowska Z. Forensic analysis of polymorphism and regional stratification of Y-chromosomal microsatellites in Belarus. Forensic Sci Int Genet. 2009;5:e17–20. doi: 10.1016/j.fsigen.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Pericic M, Lauc LB, Klaric IM, Janicijevic B, Behluli I, Rudan P. Y chromosome haplotypes in Albanian population from Kosovo. Forensic Sci Int. 2004;146:61–4. doi: 10.1016/j.forsciint.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Pericic M, Lauc LB, Klaric IM, Janicijevic B, Rudan P. Review of Croatian genetic heretige as revealed by mitochondrial DNA a Y chromosomal lineages. Croat Med J. 2005;46:502–13. [PubMed] [Google Scholar]
- 31.Barac L, Pericic M, Klaric MI, Rootsi S, Janicijevic B, Kivisild T, et al. Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet. 2003;11:535–42. doi: 10.1038/sj.ejhg.5200992. [DOI] [PubMed] [Google Scholar]
- 32.Rodig H, Grum M, Grimmecke HD. Population study and evaluation of 20 Y-chromosome STR loci in Germans. Int J Legal Med. 2007;121:24–7. doi: 10.1007/s00414-005-0075-5. [DOI] [PubMed] [Google Scholar]
- 33.Zalan A, Beres J, Pamjav H. Paternal genetic history of the Vlax Roma. Forensic Sci Int Genet. 2011;5:109–13. doi: 10.1016/j.fsigen.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Palo JU, Perttimaa M, Bengs A, Johnsson V, Ulmanen I, Lukka M, et al. The effect of number of loci on geographical structuring and forensic applicability of Y-STR data in Finland. Int J Legal Med. 2008;122:449–56. doi: 10.1007/s00414-008-0250-6. [DOI] [PubMed] [Google Scholar]
- 35.Zaharova B, Andonova S, Gilissen A, Cassiman JJ, Decorte R, Kremensky I. Y-chromosomal STR haplotypes in three major population groups in Bulgaria. Forensic Sci Int. 2001;124:182–6. doi: 10.1016/S0379-0738(01)00597-7. [DOI] [PubMed] [Google Scholar]
- 36.Lovrecic L, Ristic S, Brajenovic B, Kapovic M, Peterlin B. Human Y-specific STR haplotypes in the Western Croatian population sample. Forensic Sci Int. 2005;149:257–61. doi: 10.1016/j.forsciint.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 37.Egyed B, Furedi S, Padar Z. Population genetic study in two Transylvanian populations using forensically informative autosomal and Y-chromosomal STR markers. Forensic Sci Int. 2006;164:257–65. doi: 10.1016/j.forsciint.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Furedi S, Woller J, Padar Z, Angyal M. Y-STR haployping in two Hungarian populations. Int J Legal Med. 1999;113:38–42. doi: 10.1007/s004140050276. [DOI] [PubMed] [Google Scholar]
- 39.Pericic M, Klaric MI, Lauc LB, Janicijevic B, Dordevic D, Efremovska L, et al. Population genetics of 8 Y chromosome STR loci in Macedonians and Macedonian Romani (Gypsy) Forensic Sci Int. 2005;154:257–61. doi: 10.1016/j.forsciint.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Soltyszewski I, Pepinski W, Spolnicka M, Kartasinska W, Konarzewska M, Janica J. Y-chromosomal haplotypes for the AmpFISTR Yfiler PCR Amplification Kit in a population sample from Central Poland. Forensic Sci Int. 2007;168:61–7. doi: 10.1016/j.forsciint.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Roewer L, Willuweit S, Kruger C, Nagy M, Rychkov S, Morozowa I, et al. Analysis of Y chromosome STR haplotypes in the European part of Russia reveals high diversities but non-significant genetic distances between populations. Int J Legal Med. 2008;122:219–23. doi: 10.1007/s00414-007-0222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevanovic M, Dobricic V, Keckarevic D, Perovic A, Savic-Pavicevic D, Markovic MK, et al. Human Y-specific STR haplotypes in population of Serbia and Montenegro. Forensic Sci Int. 2007;171:216–21. doi: 10.1016/j.forsciint.2006.05.038. [DOI] [PubMed] [Google Scholar]