Abstract
Although cells undergo dramatic shape changes during cytokinesis, the role of the plasma membrane and lipids is poorly understood. We report that inactivation of glucosyl ceramide synthase (GCS), either by RNAi or with the small molecule PPMP, causes failure of cleavage furrow ingression. Using mass spectrometry-based global lipid profiling, we identify individual lipids that are enhanced or depleted due to GCS inhibition. We show that GCS inhibition results in the mis-localization of actin and the ERM proteins, key cytoskeletal proteins that connect the plasma membrane to the actin cortex. Our data suggest that ceramides participate in mediating the interactions between the membrane and the cortex.
Cytokinesis, the final step of cell division, has to achieve two crucial processes: equal sorting of cellular components between the daughter cells, and membrane ingression and closure.1 Both processes strongly rely on membrane components and properties.2,3 Although it is known that cell membranes undergo dramatic structural rearrangements during cytokinesis, and it is obvious that membrane rearrangements are needed to seal daughter cells after severing, little is known about whether (and how) specific lipids are involved in cytokinesis.4 It is much more difficult to study lipids in their biological context than proteins, because there are fewer options to manipulate and visualize lipids. For example, only four lipids (PIP2,5,6 PE,7 cholesterol8 and GM18) have been shown to localize to the cleavage furrow during cytokinesis in higher eukaryotes, mostly because more or less specific sensors exist for these lipids. We used mass spectrometry-based global lipid profiling9 coupled with small molecule perturbations to investigate which lipids participate in cytokinesis. We report here that sphingolipids are important for cytokinesis and present evidence that they affect the actin cytoskeleton.
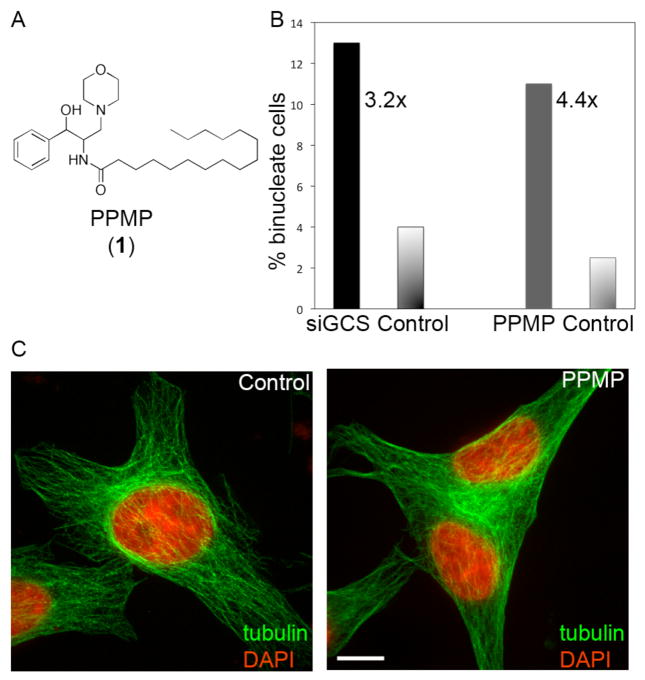
We systematically perturbed the biosynthesis of different lipid families, and evaluated if the perturbation also caused cytokinesis failure. We determined if cells had failed cytokinesis by imaging and scoring for cells with two or more nuclei, a hallmark of failed cell division. We initially tested 14 commercially available small molecule inhibitors of lipid synthesis, which included known inhibitors of different steps during fatty acid, cholesterol and sphingolipid biosynthesis (Supporting Table 1). Four of these compounds inhibited cytokinesis to different extents, with inhibitors of sphingolipid synthesis showing the most robust inhibition. We subsequently used additional small molecules (Supporting Table 1) as well as RNA interference (RNAi) to target most major steps in sphingolipid synthesis (Supporting Figure 1) and found that inhibition of many of these steps caused moderate cytokine-sis failure. Interestingly, a previous study showed that sphingolipid composition changes in dividing cells relative to interphase cells.10 We decided to focus on 1-phenyl-2-palmitoylamino-3-morpholino-1-propanol (PPMP, 1), an inhibitor of glucosyl ceramide synthase (GCS)11 because it is a relatively strong inhibitor of cytokinesis and because glucosyl ceramide synthesis is a late step during sphingolipid synthesis and we would therefore expect to see more specific effects (Figure 1). Also, PPMP had been previously shown to affect cell division in the parasite Giardia lamblia.12 Using live imaging, we found that PPMP-treated cells can assemble a cleavage furrow and ingress, but then fail during ingression and become binucleated (Supporting Figure 2). RNAi of GCS also inhibited cytokinesis, confirming that the lipids synthesized by this biosynthetic branch are important in cytokinesis.
Figure 1.
A Structure of PPMP, an inhibitor of glucosyl ceramide synthase that causes cytokinesis failure. B PPMP treatment over 24 hours and RNAi depletion of the target enzyme, GCS, over 72 hours causes an increase in the number of binucleated cells (n=300). C Representative control and binucleated HeLa cells. Microtubules are shown in green, DNA in red (Scalebar = 10 μm).
We next wanted to understand which specific lipids were changing due to GCS inhibition because these lipids are likely to be involved in cytokinesis. Lipid synthesis is complex, with many different feedback loops and branch points.13,14 The most simplistic outcome of inhibiting GCS would be the accumulation of ceramide, its substrate, and depletion of glucosyl ceramide, its product. However, ceramide is a central substrate for several other biosynthetic branches (including galactosyl ceramide, sphingomyelin and sphingosine synthesis, Supporting Figure 1) and therefore any accumulated ceramide could be channeled into these other branches. Similarly, glucosyl ceramide is a substrate in the synthesis of several other, more highly glycosylated lipid species, which could all be perturbed when GCS is inhibited. In addition, many different ceramides with differing lengths of fatty acids chains exist. We used liquid chromatography/mass spectrometry (LC/MS)-based global lipid profiling to determine which lipids were changed upon glucosyl ceramide synthase inhibition.
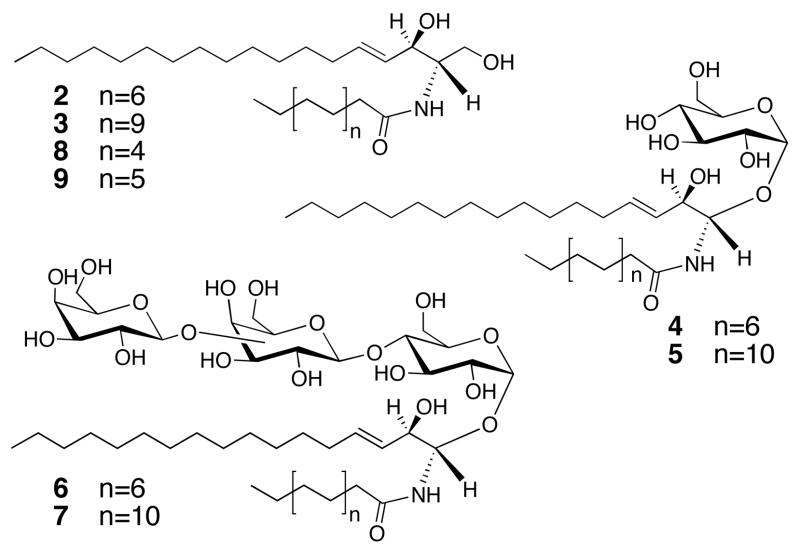
We first used untargeted global lipid profiling to compare cells treated with PPMP to untreated cells. We cultured cells in the presence or absence of PPMP for 24 hours, lysed cells in a Downs homogenizer and extracted lipids with methanol/chloroform.9,15 Triplicate samples were then injected into the LC/MS and the resulting profiles were compared using the XCMS software package.16 Lipid species that were observed in three independent experiments with greater than 4-fold changes between treated and control cells and p values <0.05 were inspected and integrated manually (See Supporting Information for detailed data analysis). As expected for an inhibitor of glucosyl ceramide synthase, we found that PPMP caused approximately 8- and 10-fold accumulations of two major ceramides, C16- and C22-ceramide (2 and 3, Figure 2), respectively (Supporting Table 2 and 4). Perhaps more surprisingly, out of the full lipidome we analyzed, these two lipids were the only ones that changed robustly and reproducibly in the presence of PPMP.
Figure 2.
Ceramides with altered levels as a result of PPMP treatment (2 and 3) and siGCS treatment (4, 5, 6 and 7). Exogenous addition of C12- and C14-ceramide (8 and 9) causes cytokinesis failure.
These findings strongly implicate ceramides in cytokinesis. Ceramides with fatty acid side chains ranging from C2 to C26 exist in cells in varying abundance,13 but we only identified two species in the global analysis. We therefore used a targeted approach to determine if other ceramide species were absent in the HeLa cells used in this study, or present, but unchanged in the presence of PPMP. We found that C2-C12 ceramides were either not present or present at very low levels. When we calculated the fold changes of different ceramide species that were present in the lipid extracts (Supporting Table 4), we found that in addition to C16- and C22-ceramides, C20-ceramide accumulated ~4 fold with PPMP treatment. We did not observe significant changes in other longer chain ceramide species upon PPMP treatment. It is possible that these other ceramide species are preferentially used by other sphingolipid synthetic branches and therefore do not accumulate when GCS is inhibited.
Because we wanted to understand better why only certain ceramides were accumulated during PPMP treatment and because both small molecule and RNAi experiments can be confounded by off-target effects, we analyzed the global lipid profiles of cells where GCS was depleted by RNAi. The experiment was similar to PPMP treatment, but cells were treated with siRNAs for 3 days rather than one day with PPMP because this length of time is needed to observe cytokinesis inhibition. We used cells treated with transfection reagent, but no siRNA, as controls to account for possible perturbations caused by lipids in the transfection reagent. PPMP and siRNA treatments resulted in complementary lipid profiles. The global profiling experiment identified four lipids that were depleted during siRNA treatment: C16- and C24-glucosyl ceramides (4 and 5) and more highly glycosylated derivatives (6 and 7) (Supplement Table 4). Even when we carried out a targeted ceramide analysis as described for PPMP, we did not observe accumulation of any ceramide species, possibly because the three-day experiment allowed these precursors to be used in other lipid biosynthetic pathways (Supporting Table 3 and 5). It was not possible to compare PPMP treatment and RNAi directly because a three-day PPMP treatment is toxic to cells. Taken together, the PPMP and RNAi experiments suggest that the correct balance between different glucosylated and non-glucosylated ceramides is required for successful cytokinesis.
Our lipid profiling experiments suggest that ceramides are involved in cytokinesis, but do not show a direct interaction. If an accumulation of ceramides or more broadly a perturbation of the balance between different glucosylated ceramide species is responsible for cytokinesis failure, we should be able to add exogenous ceramide to cells and also observe cytokinesis failure. Unfortunately, none of the long chain ceramides (2 and 3) we identified during PPMP treatment are soluble under conditions compatible with cell culture and we could therefore not test them directly. We were, however, successful in dissolving shorter chain ceramides, C12- and C14-ceramide (8 and 9) in 2% dodecane/ethanol. Exogenous addition of 8 and 9 to cells caused a ~3 fold increase in the number of binucleated cells. This experiment shows that the prediction made by the lipid profiling experiments was correct, i.e. that ceramides and their derivatives are involved in cytokinesis.
After establishing a role for these lipid species in cytokinesis, we asked how they might be involved. Lipids can have different functions, for example, they can be structural, i.e. they can participate in changing membrane curvature.17,18 They can also be signaling molecules and/or anchor proteins needed for cytokinesis. Some early clues suggest that all of these scenarios are true for different lipids during cytokinesis.19,20 To address these questions, we analyzed the localization of different cytokinesis proteins in cells where sphingolipid function had been disrupted by PPMP. None of the major protein players in cytokinesis we tested (for example, tubulin, myosin II, MKLP-1, MLCK, Aurora B, Plk1, ROCK, data not shown) showed aberrant localization patterns at cytokinetic structures. We also tested proteins that are thought to play a role in linking the plasma membrane to the actin cortex during cytokinesis, including Anillin, Septin7 and ERM (ezrin, radixin, moesin) proteins.19,21 All of these proteins localize to the cleavage furrow during cytokinesis, and their precise roles are still being investigated. While Anillin and Septin7 localized normally, we found that ERM and actin localization were perturbed in PPMP treated cells.
The ERM proteins are important structural proteins that participate in many cellular events. They are involved in stabilizing the actin cortex during mitotic rounding and have been implicated in cytokinesis, although their function is unclear.22–25 ERMs are activated upon binding to the lipid PIP2, and a recent report also suggested a role for ceramides in ERM function.19,26 In dividing PPMP-treated cells the ERMs and actin mostly localize to the cleavage furrow normally, although there appears to be a modest increase in accumulation of ERMs at the furrow. In untreated cells, the average accumulation of ERMs is ~1.3 fold higher at the cleavage furrow than the polar cortex, whereas in PPMP-treated cells it is ~1.54 fold higher (n=15, see Supporting Figure 3 for details). More broadly, in PPMP-treated cells, as well as with GCS RNAi, we found that small cellular protrusions that contain ERM proteins and actin are abolished in most cells (85%, n=33), including dividing cells (Figure 3).
Figure 3.
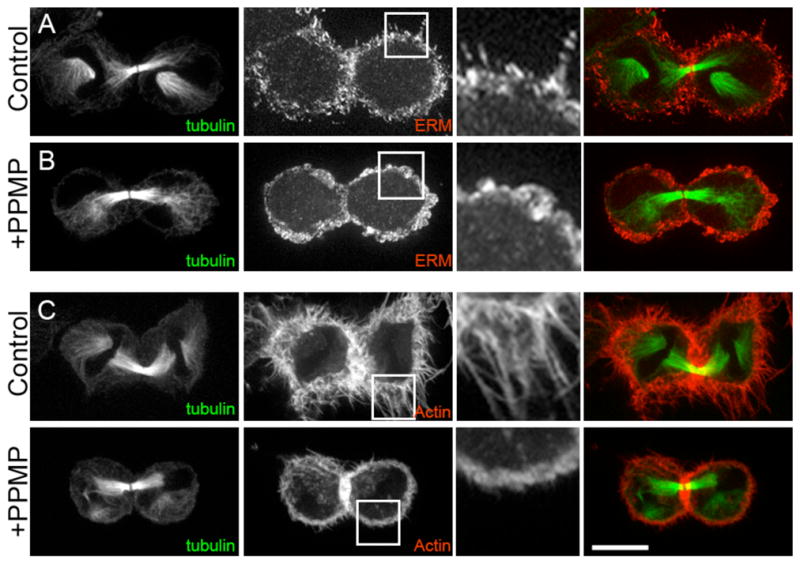
PPMP treatment changes actin and ERM localization and abolishes small cellular protrusions. A–B Control and PPMP-treated cells were stained for microtubules (anti-tubulin) and ERM (anti-ERM). C–D Control and PPMP-treated cells were stained for microtubules (anti-tubulin) and actin (phalloidin). To illustrate changes in cortical protrusions more clearly, magnified images are shown next to the grey scale ERM and actin images (Scale bar = 10 μm). Representative micrograph is shown for n= 28/33.
It is becoming increasingly clear that ring ingression during cytokinesis is much more complex than simple constriction at the furrow.27–29 It is now thought that the entire cell cortex participates, with possible mechanisms ranging from easing in cortical tensions at the cells’ poles to facilitate ingression at the furrow, to localized stiffening in the furrow region. The actin and ERM staining patterns clearly show changes in cortical topology upon PPMP treatment, and we therefore hypothesize that cytokinesis failure is due to the inability of the cortex to support deep ingression of the cleavage furrow. This hypothesis is further supported by the observation that PPMP-treated cells begin to spread inappropriately while still undergoing cytokinesis (Figure 4). Usually, cultured cells grown in monolayers round up during mitosis, divide, and then reattach to the substrate as they are completing cytokinesis. We found that ~13% of cells treated with PPMP (vs. 1% of control cells, n=30) begin to reattach at the polar regions without having completed division, suggesting that cortical regulation has been disrupted.
Figure 4.
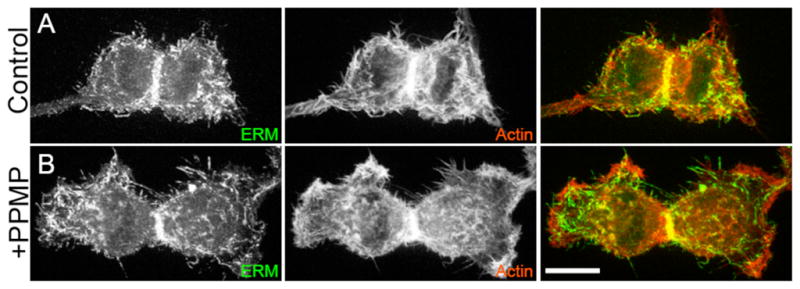
PPMP treatment causes immature spreading at the polar regions during cytokinesis, i.e. cells start to attach before completing division. Control (A) and PPMP-treated cells are (B) stained for ERM (anti-ERM) and actin (phalloidin) (Scale bar = 10 μm). Representative micrograph is shown for n=5/13.
In summary, our data provide evidence linking ceramides to cytokinesis, and add a new class of lipids to the growing list of membrane components that play a role in cell division. We show that an imbalance between ceramides and glucosylated ceramides can trigger cortical changes, possibly mediated by interactions between ceramides and ERM proteins, which can in turn cause changes in the actin cytoskeleton. Using unbiased global lipid profiling followed by a targeted analysis, we identify the specific nature of the lipids involved in this process, which allows us to link specific lipids to specific biological function. Understanding the detailed mechanism by which ceramides affect the actin cytoskeleton and ERM is a priority. While understanding the biological roles of lipids still lags behind proteins and nucleic acids, we expect that comprehensive approaches such as ours will allow us to slowly close this gap.
Supplementary Material
Acknowledgments
We thank Alan Saghatelian, Tejia Zhang, Emma Doud and members of the Eggert lab, especially Xin Zhang, for helpful discussions. LC/MS data was acquired on an Agilent 6520 Q-TOF spectrometer supported by the Taplin Funds for Discovery Program (PI, Suzanne Walker). We thank the Nikon Imaging Center at Harvard Medical School for help with light microscopy. This work was funded by the Heinrich Hertz Foundation (to S.S.), a Human Frontier Science Program Young Investigator Award (Aurélien Roux, Kaori Sakurai, co-PIs), NIH grant R01 GM082834 and the Dana-Farber Cancer Institute.
Footnotes
Supporting Information Available: A list of different inhibitors tested, experimental details for live- and fixed-cell imaging and mass spectrometry analyses as well as the identification ceramides are included in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Eggert US, Mitchison TJ, Field CM. Annu Rev Biochem. 2006;75:543. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 2.Rappaport R. Cytokinesis in Animal Cells. Cambridge University Press; Cambridge: 1996. [Google Scholar]
- 3.Mullins JM, Biesele JJ. J Cell Biol. 1977;73:672. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atilla-Gokcumen GE, Castoreno AB, Sasse S, Eggert US. ACS Chem Biol. 2010;5:79. doi: 10.1021/cb900256m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emoto K, Inadome H, Kanaho Y, Narumiya S, Umeda M. J Biol Chem. 2005;280:37901. doi: 10.1074/jbc.M504282200. [DOI] [PubMed] [Google Scholar]
- 6.Field SJ, Madson N, Kerr ML, Galbraith KA, Kennedy CE, Tahiliani M, Wilkins A, Cantley LC. Curr Biol. 2005;15:1407. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 7.Emoto K, Umeda M. J Cell Biol. 2000;149:1215. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng MM, Chang F, Burgess DR. Dev Cell. 2005;9:781. doi: 10.1016/j.devcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Biochemistry. 2004;43:14332. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama K, Suzuki M, Kawashima I, Karasawa K, Nojima S, Enomoto T, Tai T, Suzuki A, Setaka M. Eur J Biochem. 1997;249:450. doi: 10.1111/j.1432-1033.1997.00450.x. [DOI] [PubMed] [Google Scholar]
- 11.Abe A, Inokuchi J, Jimbo M, Shimeno H, Nagamatsu A, Shayman JA, Shukla GS, Radin NS. J Biochem. 1992;111:191. doi: 10.1093/oxfordjournals.jbchem.a123736. [DOI] [PubMed] [Google Scholar]
- 12.Sonda S, Stefanic S, Hehl AB. Antimicrob Agents Chemother. 2008;52:563. doi: 10.1128/AAC.01105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wennekes T, van den Berg RJ, Boot RG, van der Marel GA, Overkleeft HS, Aerts JM. Angew Chem Int Ed Engl. 2009;48:8848. doi: 10.1002/anie.200902620. [DOI] [PubMed] [Google Scholar]
- 14.Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, Obeid LM. J Lipid Res. 2010;52:68. doi: 10.1194/jlr.M009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saghatelian A, McKinney MK, Bandell M, Patapoutian A, Cravatt BF. Biochemistry. 2006;45:9007. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]
- 16.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. Anal Chem. 2006;78:779. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 17.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. EMBO J. 2005;24:1537. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham TR, Kozlov MM. Curr Opin Cell Biol. 2010;22:430. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logan MR, Mandato CA. Biol Cell. 2006;98:377. doi: 10.1042/BC20050081. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka-Takiguchi Y, Kinoshita M, Takiguchi K. Curr Biol. 2009 doi: 10.1016/j.cub.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Echard A. Biochem Soc Trans. 2008;36:395. doi: 10.1042/BST0360395. [DOI] [PubMed] [Google Scholar]
- 22.Kunda P, Pelling AE, Liu T, Baum B. Curr Biol. 2008;18:91. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- 23.Canals D, Jenkins RW, Roddy P, Hernandez-Corbacho MJ, Obeid LM, Hannun YA. J Biol Chem. 2010;285:32476. doi: 10.1074/jbc.M110.141028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F. J Cell Biol. 2008;180:739. doi: 10.1083/jcb.200709161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama T, Goto H, Izawa I, Mizutani H, Inagaki M. Genes Cells. 2005;10:127. doi: 10.1111/j.1365-2443.2005.00824.x. [DOI] [PubMed] [Google Scholar]
- 26.Zeidan YH, Jenkins RW, Hannun YA. J Cell Biol. 2008;181:335. doi: 10.1083/jcb.200705060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YL. Trends Cell Biol. 2005;15:581. doi: 10.1016/j.tcb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 28.O’Connell CB, Warner AK, Wang Y. Curr Biol. 2001;11:702. doi: 10.1016/s0960-9822(01)00181-6. [DOI] [PubMed] [Google Scholar]
- 29.Guha M, Zhou M, Wang YL. Curr Biol. 2005;15:732. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.