Abstract
Innate immune cells respond to invading microbes upon detection of pathogen-associated molecular patterns (PAMPS). PAMP-recognition machinery is evolutionarily conserved, allowing for characterization in model organisms. The model organism Dictyostelium discoideum can exist as single-celled amoebae, which phagocytize bacteria for nutrients. Although D. discoideum is used extensively to study phagocytosis, it has not been determined if D. discoideum detects bacterial PAMPs using pattern-recognition machinery. Here we show that D. discoideum mounts responses against the bacterial cell wall PAMP, lipopolysaccharide (LPS). Upon treatment with LPS or its active component Lipid A, D. discoideum cells more efficiently clear phagocytized bacteria. LPS-enhanced bactericidal activity appears dependent both on MAPK signaling pathways as well as on the D. discoideum toll/interleukin-1 receptor domain-containing protein, TirA. These findings indicate that pattern-recognition machinery required to detect and respond to bacterial PAMPs may be conserved in D. discoideum.
Keywords: Dictyostelium discoideum, innate immunity, PAMP, lipopolysaccharide, MAPK, TirA, phagocytosis, pattern recognition, bacteria
Introduction
Innate immune cells detect invading pathogens by recognizing highly-conserved pathogen-associated molecular patterns (PAMPs) expressed by bacteria, viruses and fungi (Akira et al., 2006; Janeway, Jr. and Medzhitov, 2002). These PAMPs include important structural features of the pathogens that are absent on host cells. Among the most well-known PAMPs is the gram-negative bacterial cell wall component lipopolysaccharide (LPS). LPS, which is used by gram-negative bacteria for protection against detergents and antibiotics (Munford and Varley, 2006), has long been recognized as a potent stimulant of mammalian immune responses. The mechanisms by which the mammalian immune systems recognize LPS and the role that pattern-recognition receptors (PRRs) play in this recognition only recently have been appreciated (Poltorak et al., 1998).
Recent evidence has indicated that a group of evolutionarily conserved PRRs is used by innate immune systems to detect microbial PAMPs, including LPS (Akira et al., 2006; Janeway, Jr. and Medzhitov, 2002). The best characterized family of PRRs includes the mammalian toll-like receptors (TLRs) (Akira et al., 2006; Janeway, Jr. and Medzhitov, 2002). Upon recognition of PAMPs, the TLRs signal for increased innate immune responses. These signaling events result in transcription of immunologically important cytokines and antimicrobial peptides, production of reactive oxygen species and enhancement of phagocytic responses (Brikos and O’Neill, 2008). Each of these effects, in turn, aids in the elimination of invading pathogens from the host.
The first mammalian TLRs were discovered based on homology to Drosophila toll (Medzhitov et al., 1997), and researchers have since found that pattern-recognition receptors are remarkably conserved throughout evolution. Receptors homologous to the mammalian TLRs and a family of cytosolic PRRs, the nucleotide binding and oligomerization domain-like receptors (Nod-like receptors or NLRs), for example, have been characterized in organisms as diverse as horseshoe crabs, sponges and plants (DeYoung and Innes, 2006; Kurata et al., 2006; Leulier and Lemaitre, 2008; Rosenstiel et al., 2008). For this reason, simple model organisms often are used to identify and characterize new pattern-recognition receptors (Kurata et al., 2006; Lemaitre et al., 1996; Rosenstiel et al., 2008).
The soil amoeba Dictyostelium discoideum is a unique model organism that exists for part of its life-cycle as unicellular amoebae, but is induced to form a multicellular structure upon starvation (Newell, 1971). Living within the soil, D. discoideum encounters various types of soil bacteria (Cosson and Soldati, 2008). D. discoideum amoeboid cells are able to phagocytize several classes of bacteria for nutritional purposes (Clarke and Maddera, 2006). There do exist, however, some types of bacteria that can evade phagocytosis and infect D. discoideum (Cosson and Soldati, 2008; Skriwan et al., 2002). To survive infection, D. discoideum has evolved various mechanisms, including the development of specialized sentinel cells that protect the multicellular body and expression of gene products that have homology to those known in other organisms to be involved in bacterial killing (Benghezal et al., 2006; Chen et al., 2007; Eichinger et al., 2005; Sillo et al., 2008). Among the gene products whose expression in sentinel cells are regulated are some with homology to known PRRs, including the D. discoideum toll/interleukin-1 receptor domain-containing protein TirA, which has homology to mammalian TLRs and appears required for survival of D. discoideum in the presence of bacteria (Chen et al., 2007; Sillo et al., 2008). It has not been appreciated, however, to what extent D. discoideum amoeboid cells use PRRs to detect bacteria for nutritional or defense purposes.
If D. discoideum indeed uses PRRs to detect microbial patterns on bacteria, then we would anticipate that D. discoideum cells would respond to treatment with known PAMPs, such as LPS. Here we show that treatment of D. discoideum amoeboid cells with LPS, and with its active lipid component, Lipid A, enhances their ability to clear phagocytized bacteria. These responses are dependent on the MAPK ErkB, indicating that LPS recognition by D. discoideum is at least partly mediated by MAPK signaling pathways. In addition, LPS-dependent enhanced bactericidal activity of D. discoideum cells appears dependent on TirA, suggesting that elements of pattern recognition pathways may be conserved in D. discoideum cells.
Material and methods
Cell culture
D. discoideum AX2 cells were received from T. Jin (NIAID, NIH, Rockville, MD), TirA-deficient (TirA−, Chen et al., 2007) cells were received from A. Kuspa (Baylor College of Medicine, Houston, TX). ErkB-deficient (ErkB−) and Nramp-deficient (Nramp−) cells, originally created as null strain by homologous recombination (Segall et al., 1995), were received from the Dicty Stock Center. Cells were grown axenically at 22°C in HL-5 media supplemented with vitamins (400 μg/L biotin, 100 μg/L cyanocobalamin, 4 mg/L folic acid, 8 mg/L lipoic acid, 10 mg/L riboflavin and 1 mg/L thiamine).
Bacterial cultures of Klebsiella pneumoniae, Staphylococcus aureus, Enterococcus faecalis, Bacillus subtilis and Mycobacterium smegmatis were obtained from Carolina Biological Supply Co (Burlington, NC). Salmonella typhimurium (ATCC 29631) and Escherichia coli HB101 were obtained from the American Type Culture Collection (Manassas, VA).
Bacterial Clearance and Phagocytosis Assays
Overnight bacterial cultures were harvested and labeled with STYO-9® (Invitrogen, Carlsbad, CA). The labeled bacterial cells were washed, resuspended in PBS and a 22.5 gauge needle was used to break up bacterial clumps.
D. discoideum cells were harvested from log-phase shaking culture, washed and resuspended in shaking culture at 2 × 106 cells/mL in HL-5 media without streptomycin. E. coli O55:B5 LPS (1μg/mL, Sigma, St. Louis, MO, L2880), ultra pure E. coli 055:B5 LPS (1μg/mL, List Biological Laboratories, Campbell, CA), or Kdo2-Lipid A from E. coli (1μg/mL, Enzo Life Sciences, Plymouth Meeting, MA) was added and the cells were incubated at 22°C for 1 h. Fluorescently-labeled bacteria (prepared as described above) were added to cells at a ratio of 75:1 and the samples were incubated at 22°C in the dark in shaking culture for 45 min to allow D. discoideum cells to take up the bacteria by phagocytosis. D. discoideum cells were then centrifuged at 150 × g to remove excess bacteria, and cells were resuspended in HL-5. To assay the clearance of fluorescently-labeled phagocytized bacteria from the cells, samples were collected at time points between 0 and 6 h. Harvested samples were washed three times by centrifugation at 150 × g with PBS and resuspended in PBS containing 1% paraformaldehyde. Samples were analyzed by flow cytometry, measuring the mean fluorescence of Syto9-labeled bacteria inside D. discoideum cells.
For phagocytosis assays, AX2 cells as prepared above and treated with or without 1 μg/mL LPS for 1 h were incubated with Alexa-fluor 488-labeled gram-negative E. coli and gram-positive S. aureus particles (Invitrogen, Carlsbad, CA) at a ratio of 75 particles per cell. After 45 min at 22°C, cells were harvested by layering 1 × 106 cells over a cushion of polyethylene glycol 3500 and centrifuging to separate D. discoideum cells from non-internalized bacterial particles (Vogel et al., 1980). The levels of phagocytosis of the fluorescent bacterial particles were measured using flow cytometry.
Bacterial Intracellular Survival Assay
AX2 cells were harvested at log phase from shaking suspension and 3 × 105 cells were plated in 24-well plates with HL-5 media without glucose or streptomycin. Cells were treated with or without 1 μg/mL LPS for 1 h at RT and mixed at a 1:10 ratio with live S. aureus harvested from overnight cultures. After 20 min, cells were washed of non-internalized bacteria and treated with 5 μg/mL streptomycin to kill any bacteria remaining outside the cells.
The viability of S. aureus inside D. discoideum phagosomes was assessed between 20 and 200 min after phagocytosis by lysing cells with 0.1% Triton X-100 in PBS and plating released bacteria on nutrient agar. Viable colonies were counted 24 h after incubation at 37° C. Percent survival of phagocytized bacteria was assessed by comparing the number of viable bacterial colonies at each time point with the total amount of bacteria phagocytized, as determined by subtracting the number of bacteria washed from the cells after the initial 20 min incubation time from the total number of bacteria originally added to the cultures (Hampton et al., 1994).
Results
We have hypothesized that D. discoideum amoeboid cells use PRRs to detect microbial patterns displayed by their bacterial prey. To test this hypothesis, we determined whether incubation of D. discoideum amoeboid cells with the microbial pattern LPS or its lipid component Lipid A altered the response of D. discoideum to bacterial cells.
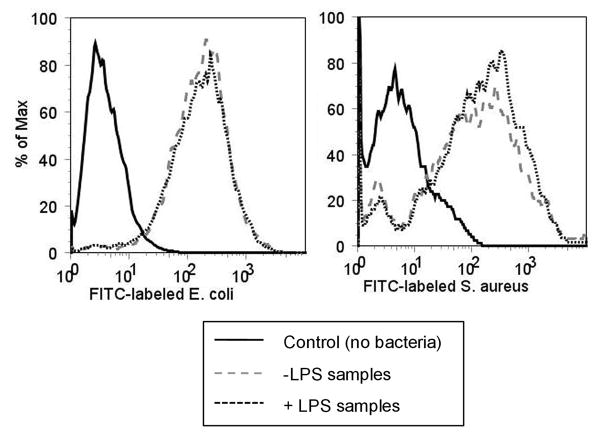
First, the levels of phagocytosis of gram-negative and gram-positive bacterial particles were measured in cells treated with or without LPS. D. discoideum AX2 cells were incubated with fluorescently-labeled E. coli and S. aureus bacterial particles for 45 min and the levels of phagocytosis were measured using flow cytometry. As shown in Fig 1, the levels of phagocytosis of E. coli and S. aureus particles were unaffected by treatment of cells with LPS.
Fig. 1.
D. discoideum phagocytosis of bacterial particles is unaffected by LPS treatment. AX2 cells were treated with 1 μg/mL LPS and mixed with Alexa-fluor 488-labeled S. aureus or E. coli bacterial particles for 45 min. Cells were washed and the levels of internalization of the fluorescently-labeled bacteria were measured by flow cytometry. Shown is a histogram that is representative of the data collected in at least three independent experiments.
Although LPS treatment did not affect the initial internalization of bacterial particles, it was possible that LPS might alter the clearance of phagocytized bacteria once they were internalized to degradative compartments. To determine the levels of bacterial clearance in the presence and absence of LPS, D. discoideum AX2 cells were incubated with or without LPS or Lipid A along with live fluorescently-labeled bacteria for 45 min to allow for phagocytosis. The cells were washed to eliminate non-internalized bacteria and then incubated 0–360 min further to allow for clearance of the bacteria, as measured by flow cytometric monitoring of the loss of fluorescence from the cells.
As shown in Fig. 2A, D. discoideum cells treated with LPS cleared significantly more fluorescently-labeled S. aureus between 45–90 min than did untreated cells. While nearly 90% of the fluorescently-labeled bacteria remained associated with untreated cells at 45 min after phagocytosis and 77% remained associated at 90 min, in cells treated with LPS, only 73% of the bacteria remained at 45 min following phagocytosis and 64% after 90 min. Since commercial preparations of LPS vary in their composition and purity, we repeated our observations using a highly purified preparation of LPS (ultra pure LPS) as well as with the purified Lipid A moiety from E. coli LPS (Kdo2-LipidA). We found that clearance of fluorescently-labeled S. aureus was enhanced both in cells treated with ultra pure LPS as well as in cells treated with Lipid A (Fig 2B).
Fig. 2.
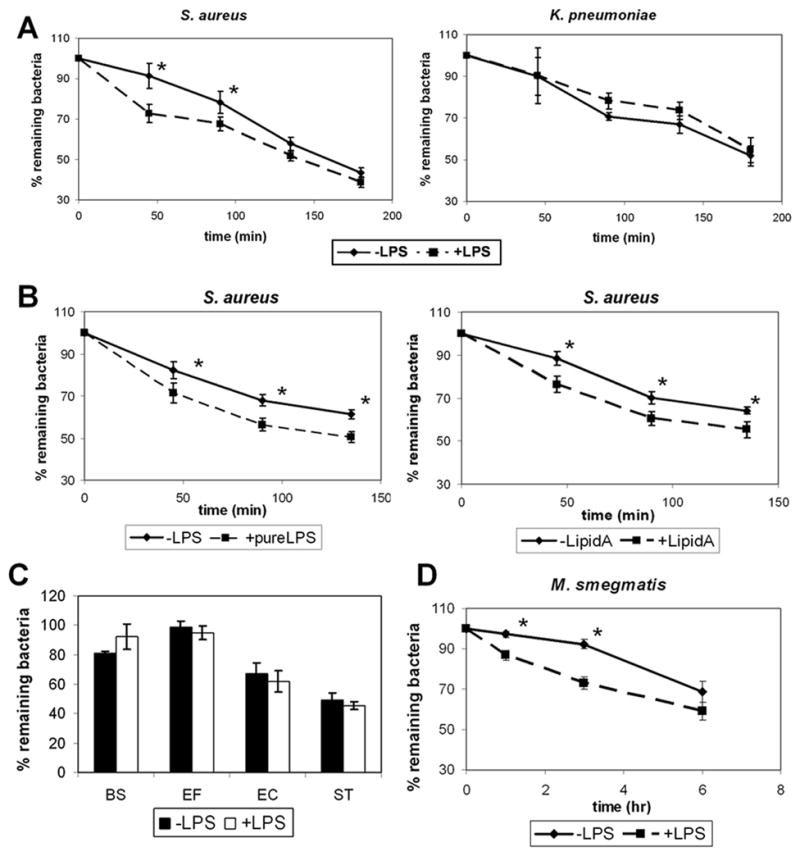
LPS treatment promotes D. discoideum bactericidal activity against specific bacterial species. a. D. discoideum AX2 cells were treated with 1 μg/mL LPS and mixed with Syto9-fluorescently labeled S. aureus or K. pneumoniae for 45 min. Cells were washed and the percent bacteria remaining inside cells between 0 and 180 min after phagocytosis was determined by flow cytometry, measuring mean fluorescence in cells. b. The clearance of Syto9-fluorescently labeled S. aureus was measured in cells treated or not with 1 μg/mL ultra pure LPS or 1 μg/mL Lipid A. c. The clearance of Syto9-fluorescently labeled B. subtilis (BS), E. faecalis (EF), E. coli (EC) or S. typhimurium (ST) was measured in cells treated or not with 1 μg/mL ultra-pure LPS. d. The clearance of Syto9-fluorescently labeled M. smegmatis between 0 and 6 h following phagocytosis was measured in cells treated or not with 1 μg/mL LPS. Shown are means and SEM from at least three independent experiments for each figure. * refers to statistically significant differences (p<0.05, t-test) between LPS-treated and untreated cells.
Interestingly, clearance of fluorescently-labeled K. pneumoniae was not significantly enhanced by treatment with LPS (Fig. 2A). We performed experiments with additional strains of both gram-negative and gram-positive bacteria. We found that, like with K. pneumoniae, clearance of the gram-positive species, B. subtilis and E. faecalis and of the gram-negative species, E. coli and S. typhimurium, was not affected by treatment with LPS (Fig. 2C).
Clearance of the mycobacterial species, M. smegmatis, however, was enhanced in cells treated with LPS (2D). Fluorescently-labeled M. smegmatis were cleared more slowly from D. discoideum amoeboid cells than were the other bacterial cell types tested (Fig 2A–D). No observable decrease in fluorescently-labeled internal M. smegmatis was seen in cells untreated with LPS until 6 h following phagocytosis. On the other hand, only 87% of phagocytized fluorescently-labeled bacteria remained associated with D. discoideum cells treated with LPS 1 h following phagocytosis and only 73% remained 3 h after phagocytosis (Fig 2D). Thus, treatment with LPS significantly enhanced the clearance of M. smegmatis from D. discoideum 1–3 h following phagocytosis. By 6 h following phagocytosis, there was no significant difference found between the levels of fluorescently-labeled bacteria remaining associated with untreated vs. treated cells.
In order to verify that enhanced clearance of fluorescently-labeled S. aureus upon treatment with LPS represented enhanced killing of phagocytized S. aureus, we performed an intracellular survival assay to monitor the survival of bacteria that had been phagocytized by D. discoideum cells. D. discoideum cells were untreated or treated with LPS and incubated with live S. aureus. After an initial incubation time of 20 min, the S. aureus that had not been phagocytized were washed from the cells and the cells treated with streptomycin to kill extracellular bacteria that remained after washing. At various time-points following the wash, the D. discoideum cells were lysed to release phagocytized bacteria and the survival of these bacteria was assessed by plating on nutrient agar. As shown in Fig. 3, treatment with LPS significantly enhanced the killing of phagocytized S. aureus between 20 and 40 min. after phagocytosis. While approximately 17% of phagocytized bacteria remained viable 20 min after phagocytosis in untreated cells, only 10% survived in cells treated with LPS 20 min after phagocytosis. After 40 min, almost 8% of phagocytized bacteria remained viable in untreated cells, while only 5% survived in cells treated with LPS. By 60 min after phagocytosis only a small percentage of bacteria remained viable in both untreated and treated cells. These results indicating that killing of phagocytized S. aureus is increased in cells treated with LPS are consistent with our results from the flow cytometric clearance assay.
Fig. 3.
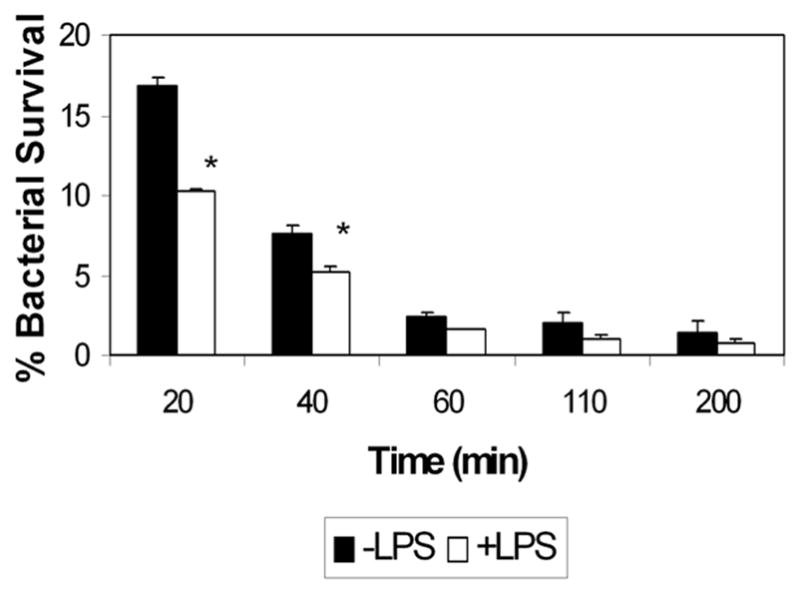
S. aureus is killed more rapidly in cells treated with LPS. AX2 cells were treated with 1 μg/mL LPS and mixed with live S. aureus. At various time points after phagocytosis, cells were lysed and released bacteria were plated on nutrient agar to test for viability. Shown are means and SEM from at least three independent trials for each experiment. * refers to statistically significant differences (p<0.05) between LPS-treated and untreated cells as measured by the Student’s t-test.
Pattern-recognition receptors in various organisms commonly use NF-κB, caspase-dependent, and MAPK pathways for signaling (Akira et al., 2006; Brikos and O’Neill, 2008; Janeway, Jr. and Medzhitov, 2002). Although the D. discoideum genome does not contain NF-κB or caspases (Eichinger et al., 2005), MAPK signaling pathways are used by D. discoideum for various cellular processes. Among the best characterized MAPK molecules in D. discoideum is the extracellular signal-regulated kinase, ErkB (Segall et al., 1995). We used the flow cytometric clearance assay to test the effect of ErkB deficiency on the LPS-induced clearance of phagocytized bacteria from D. discoideum amoeboid cells. As shown in Fig. 4A, treatment with LPS did not affect clearance of gram-positive S. aureus from ErkB-deficient cells. This is in contrast to the results seen in wild-type AX2 cells in which LPS treatment significantly enhanced clearance of fluorescently-labeled S. aureus (Fig 2A). As expected, LPS treatment, which did not affect clearance of phagocytized gram-negative K. pneumoniae from wild-type AX2 cells (Fig 2A), also did not alter the clearance of K. pneumoniae in ErkB-deficient cells (Fig 4B).
Fig. 4.
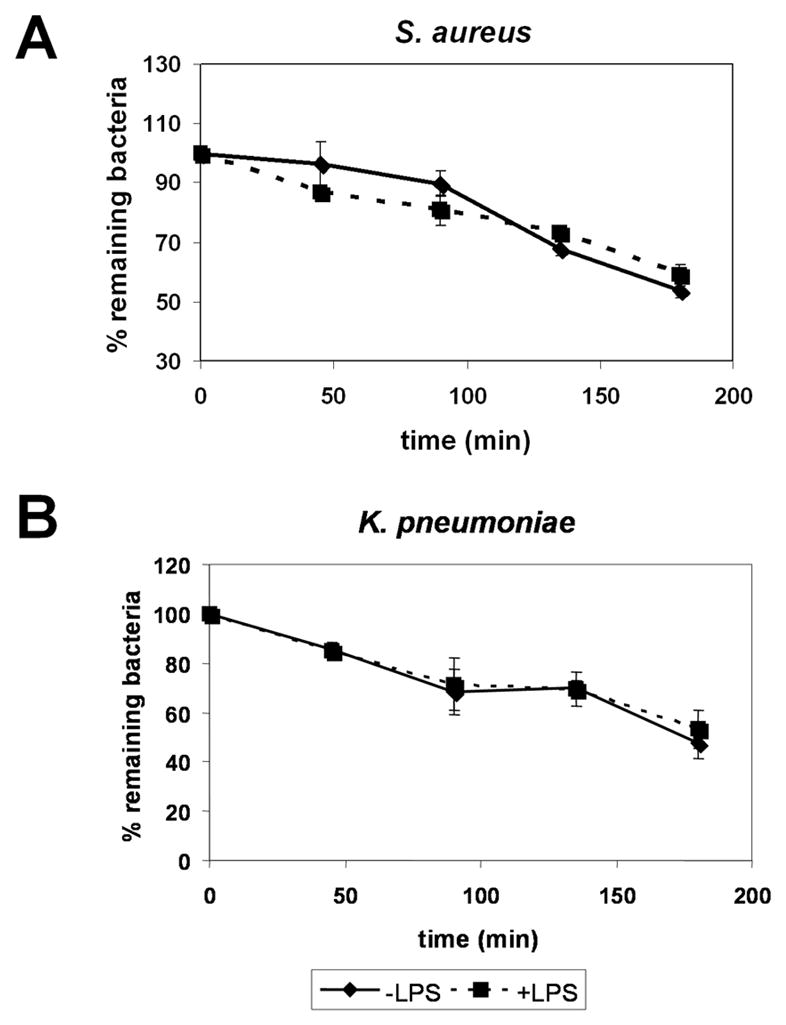
LPS-enhanced clearance of phagocytized bacteria is dependent on the MAPK ErkB. D. discoideum ErkB− cells were treated with 1 μg/mL LPS and mixed with Syto9-labeled S. aureus (a) or K. pneumoniae (b) for 45 min. Cells were washed and the levels of fluorescently-labeled bacteria remaining in cells 0 to 180 min. after phagocytosis were determined by flow cytometry, measuring the mean fluorescence of cells. Shown are means and SEM from at least three independent experiments.
To determine if LPS-enhanced bactericidal activity in D. discoideum is mediated through Tir domains, which play important roles in initiating signal pathways downstream of PAMPs in mammalian innate immune cells, we used the flow cytometric clearance assay to analyze bactericidal activity in D. discoideum cells deficient for expression of TirA. We also looked at bactericidal activity in cells deficient for Nramp, a phagosomal protein that has been implicated in bactericidal activity in D. discoideum (Peracino et al., 2006). We found that, just as in wild-type AX2 cells, treatment with either LPS or Lipid A significantly enhanced bacterial clearance in cells deficient for Nramp (Fig. 5). In contrast, similar to results in ErkB-deficient cells, neither LPS nor Lipid A treatment enhanced bacterial clearance in TirA-deficient cells (Fig 5).
Fig. 5.
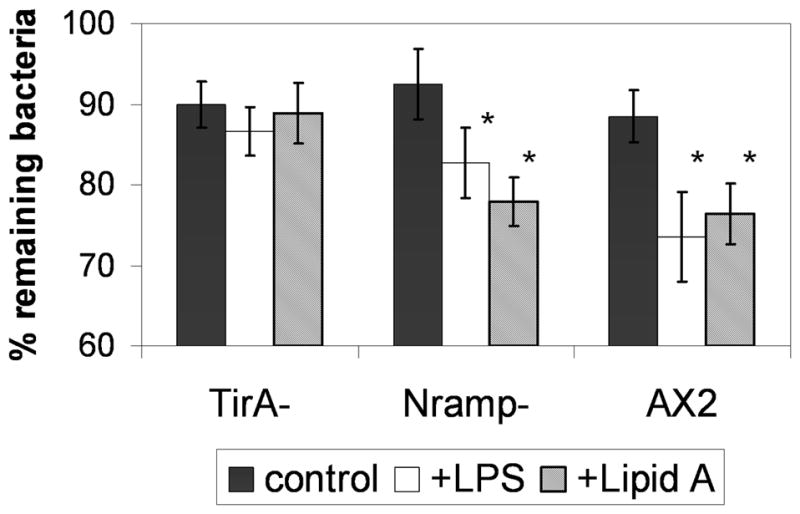
LPS-enhanced clearance of phagocytized bacteria appears dependent on TirA. The clearance of Syto9-fluorescently labeled S. aureus 45 min following phagocytosis in AX2, TirA-deficient and Nramp-deficient cells treated with or without 1 μg/mL ultra-pure LPS or Lipid A was measured as in Fig 2. Shown are means and SEM from at least three independent experiments. * refers to a statistically-significant difference (p<0.05, t-test) between LPS or Lipid A-treated and untreated cells.
Discussion
Innate immune systems of both vertebrates and invertebrates rely on PRRs to detect PAMPs present on microbial invaders, such as bacterial pathogens, but absent from host organisms (Akira et al., 2006; Janeway, Jr. and Medzhitov, 2002). D. discoideum amoeboid cells also must respond to bacteria for both nutritional and defense purposes (Cosson and Soldati, 2008). If D. discoideum cells use the same types of pattern-recognition machinery to detect bacteria as do innate immune cells, then we would expect that D. discoideum cells would mount a response to bacterial PAMPs. Here we have shown that D. discoideum amoeboid cells are able to respond to a known PAMP, the gram-negative bacterial cell wall component LPS, as well as to its lipid component Lipid A. D. discoideum cells treated with LPS or Lipid A cleared certain strains of phagocytized bacteria more efficiently than did untreated cells. Interestingly, the response of D. discoideum cells to LPS appears to be mediated through both TirA and MAPK ErkB. Taken together, these results suggest that there exists some type of conserved pattern-recognition mechanism specific for LPS in D. discoideum.
PRRs are highly conserved among both vertebrate and invertebrate species. Analysis of the completed D. discoideum genome sequence reveals the existence of putative D. discoideum genes homologous to known PRRs (Eichinger et al., 2005), including those that contain Tir domains and LRRs (Chen et al., 2007; Sillo et al., 2008). The expression of both D. discoideum TirA, as well as of the LRR-containing slrA, are enriched in D. discoideum sentinel cells (Chen et al., 2007) and upregulated upon infection of cells with Legionella pneumophila (Li et al., 2009),while TirC expression has been found to be upregulated upon phagocytosis of bacteria (Sillo et al., 2008). The most compelling evidence for a role for Tir domain-containing proteins in the detection of bacterial prey by D. discoideum comes from studies of amoeboid cells deficient for TirA. Although TirA-deficient cells grow normally in axenic media, the cells do not grow efficiently in the presence of bacteria, indicating that TirA is necessary for D. discoideum interactions with bacteria (Chen et al., 2007). Mammalian Tir domain-containing receptors and cytoplasmic adaptors are important mediators of pattern recognition responses to microbial PAMPs (Akira et al., 2006; Janeway, Jr. and Medzhitov, 2002). Here we have shown that LPS and Lipid A fail to enhance bactericidal activity in cells deficient for TirA. Thus, just as Tir domains play important roles in PAMP recognition in mammals, D. discoideum Tir domain-containing proteins, such as TirA, may mediate pattern-recognition responses to bacteria in D. discoideum.
Our results also begin to shed light on downstream signaling molecules that may be involved in LPS recognition. Signaling downstream of pattern recognition receptors commonly involve pathways including the NF-κB-mediated, caspase-dependent and MAPK signaling pathways (Brikos and O’Neill, 2008). Analysis of the D. discoideum genome reveals the absence of any NF-κB homologue (Eichinger et al., 2005). In addition, no caspases or metacaspases are encoded by the D. discoideum genome (Eichinger et al., 2005). On the other hand, the MAPK proteins ErkA and ErkB have been implicated in various growth, developmental, and defense functions in the D. discoideum life-cycle (Sawai et al., 2007; Segall et al., 1995). Interestingly, recent evidence indicates that the D. discoideum MAPK response is also involved in host responses to the pathogen L. pneumophila (Li et al., 2009). Using ErkB-deficient cells, we have shown here that the D. discoideum response to LPS is dependent on signaling through ErkB.
Our findings that the MAPK ErkB is involved in signaling downstream of LPS detection in D. discoideum are consistent with results from studies of LPS-mediated signaling in various organisms. Treatment of mammalian monocytes with LPS has been shown to activate Erk, JNK and p38 MAPK signaling pathways (Guha and Mackman, 2001), and moreover, LPS has been shown to upregulate expression of TLRs in mammalian macrophages and dendritic cells via Erk and p38 MAPK signaling pathways (An et al., 2002). LPS treatment of chicken heterophils (which are functionally equivalent to mammalian neutrophils) activates both Erk and p38 MAPK signaling pathways downstream of TLRs to induce the secretion of pro-inflammatory cytokines (Kogut et al., 2008). Finally, studies with mutant C. elegans have shown that both Erk and p38 MAPK signaling pathways mediate defense against pathogens in this nematode species. (Aballay et al., 2003; Kim et al., 2002; Nicholas and Hodgkin, 2004). Thus, our results implicating ErkB as an important player in mediating responses to LPS in D. discoideum cells give additional evidence for a conserved role for Erk in conferring resistance to bacterial pathogens downstream of pattern-recognition receptors in a variety of vertebrate and invertebrate species.
Our studies here suggest a role in D. discoideum for pattern-recognition machinery in detecting bacteria in the environment. D. discoideum amoeboid cells in the soil prey on bacterial species for nutritional purposes. If bacteria in the soil evolved mechanisms to evade amoeboid phagocytosis, they would enjoy distinct selective advantages in such an environment (Cosson and Soldati, 2008). In fact, it has been shown that various bacterial species use similar virulence mechanisms to evade phagocytic killing by D. discoideum as do bacterial pathogens in evading killing by mammalian macrophages and neutrophils (Cosson and Soldati, 2008; Skriwan et al., 2002). In order to compete with bacterial virulence mechanisms, D. discoideum may have evolved pattern-recognition mechanisms to increase efficiency of amoeboid phagocytosis by concentrating bacteria at the amoeboid cell surface (Clarke and Maddera, 2006) or by initiating downstream signaling pathways for enhanced phagocytic and bactericidal activity.
Although our results here indicate that D. discoideum cells treated with LPS and Lipid A are better able to clear phagocytized bacteria, it is not evident by what mechanisms potential pattern-recognition machinery in D. discoideum might modify the cells for more enhanced killing. One possible outcome of signaling downstream of LPS recognition might be a modification of phagosomal compartments. Further characterization of the phagosomsal-lysosomal pathway in LPS-treated D. discoideum cells may provide added insight into the cellular mechanisms induced upon LPS activation.
In conclusion, we have shown here that treatment of D. discoideum amoeboid cells with the known PAMPs LPS and Lipid A accelerates killing of phagocytized bacteria. These responses are dependent on Erk-mediated signaling, indicating a role for receptor-mediated processes in these responses. In addition LPS-enhanced killing of phagocytized bacteria appears dependent on TirA, pointing to a conserved role for Tir domain-mediated responses to PAMPs in D. discoideum. Thus, our results which reveal that D. discoideum cells are able to respond to the known PAMP LPS are consistent with our hypothesis that D. discoideum use pattern-recognition receptors to detect bacteria via bacterial PAMPS. Further investigation of putative pattern-recognition receptors and downstream signaling effectors encoded by the D. discoideum genome should reveal a greater understanding of the pattern-recognition machinery in D. discoideum and may also lend insight into pattern-recognition mechanisms used by innate immune cells in higher-level organisms.
Acknowledgments
We would like to thank Drs. Tian Jin and Susan K. Pierce for careful reading of the manuscript. AX2 cells were kindly provided by Dr. Tian Jin, TirA-deficient cells were kindly provided by Dr. Adam Kuspa, and ErkB-deficient and Nramp-deficient cells were obtained from the Dicty Stock Center. The project described was supported by Award Number R15AI085503 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. The project was also supported by grants to M. Snyder from the TU Faculty Development and Research Committee and to J. Leuschner, D. Samaroo and D. Cassilly from the TU Undergraduate Research Committee and The Jess and Mildred Fisher College of Science and Mathematics Undergraduate Research Committee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52. doi: 10.1016/s0960-9822(02)01396-9. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- An H, Yu Y, Zhang M, Xu H, Qi R, Yan X, Liu S, Wang W, Guo Z, Guo J, Qin Z, Cao X. Involvement of ERK, p38 and NF-kappaB signal transduction in regulation of TLR2, TLR4 and TLR9 gene expression induced by lipopolysaccharide in mouse dendritic cells. Immunology. 2002;106:38–45. doi: 10.1046/j.1365-2567.2002.01401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, Lardy B, Klein G, Sansonetti P, Charette SJ, Cosson P. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- Brikos C, O’Neill LA. Signalling of toll-like receptors. Handb Exp Pharmacol. 2008:21–50. doi: 10.1007/978-3-540-72167-3_2. [DOI] [PubMed] [Google Scholar]
- Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Maddera L. Phagocyte meets prey: uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur J Cell Biol. 2006;85:1001–1010. doi: 10.1016/j.ejcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van DN, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, James K, Quiles M, Madan BM, Saito T, Buchrieser C, Wardroper A, Felder M, Thangavelu M, Johnson D, Knights A, Loulseged H, Mungall K, Oliver K, Price C, Quail MA, Urushihara H, Hernandez J, Rabbinowitsch E, Steffen D, Sanders M, Ma J, Kohara Y, Sharp S, Simmonds M, Spiegler S, Tivey A, Sugano S, White B, Walker D, Woodward J, Winckler T, Tanaka Y, Shaulsky G, Schleicher M, Weinstock G, Rosenthal A, Cox EC, Chisholm RL, Gibbs R, Loomis WF, Platzer M, Kay RR, Williams J, Dear PH, Noegel AA, Barrell B, Kuspa A. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal. 2001;13:85–94. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- Hampton MB, Vissers MC, Winterbourn CC. A single assay for measuring the rates of phagocytosis and bacterial killing by neutrophils. J Leukoc Biol. 1994;55:147–152. doi: 10.1002/jlb.55.2.147. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kogut MH, Genovese KJ, He H, Kaiser P. Flagellin and lipopolysaccharide up-regulation of IL-6 and CXCLi2 gene expression in chicken heterophils is mediated by ERK1/2-dependent activation of AP-1 and NF-kappaB signaling pathways. Innate Immun. 2008;14:213–222. doi: 10.1177/1753425908094416. [DOI] [PubMed] [Google Scholar]
- Kurata S, Ariki S, Kawabata S. Recognition of pathogens and activation of immune responses in Drosophila and horseshoe crab innate immunity. Immunobiology. 2006;211:237–249. doi: 10.1016/j.imbio.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Leulier F, Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- Li Z, Dugan AS, Bloomfield G, Skelton J, Ivens A, Losick V, Isberg RR. The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA. Cell Host Microbe. 2009;6:253–267. doi: 10.1016/j.chom.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Munford RS, Varley AW. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2006;2:e67. doi: 10.1371/journal.ppat.0020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PC. The development of the cellular slime mould Dictyostelium discoideum: a model system for the study of cellular differentiation. Essays Biochem. 1971;7:87–126. [PubMed] [Google Scholar]
- Nicholas HR, Hodgkin J. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr Biol. 2004;14:1256–1261. doi: 10.1016/j.cub.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Peracino B, Wagner C, Balest A, Balbo A, Pergolizzi B, Noegel AA, Steinert M, Bozzaro S. Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic. 2006;7:22–38. doi: 10.1111/j.1600-0854.2005.00356.x. [DOI] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van HC, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Rosenstiel P, Jacobs G, Till A, Schreiber S. NOD-like receptors: ancient sentinels of the innate immune system. Cell Mol Life Sci. 2008;65:1361–1377. doi: 10.1007/s00018-008-7502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S, Guan XJ, Kuspa A, Cox EC. High-throughput analysis of spatio-temporal dynamics in Dictyostelium. Genome Biol. 2007;8:R144. doi: 10.1186/gb-2007-8-7-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JE, Kuspa A, Shaulsky G, Ecke M, Maeda M, Gaskins C, Firtel RA, Loomis WF. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1995;128:405–413. doi: 10.1083/jcb.128.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillo A, Bloomfield G, Balest A, Balbo A, Pergolizzi B, Peracino B, Skelton J, Ivens A, Bozzaro S. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics. 2008;9:291. doi: 10.1186/1471-2164-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriwan C, Fajardo M, Hagele S, Horn M, Wagner M, Michel R, Krohne G, Schleicher M, Hacker J, Steinert M. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol. 2002;291:615–624. doi: 10.1078/1438-4221-00177. [DOI] [PubMed] [Google Scholar]
- Vogel G, Thilo L, Schwarz H, Steinhart R. Mechanism of phagocytosis in Dictyostelium discoideum: phagocytosis is mediated by different recognition sites as disclosed by mutants with altered phagocytotic properties. J Cell Biol. 1980;86:456–465. doi: 10.1083/jcb.86.2.456. [DOI] [PMC free article] [PubMed] [Google Scholar]