Abstract
MicroRNAs have been shown to regulate gene expression both transcriptionally and translationally. Here, we examine evidence that various stresses regulate miRNAs which, in turn, regulate immune gene levels. Multiple studies are reviewed showing altered microRNA levels in normal cells under stress and in various disease states, including cancer. Unexpected was the finding that Dicer expression is altered by treatments with several agents, such as interferons and cortisone, employed in the treatment of immune disorders. Potential signal transduction pathways, including JAK/Stat, PI3K and PKR, that may regulate Dicer and microRNA levels in normal and stressed mammalian cells are discussed.
Keywords: MHC, Epigenetic, MicroRNA, Stress, Senescence, Immunity, Gene regulation
The existence of the human race depends on the ability of the semi-allogeneic fetus to persist during a normal pregnancy. Prominent among the proposed mechanisms is that normal trophoblasts ‘escape’ maternal immunity by epigenetically repressing the expression of immune factors such as MHC and costimulatory molecules [1–4]. A generally similar concept for the escape of tumors from host immunity has also been suggested and the potential mechanisms involved in evasion of immunity have been reviewed [5]. Histone deacetylase inhibitors (HDACi), such as trichostatin A (TSA) and valproic acid, which have broad HDAC specificity can directly alter the chromatin at immune genes and induce MHC class II, CD40, MICA, and MICB genes [6–8]. This induction of MHC and costimulatory molecules on tumors has been shown to elicit effective antigen presentation in vitro and durable immunity, mediated by both T and NK cells, in vivo [7–10].
In mammalian cells, histone H3 is phosphorylated following exposure to stresses that activate the MAPK and stress-activated protein kinases (SAPK). Both pathways lead to the downstream activation of MSK1 and MSK2 kinases which phosphorylate serine 10 and 28 of histone H3. These phosphorylations are associated with changes in chromatin accessibility. Histones that are phosphorylated at H3S10 are much more sensitive to lysine acetylation at H3K9 in response to the histone deacetylase inhibitor TSA [11]. This suggests that the various histone modifications may be synergistically coupled. Terminal kinases may also directly phosphorylate transcription factors and co-factors, and p38 MAPK can be recruited by transcription factors and co-factors to promoter complexes [11]. Such recruitment could, therefore, lead to direct phosphorylation of histones, transcription factors and co-factors, as well as enhancement of histone acetylation, methylation, and chromatin remodeling.
Our initial studies focused on the mechanism of suppression of the MHC class II locus in normal and cancer cells and recognized four distinct types of cellular responses to HDACi and IFN-γ [6, 8]. (1) Cells such as HeLa which produce high levels of class II in response to IFN-γ but low levels following treatment with HDACi. (2) Plasma cells which were not responsive to IFN-γ. In these cells, HDACi treatments led to the expression of MHC class II in the absence of CIITA. (3) Trophoblast cells, and some tumor cells, which do not respond to IFN-γ but expressed class II after HDACi treatment. Interestingly, TSA activation of class II expression in JAR, JEG-3, and several other trophoblast as well as many cancer cells, did not, or only minimally, enhance CIITA expression. This repression of CIITA is a result of epigenetic silencing, including DNA methylation and/or possibly miRNA inhibition as discussed below. (4) Several tumor cells, such as colon 26, showed a vigorous IFN-γ activation of MHC class II requiring CIITA and also TSA-activated class II in the absence of CIITA, i.e., both CIITA-dependent and CIITA-independent pathways.
During the course of these studies, we employed chromatin immunoprecipitation (ChIP) assays to systematically analyze chromatin modifications and protein-DNA interactions at the HLA-DRα promoter, structural gene, and the LCR [12]. These studies, together with data from the literature (reviewed in 5, 13), have led to a model for the participation of each modification during the various phases of MHC class II induction. Upon receiving an appropriate activation signal from IFN-γ, the MHC class II enhanceosome, composed of transcription factors, co-factors, and chromatin modifiers, is assembled on the promoter region as described [5, 13–15]. Enhanced histone acetylation at the LCR is an early event and results in a more “open” chromatin structure that allows further recruitment of the RNA pol II complex. As transcription is initiated, the RNA pol II complex is assembled and Set1 and Dot1 histone methyltransferases (HMTs) are recruited to the promoter and H3K4 and H3K79 are methylated. As the pol II complex proceeds downstream, the C-terminal domain (CTD) heptapeptide serine 5 residues are dephosphorylated, while the serine 2 residues are phosphorylated. The H3K4 HMT is released, while the H3K36 HMT (SET2) is recruited to the RNA pol II complex as the elongation process is continued. The H3K79 HMT remains bound to the RNA pol II complex during early elongation, but is discharged during the later stages of elongation. However, SET2 continues to travel with the pol II until it reaches the termination signal. The localization of each histone marker at its unique sites may serve as a checkpoint for monitoring the transcriptional events occurring during MHC class II gene expression. It is important to recognize that the immune genes of different tissues may respond differently to treatment with HDACi and IFN-γ. These variations may be related to cellular differences in the expression of the three cell type specific CIITA promoters: CIITA PI in macrophages/DCs, PIII in B cells, and PIV in T cells [13, 16, 17].
Cellular senescence is induced in non-transformed as well as cancer cells by various cell stresses, including DNA damage, ROS (reactive oxygen species), irradiation, hypoxia, and by the activation of oncogenes, such as Ras [18]. Ras proteins elevate intracellular H2O2 which is thought to be a significant factor in mediating senescence [19] and Ras-induced senescence is associated with activation of Raf/MEK/MAPK [20] and PI3K pathways [21]. Noteworthy are reports that the senescence pathway must be bypassed for carcinogenesis to occur [18, 22]. Therefore senescence, like apoptosis, is thought to represent a barrier to tumorigenesis [23]. A role for chromatin in the initiation and maintenance of the senescent state has been suggested [23–26] and HDACi have been shown to induce senescence [24]. HDACi can directly activate stress pathways, including MAPK, PI3K/TOR, and NF-κB [27, 28], and these pathways are often associated with induction of senescence.
An important development in our understanding of immune gene expression was the recent finding that agents that induce cellular stress genes, when administered prior to TSA or IFN-γ, substantially enhance expression of MHC class II and certain other costimulatory genes (CD40, MICA, MICB) [29]. The pretreatment of trophoblast, and certain cancer cells having the normal repressed phenotype, with PMA/IM or H2O2 restores a vigorous MHC response to IFN-γ in poor responder cells. Moreover, transfection of the Ras oncogene into human fibroblast lines (MRC-5 and IMR90) substantially enhances the subsequent expression of MHC genes by IFN-γ. It is important to note that stresses that activate the MAPK pathway, and enhance IFN-γ activation, also promote senescence. HDACis have also been shown to induce stress pathways involving MAPK, PI3K/TOR, and NF-κB [27, 28] which are known to be associated with senescence induction. In future studies, it will be important to determine if the immune genes of senescent cells, induced by chemotherapeutic agents, are more readily activated by HDACi and IFN-γ. Would it be advantageous to combine cancer chemotherapeutic agents that activate stress pathways and induce senescence with chromatin active agents? An interesting question is whether treatment with stress inducing agents, including current chemotherapeutic drugs, have enhancing effects on endogenous chromatin factors, such as CBP/p300 and PCAF which have HAT activity.
Several years ago, following the description of the microRNA system, we became interested in whether these agents, known to repress genes, were involved in the epigenetic silencing of immune genes in trophoblast and cancer cells. MicroRNAs (miRs) are small, highly conserved, 20–24 nt non-coding RNAs which function in developmental timing, cell differentiation, and development. Currently ~ 800 miRs have been identified and it is estimated that there may be more than 1500 miRs [30]. MiRs are generated in the nucleus by Drosha/Pasha ‘microprocessor’ mediated cleavage of long primary transcripts to ~ 70 nt stem-loop pre-miRs. The pre-miRs are transported to the cytoplasm by the Ran-CTP-dependent exportin-5 pathway and cleaved to the 22 nt miRs by the RNase III enzyme Dicer. The 22 nt miR duplex is unwound, one strand (passenger strand) is digested by Argonaute 2 (Ago 2) and a single strand is guided to its target by the RNA-induced silencing complex (RISC) containing four Argonaute proteins [31, 32]. The targets for the majority of mammalian miRs are located in the 3′ UTR of the messenger RNA (mRNA) and each miR binds, on average, ~ 200 different mRNA [33]. It is estimated that miRs target about 30% of the genome and therefore regulate on the order of 7–8,000 genes. In animals, miRs often repress primarily by inhibiting translational initiation but not uncommonly by degrading RNA or by a combination of both mechanisms. The role of miRs in cytoplasmic structures, called P-bodies, and their role in message stability has recently been reviewed [34]. MiRs are involved in heterochromatic function, although it is unclear whether they bind directly to DNA or to nascent nuclear transcripts to exert these effects [35, 36]. MicroRNAs may recruit HDACs to repressed sites [36] and thus transcriptional gene silencing by miRs in heterochromatin is potentially reversible by agents that remodel chromatin. Little is currently known of the role of miRs in the regulation of the heterochromatic-like foci, with H3K9 methylation and HP-1 binding, that are found in euchromatin [37].
Substantial evidence indicates a cell-type specific signature of miRs and that, in general, cells are enriched for mRNAs that do not have binding sites for the miRs expressed in the tissue [31, 38]. These mutually exclusive cell expression patterns, together with data on miR expression during developmental processes, have suggested that miRs regulate the fidelity of gene expression during differentiation and development rather than acting as acute switches from one state to another. For example, transfection of miR-124, which is preferentially expressed in brain, causes the miR profile in HeLa (cervical carcinoma cell line) to shift toward that of brain and one hundred or more mRNAs are downregulated. The downregulated genes are found in low levels in normal brain tissues [39]. There is also increasing data that alteration, either by transfection or knockdown, of a single miR can have dramatic effects on cellular function including immunity. One recent example is miR-155 which is encoded in the B cell integration cluster whose knockdown produces B cell defects and a failure of immunoglobulin switched plasma cells. The transcription factor PU.1 is a validated target of miR-155 [40]. Our ongoing studies explore the possibility that one miR, or more likely a restricted number of miRs, controls trophoblast differentiation.
Recent work has suggested that miRs may also be involved in acute processes that require dynamic responses to specific environmental changes such as amino acid depletion and osmotic stress [41, 42]. A recent report [43] demonstrates that miRs respond to bacterial components, such as LPS, which activate innate Toll-like receptors (TLRs). Activation of one miR (miR-146) was shown to be NF-κB-dependent supporting the view that miRs may be induced in acute stress responses including stimuli that induce an immune response. The involvement of miRs in immunity is also suggested by studies of Dicer knockdown mice showing defects in T and B cell differentiation [44] and in T-regulatory cell development [45]. As detailed in a recent publication [46], we provide evidence that miRs regulate ~ 285 immune genes and that specific miRs targeting CIITA are involved in the regulation of MHC class II by IFN-γ.
We previously characterized a negative regulatory element (NRE) upstream of the MHC class II (IAα) promoter and identified two transcriptional repressor proteins that bound the NRE [47]. In assessing repressive mechanisms, we noted that the NRE has an AU-rich composition and sites similar to those described in the AU-rich elements (AREs) classically located in the 3′ UTR of many genes which regulate message stability. The 5′ NRE sequence is, in fact, a class III ARE and similar AREs have been identified in the 5′ region of other genes [48]. Recent evidence indicates that miRs can also regulate gene sequences when miR target sites are inserted in 5′ regions [49]. We found that AREs are not present in the 3′ UTR of 21 MHC class I and II genes but CIITA has two 3′ UTR ARE sites. These findings prompted studies of ARE binding proteins and microRNAs, which may regulate message stability in trophoblast, cancer, and other cells. Thus the data reported by Asirvatham et al. [46] describes a new interface between AREs, miRs, and message stability in immune gene regulation.
MicroRNAs predicted to regulate immune genes
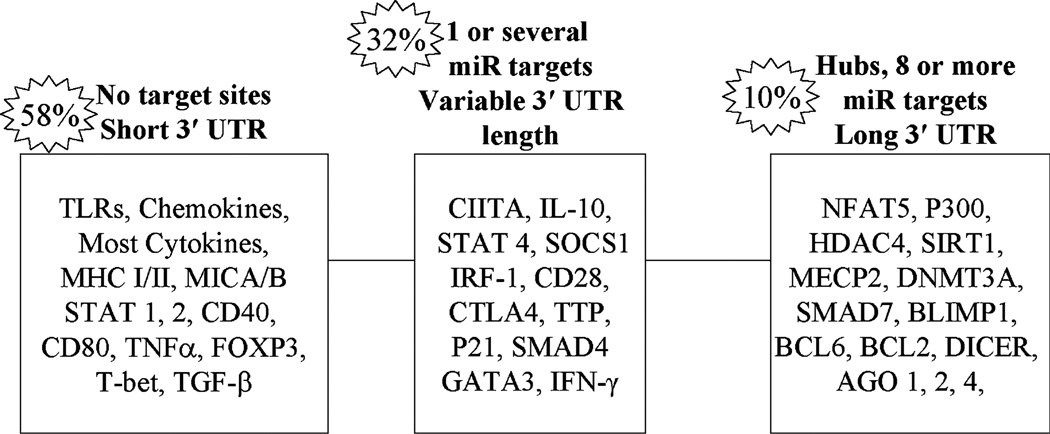
When we began this work, little was known of the role of miRs in immunity, we first initiated a broad-based analysis of miR targets in immune genes. The complete data set resulting from this analysis can be found in the supplemental data of a recent publication [46]. Using a stringent computational approach requiring the concordance of several algorithms, we determined the miRs that target 613 genes selected by data mining of immune pathways and gene ontology (GO) annotations. Analyses of 31 functionally annotated gene–miR interactions revealed a >90% predictive value of our computational approach. Statistical analysis of the miR targets demonstrated a highly significant selective targeting of immune genes by miRs relative to a genome-wide set of 20,232 genes (p < 0.001). Of the 613 genes, ~ 46% (285) were found to be potential targets of miRs. Of these, ~ 250 are immune genes not previously reported to be miR targets at the time of this publication. Selected examples of targets and non-targets are depicted in Fig. 1. This figure analyzes the distribution of genes in three categories depending on the number of miRs targeting each 3′ UTR. We define hubs as genes with multiple (≥8) different miR binding sites. The number of 3′ UTR binding sites is an important determinant of the level of inhibition and data obtained for the same miR binding to multiple UTR sites shows synergism [34, 50]. In addition, exponential enhancement of gene repression is noted when different miRs target different sites on the same UTR [51]. It should be emphasized that our data set does not encompass all immune genes and that the multiple algorithm approach [46], although highly predictive, may miss some functionally relevant miR–gene interactions. Therefore, we annotated all of the miRs which target the 613 genes employing single as well as multiple algorithms. We also defined the miRs that potentially target about two dozen immune pathways, such as the JAK/Stat, TGF-β, and others, as well as miRs that target general cellular components, such as transcription factors, cofactors, signaling, and chromatin factors, known to influence immune gene expression. For example, 17 of the 24 factors identified as participating in the TGF-β pathway were miR targets [46].
Fig. 1.
Computational analysis of selected immune gene 3′ UTRs for microRNA (miR) binding sites. Distribution of 613 miRs that target genes important in immunity were analyzed. 58% had no predicted targets, 32% one or a few targets and 10% appear to be hubs with multiple (8 or more) miR targets. Examples are provided in each category
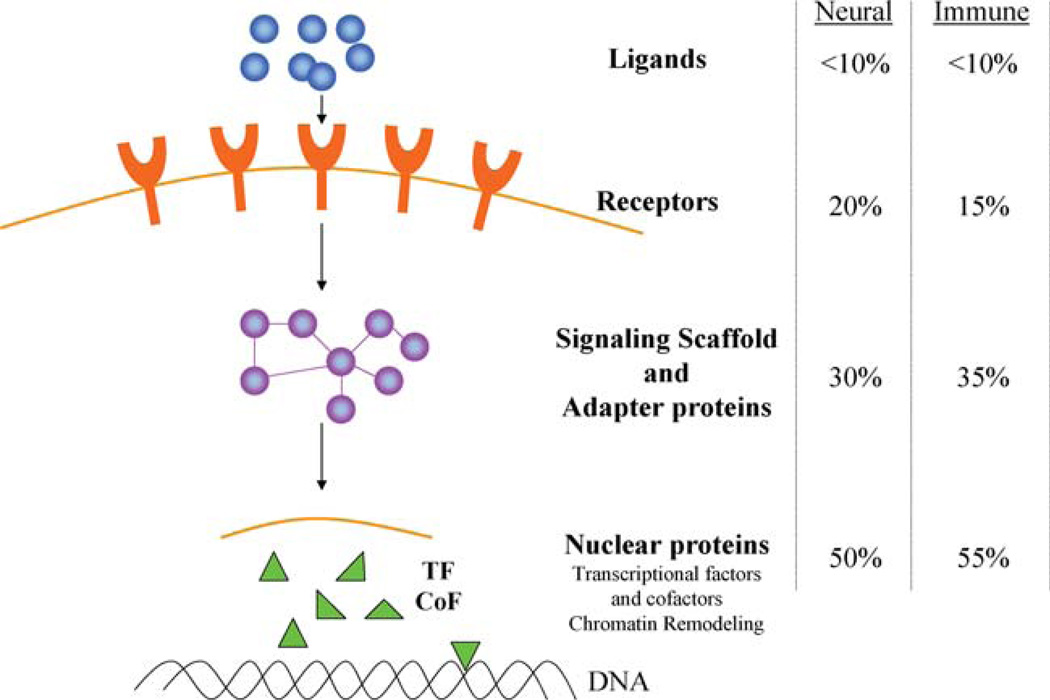
From our analysis, it appears that miRs preferentially regulate nuclear factors which have multiple downstream connections and upstream factors, such as ligands and receptors of cytokines, chemokines, TLRs, are generally poor targets (Fig. 2) [52]. Highly connected transcription factors (e.g., NFAT), cofactors (e.g., mSin3), scaffolding factors (e.g., caveolin-1), adaptor proteins (e.g., SARA), and chromatin pathway components (e.g., MeCP2) are examples of major targets. Thus, surface receptors and their ligands are generally poor targets, while nuclear factors are rich in potential targets.
Fig. 2.
Cellular networks and the distribution of miR targeted genes. This schematic representation of the network of cellular components describes the frequency of miR targeted genes at multiple levels of cellular organization. A similar distribution has been published for neural genes [52] and quite closely parallels our analysis of immune genes
MicroRNAs functionally regulate MHC class II expression
We have employed miR arrays in trophoblast (JAR) and other selected cell types (HeLa, Raji) and showed that cells differ in their ‘immunomiR’ profiles, as shown by others for ‘oncomiRs’ in cancer cells [53]. Using miR arrays to study the effects of IFN-γ, we have found that the induction of miRs is strikingly different in various cell types (data not shown). This is in marked contrast to well-established gene expression patterns where a common signaling pathway (e.g., JAK/Stat) activates a generally similar set of IFN-γ inducible genes in different cell types.
Computational analyses suggest that MHC class I and II genes are non-targets of miRs. However, CIITA has three potential sites in its 3′ UTR. The target sites for miR-145 (1 site) and miR-198 (2 sites) in the CIITA 3′ UTR, together with ARE sites are described in detail (see Fig. 6 in ref 46). Based on algorithm predictions, we carried out transfections with miR-145 and -198 or their specific antagonists (anti-miRs). In experiments using HeLa cells transfected with miR-145 activation of HLA-DR by IFN-γ was inhibited [46]. Transfection of HeLa cells with luciferase reporter constructs carrying the entire 3′ UTR of CIITA or fragments of the 3′ UTR that include the predicted miR target sites support the conclusion that miRNAs (miR-145, -198) specifically inhibit CIITA [46]. Of note is that the miR-145 immunomiR has also been identified as an oncomiR [53].
In addition to targeting CIITA and possibly other components of the class II enhanceosome, miRs may regulate other factors that modulate the JAK/Stat signaling pathway. We have identified numerous miRs which target the JAK/Stat pathway and the class II enhanceosome. While the miRs that target the enhanceosome components would be expected to generally repress transcription of class II, those which target repressors potentially enhance gene expression. Thus, the balance of these positive and negative miRs may affect the level of constitutive expression of immune genes as well as the IFN-γ inducibility in various cell types. This data suggests an additional level of control of MHC expression not previously appreciated, and a potential mechanism by which IFN-γ inducible genes may be repressed in trophoblasts and cancer cells. Importantly, miR repression may focus on a single component in a signaling cascade (e.g., CIITA). Such a system could add significant plasticity to networks. We hypothesized that tumor cells and trophoblasts utilize similar general mechanisms of silencing genes, e.g., chromatin, DNA methylation, at the transcription level and now miRs, by inhibiting translational initiation and/or enhancing mRNA degradation may contribute to immune escape.
MiRNA regulation during trophoblast differentiation in vitro
Trophoblast stem cells, derived from blastocysts, can differentiate in vitro and trophoblast stem cell lines have been established as tools for studying stem cell differentiation and its regulation. We selected the Rcho-1 rat trophoblast stem cell line [54] for an investigation of miRNA regulation during trophoblast differentiation. This well-characterized cell line can be maintained as proliferating stem cells in culture and, in the absence of mitogens, will differentiate into predominantly trophoblast giant cells. Rcho-1 cultures were plated at near confluence in differentiation media and, after 6 days, trophoblast giant cell differentiation was assessed by measurement of cell size, morphology, and DNA content. Harvested cells were stained with DRAQ5 (DNA stain) and analyzed on the ImageStream imaging flow cytometer. Fluorescence and bright field images of individual cells during flow were analyzed for DNA content (DRAQ5 fluorescence intensity) and cell size (cell perimeter from bright field images) to quantify the endoreduplication and morphological changes that accompany Rcho-1 differentiation (Magner and Tomasi, unpublished data) [55, 56]. RT–PCR analysis of differentiation markers (PL-1, PLP-A, Snai1) confirmed the differentiation under mitogen withdrawal.
As an initial test of the hypothesis that miR expression patterns will change during trophoblast differentiation, we analyzed the expression of 15 miRs in our proliferating and differentiated Rcho-1 RNA samples. We found that mitogen withdrawal increased the levels of four miRs by at least 2 Ct (≥4-fold) and induced the expression of miR-153 which was undetectable in the proliferating cells. It has recently been shown that certain stresses, such as folate deficiency, can induce specific miRNA expression [57]. Since Rcho-1 differentiation in vitro involves withdrawal of mitogens, it is possible that a stress-like response may also induce changes in the Rcho-1 miR expression profile. Thus, some of the miRNA alterations may be similar to changes seen in cells after amino acid starvation or other stresses while certain changes could be directly related to the differentiation process.
Chromatin regulation and miRNA
Relevant to the trophoblast studies is the observation that genes that are involved in chromatin remodeling and DNA methylation are rich in miR target sites [46]. This includes genes for several HATs (CBP, p300, PCAF, GCN5, and others), the class II HDACs (4, 5 and 9), the DnmT3a DNA methyltransferase, the MeCP2 methyl-binding protein and several of the recently described JMJ domain histone lysine demethylase proteins. However, many other genes known to have chromatin effects are non-targets (e.g., HDAC 1, 2, 3). MiRs could either activate genes (e.g., by repressing HDACs) or inhibit (e.g., by repressing HATs) and thereby influence epigenetic regulation. Thus far we have found that the set of miRs upregulated by TSA differs substantially between various cell types. However, some miRs (122a, 127, 138) appear to respond to epigenetic control in most, although not all, cell types. CpG islands have been found in the miR-127 5′ region and are presumably responsible for the susceptibility of miR-127 to activation by 5-azacytidine (5-aza) [58]. In this report from the Peter Jones laboratory [58], 4-phenyl butyric acid (PBA), an HDACi in clinical use, did not activate miR-127 in the absence of 5-aza. We have verified this finding but also show that TSA induces a robust response in HeLa exceeding that of PBA and 5-aza. This data illustrates the differences in response depending on the HDACi. Also of note, is that closely spaced miRs (−127 and −432), which are components of a microRNA cluster, can maintain unique miR specific epigenetic regulation (Gregorie and Tomasi, unpublished data). This may result from separate promoters or distinct splicing mechanisms that operate at clustered miR genes.
MicroRNAs regulate cytokines via ARE machinery components
Since inflammation is a current topic of interest in trophoblast biology as well as in immunity, we searched for miR target genes in our data set that are known regulators of inflammation. We first focused on inflammatory cytokines that are regulated, in significant part, by message stability via AU-rich elements (ARE) in their 3′ UTR. We found that with few exceptions most cytokine genes do not have miR binding sites [46]. Notable is the finding that IL-1 and IL-6, reported to decrease during cytotrophoblast differentiation [59], were selected as good targets. Unanticipated, however, was the finding that several translational components (TTP, AuF1, and HUR) which are factors involved in the regulation of ARE mediated message stability have high probability miR sites. This implies that message levels of TNF-α, GM-CSF, IL-1, -6, and -8, and possibly other inflammatory genes, may be controlled indirectly by miR via their effects on ARE machinery components.
MicroRNAs regulate Dicer and other machinery components of the miR pathway
Unexpectedly, in pursuing the effects of miR on immune gene expression, we discovered that certain components responsible for generating, transporting, and executing miR-induced gene repression have multiple high probability 3′ UTR miR binding sites and may themselves be regulated by miRs. This suggests the possibility of feedback loops where a set of miRs could determine the cellular levels of other miRs. It will be important, therefore, to determine if this set of miRs could act as a rheostat which regulates the ~ 30% of the genome known to be targeted by miRs. Speculatively, the genes that are targeted by miRs could have evolved initially to regulate the increasing complexities of cell differentiation and development in early metazoan organisms. Subsequently in evolution, certain miRs may have been commandeered to adapt to environmental stresses. MicroRNAs are continuing to evolve in humans and ‘stress miRs’ may be evolving more rapidly in recent evolution.
The RNase III enzyme, Drosha, and its RNA binding partner, Pasha/DGCR8, were found to be non-targets of miRs. The Ran exportin-5, a CTP factor required for pre-miR transport from the nucleus to the cytoplasm, is the target of two miRs. Strikingly, the Dicer 3′ UTR was found to have 28 potential miR binding sites of which six were selected as high probability by all three algorithms involved in these studies [46]. Notably, the most intense focus of miRs, in all our data occurred with Argonaute1 with >60 individual miR binding sites predicted in its 5682 bp 3′ UTR. Of these, 20 sites were selected by all three algorithms to be miR targets. Argonaute-2, 3, and 4 have 4, 1, and 12 miR binding sites, respectively. Thus, a total of ~ 100 miRs were predicted to target the Dicer/Argonaute machinery components. The let-7 miR was predicted to bind to the 3′ UTRs of all four Argonaute family members and also Dicer. Analyses of the frequency of occurrence of let7 targets in our data set shows that it is highly unlikely that let7 targets Dicer and all four Argonaute genes by chance alone. Of the 11 different let7 species, we have identified let7d (on ch 9q22.32) as targeting Dicer, while let7b (ch 22q13.31) is predicted to bind to the 3′ UTR of all four Argonaute genes. Other miRs that have common targets in Dicer and Argonaute1 are miR-122a, -103, -107, -130, -29. Using transfection with single miRs, we have preliminary evidence in HeLa that miR-122a and -29 may repress Dicer. However, not all miRs predicted to target Dicer repress as illustrated by preliminary transfection studies in which miR-107, similar to control miR-124, not predicted to target Dicer, did not alter protein levels (Asirvatham, Wiesen and Tomasi, unpublished data).
Dicer is regulated by cellular stresses and Type I and II interferons
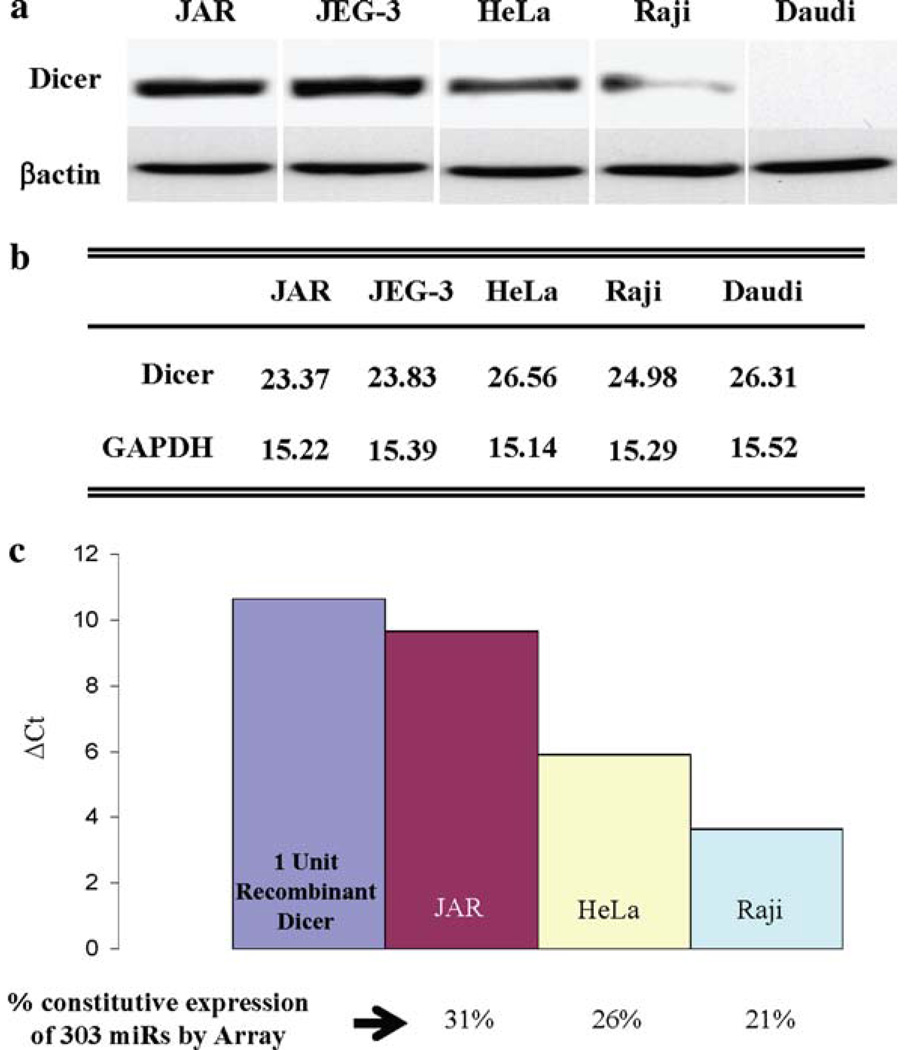
We initially studied three types of human cells varying in their immune gene expression patterns—JAR and JEG-3 trophoblast cells, in which MHC class II and costimulatory genes are silenced; HeLa cervical carcinoma cells, in which immune genes are not constitutively expressed but are IFN-γ inducible; and Raji and Daudi B cells, which, like normal B cells, have high constitutive expression of MHC class II and multiple other immune genes [6, 8]. Figure 3 illustrates the high levels of Dicer protein in JAR and JEG-3 cells, intermediate levels in HeLa and low to absent in Raji and Daudi cells. As shown in Fig. 3b, the cellular Dicer mRNA levels measured by real time RT–PCR are not well correlated with Dicer protein expression. To determine the functional levels of Dicer in these cells, we developed an activity assay employing a pre-miRNA which, on treatment with S100 cell extracts containing Dicer, was sliced to the mature 22 nt species. Quantitative RT–PCR using primers specific for the 22 nt product determined Dicer activity in cell extracts. The Dicer activity of cell extracts was assayed by comparison of the level of the 22 nt sliced product with the level generated by recombinant Dicer as shown in Fig. 3c. This figure also illustrates the number of constitutively expressed miRNAs in each of the cell types as determined by miRNA microarrays. Notable are the cell type variations in Dicer levels and the correlation of its expression and activity with the numbers of constitutively expressed miRNAs in each cell type. These data, although limited, suggest that global differences in Dicer protein expression levels may be reflected in the cellular expression of miRNAs. The above studies led us to further examine how Dicer is regulated and differentially expressed. Unexpectedly, rather than enhancing Dicer, TSA substantially inhibited Dicer protein levels in JAR cells. A similar inhibition of Dicer protein with no change in mRNA levels was noted in JEG-3 trophoblasts, the B16 melanoma, as well as in HeLa cells (Wiesen and Tomasi, unpublished data). One possibility is that TSA enhanced the acetylation of Dicer protein and increased its proteolysis as has been noted for several proteins, for example, IRF7 [60]. However, immunoprecipitation of Dicer from TSA-treated JAR extracts followed by Western analysis for acetylated lysines did not detect acetylation of Dicer (data not shown). Moreover, although JAR extracts contained Dicer activity, adding TSA directly to cell extracts did not alter Dicer in a functional assay. In addition to their affect on chromatin, certain HDACi, such as TSA, can activate cellular stress pathways, including NF-κB, MAPK, and PI3K [27, 28, 61, 62] and therefore, TSA’s affect on Dicer expression could possibly be related to these pathways and additional studies are in progress to elucidate this point.
Fig. 3.
a Dicer protein level differences in three different cell types. Untreated JAR and JEG-3 trophoblast cells, HeLa cervical carcinoma cells, and Raji and Daudi B cell lysates were analyzed by western blotting for Dicer and β-actin levels. b Dicer levels assessed by quantitative real time RT–PCR. c Differential Dicer activity between cell types. Constitutively expressed miRs were determined by miRNA microarray analyses and are presented as a percentage of miRs analyzed
Toll-like receptors (TLRs) are sensors of various environmental stresses, including the dsRNA pathway that activates TLR3, and Type I interferon is a major mediator of the TLR3 response [63–65]. Importantly, dsRNA (Poly I:C), as well as IFN-α, inhibited Dicer protein expression [66]. Additionally, recombinant IFN-α2A and IFN-β repressed Dicer in JAR cells (data not shown). Therefore, it seems that several different types of stresses share the common property of repressing Dicer protein expression and that this may be largely post-transcriptional, possibly at the level of translation which is known to be a major focus of IFN-α/β regulation [65, 67, 68]. However, IFN-γ enhanced Dicer expression in JAR and HeLa cells and this has been shown to be largely transcriptional via the JAK/Stat pathway (Wiesen and Tomasi, unpublished data).
A fundamental thesis in the response of organisms to stress and apoptosis is a global shutdown of protein synthesis as a cellular conservative measure, while the expression of a select group of ‘survival’ proteins, largely those that have internal ribosome entry sites (IRES), continue to be produced [69]. In this regard, multiple types of stresses globally inhibit translation [70]. However, in our data, the inhibition of protein expression does not appear to be widespread, although we cannot exclude small changes in protein levels not seen by relatively insensitive western analysis.
Type I interferon signals are known to be transduced by multiple complex pathways, including the JAK/Stat, MAPK, and PI3K/TOR [71] and the PI3K pathway also plays an essential role in TLR3 gene activation [72]. At present little is known of the pathways involved in Dicer regulation by these stresses and this is currently a major project in our laboratory.
As noted above, the stresses employed here and the MAPK/PI3K/TOR pathways are involved in three complex cellular homeostasis processes which include cellular senescence, autophagy, and apoptosis. The extensive interconnections between autophagy and apoptosis make it difficult to definitely assign Dicer regulation to one (or both) pathway. Each of these are distinct and identifiable processes, and it is feasible that Dicer may be regulated in one (or several) of these pathways. A very recent study suggests that Dicer is a target of caspases in HeLa during apoptosis [73]. We believe that Dicer levels may be altered in apoptosis, but in our studies on JAR, the cells employed were adherent, viable by vital dye staining, >95% annexin V negative, and replateable. These data do suggest that the stresses and IFNα/β that repress Dicer may occur in cells that are ‘on the road’ to senescence, autophagy, and/or apoptosis. However, whether Dicer repression is a mark of any or all of these processes is, as yet, unknown.
Acknowledgements
Supported by National Institutes of Health grants HD 17013, CA 124971 and utilized core facilities of Roswell Park Cancer Institute’s NCI Cancer Center Support Grant CA16056.
Contributor Information
Thomas B. Tomasi, Email: thomas.tomasi@roswellpark.org, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA; Departments of Medicine and Microbiology and Immunology, State University of New York, School of Medicine and Biomedical Sciences, Buffalo, NY 14214, USA.
William J. Magner, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA
Jennifer L. Wiesen, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA
Julian Z. Oshlag, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA
Felicia Cao, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA.
Alex N. Pontikos, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA
Christopher J. Gregorie, Laboratory of Molecular Medicine, Department of Immunology, Roswell Park Cancer Institute, Elm and Carlton Streets, Buffalo, NY 14263, USA
References
- 1.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;9:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 2.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 3.Trowsdale J, Betz AG. Mother’s little helpers: mechanisms of maternal-fetal tolerance. Nat Immunol. 2006;7:241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 4.Guleria I, Sayegh MH. Maternal acceptance of the fetus: true human tolerance. J Immunol. 2007;178:3345–3351. doi: 10.4049/jimmunol.178.6.3345. [DOI] [PubMed] [Google Scholar]
- 5.Tomasi TB, Khan ANH, Magner WJ. Epigenetic regulation of immune escape genes in cancer. Cancer Immunol Immunother. 2006;55:1159–1184. doi: 10.1007/s00262-006-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magner WJ, Kazim AL, Stewart C, Romano MA, Catalano G, Grande C, et al. Activation of MHC class I, II and CD40 gene expression by histone deacetylase inhibitors. J Immunol. 2000;165:7017–7024. doi: 10.4049/jimmunol.165.12.7017. [DOI] [PubMed] [Google Scholar]
- 7.Khan ANH, Magner WJ, Tomasi TB. An epigenetically altered tumor cell vaccine. Cancer Immunol Immunother. 2004;53:748–754. doi: 10.1007/s00262-004-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou SD, Khan AN, Magner WJ, Tomasi TB. Histone acetylation regulates the cell type specific CIITA promoters. MHC class II expression and antigen presentation in tumor cells. Int Immunol. 2005;17:1483–1494. doi: 10.1093/intimm/dxh326. [DOI] [PubMed] [Google Scholar]
- 9.Khan A, Magner WJ, Tomasi TB. An epigenetic vaccine model active in the prevention and treatment of melanoma. J Transl Med. 2007;5:64–75. doi: 10.1186/1479-5876-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan ANH, Gregorie CJ, Tomasi TB. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother. 2008;57:647–654. doi: 10.1007/s00262-007-0402-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. doi: 10.1016/s0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- 12.Chou S-D, Tomasi TB. Spatial distribution of histone methylation during MHC class II expression. Mol Immunol. 2008;45:971–980. doi: 10.1016/j.molimm.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting JP, Trowsdale J. Genetic control of MHC Class II expression. Cell. 2002;109:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 14.Masternak K, Mottet-Muhlethaler A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC Class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 15.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 16.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 17.Kern I, Steimle V, Siegrist CA, Mach B. The two novel MHC class II transactivators RFX5 and CIITA both control expression of HLA-DM genes. Int Immunol. 1995;7:1295–1299. doi: 10.1093/intimm/7.8.1295. [DOI] [PubMed] [Google Scholar]
- 18.Bihani T, Chicas A, Lo CP, Lin AW. Dissecting the senescence-like program in tumor cells activated by Ras signaling. J Biol Chem. 2007;282:2666–2675. doi: 10.1074/jbc.M608127200. [DOI] [PubMed] [Google Scholar]
- 19.Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, et al. RAS protein induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 20.Lin AW, Barradas M, Stone J, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 22.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 23.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AHFM, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 24.Ogryzko VV, Hirai TH, Russanova VR, Barbie DA, Howard BH. Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol Cell Biol. 1996;16:5210–5218. doi: 10.1128/mcb.16.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay D, Okan NA, Bales E, Nascimento L, Cole PA, Medrano EE. Down-regulation of p300/CBP histone acetyltransferase activates a senescence checkpoint in human melanocytes. Cancer Res. 2002;62:6231–6239. [PubMed] [Google Scholar]
- 26.Munro J, Barr NI, Ireland H, Morrison V, Parkinson EK. Histone deacetylase inhibitors induce a senescence-like state in human cells by a p16-dependent mechanism that is independent of a mitotic clock. Exp Cell Res. 2004;295:525–538. doi: 10.1016/j.yexcr.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, Kofler R, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98:10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayo MW, Denlinger CE, Broad RM, Yeung F, Reilly ET, Shi Y, et al. Ineffectiveness of histone deacetylase inhibitors to induce apoptosis involves the transcriptional activation of NF-κB through the Akt pathway. J Biol Chem. 2003;278:18980–18989. doi: 10.1074/jbc.M211695200. [DOI] [PubMed] [Google Scholar]
- 29.Gregorie CJ, Wiesen JG, Magner WJ, Lin A, Tomasi TB. Restoration of immune gene expression in trophoblast and tumor cells associated with cellular senescence. J Reprod Immunol. 2009;81:25–33. doi: 10.1016/j.jri.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 34.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 36.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 37.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., III SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 39.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 40.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhass S, et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Lee HJ, Palkovits M, Young SW., III MiR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci USA. 2006;103:15669–15674. doi: 10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy SP, Gollnick SO, Pazmany T, Maier P, Elkin G, Tomasi TB. Repression of MHC class II gene transcription in trophoblast cells by novel single-stranded DNA binding proteins. Mol Reprod Dev. 1997;47:390–403. doi: 10.1002/(SICI)1098-2795(199708)47:4<390::AID-MRD5>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fahling M, Mrowka R, Steege A, Nebrich G, Perlewitz A, Persson PB, et al. Translational control of collagen prolyl 4-hydroxylase-a1 gene expression under hypoxia. J Biol Chem. 2006;281:26089–26101. doi: 10.1074/jbc.M604939200. [DOI] [PubMed] [Google Scholar]
- 49.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 54.Faria TN, Soares MJ. Trophoblast cell differentiation: establishment, characterization and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology. 1991;129:2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- 55.George TC, Basiji DA, Hall BE, Lynch DH, Ortyn WE, Perry DJ, et al. Distinguishing modes of cell death using the ImageStream® multispectral imaging flow cytometer. Cytometry. 2004;59A:237–245. doi: 10.1002/cyto.a.20048. [DOI] [PubMed] [Google Scholar]
- 56.Arechiga AF, Bell BD, Solomon JC, Chu IH, Dubois CL, Hall BE, et al. Cutting edge: FADD is not required for antigen receptor-mediated NF-κB activation. J Immunol. 2005;175:7800–7804. doi: 10.4049/jimmunol.175.12.7800. [DOI] [PubMed] [Google Scholar]
- 57.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 58.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Stephanou A, Eis ML, ALW SN, Jikhara H, Handwerger S. Ontogeny of the expression and regulation of interleukin-6 (IL-6) and IL-1 mRNAs by human trophoblast cells during differentiation in vitro. J Endocrinol. 1995;147:487–496. doi: 10.1677/joe.0.1470487. [DOI] [PubMed] [Google Scholar]
- 60.Caillaud A, Prakash A, Smith E, Masumi A, Hovanessian AG, Levy DE, et al. Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J Biol Chem. 2002;277:49417–49421. doi: 10.1074/jbc.M207484200. [DOI] [PubMed] [Google Scholar]
- 61.Yu C, Friday BB, Lai J-P, McCollum A, Atadja P, Roberts LR, et al. Abrogation of MAPK and Akt signaling by AEE788 synergistically potentiates histone deacetylase inhibitor-induced apoptosis through reactive oxygen species generation. Clin Cancer Res. 2007;13:1140–1148. doi: 10.1158/1078-0432.CCR-06-1751. [DOI] [PubMed] [Google Scholar]
- 62.Ozaki K, Minoda A, Kishikawa F, Kohno M. Blockade of the ERK pathway markedly sensitizes tumor cells to HDAC inhibitor-induced cell death. Biochem Biophys Res Commun. 2006;339:1171–1177. doi: 10.1016/j.bbrc.2005.11.131. [DOI] [PubMed] [Google Scholar]
- 63.Akira S, Takeda K. Toll-like receptor signaling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 64.Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 65.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and type I interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 68.van Boxel-Dezaire AHH, Rani MRS, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 69.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 70.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 71.Thyrell L, Hjortsberg L, Arulampalam V, Panaretakis T, Uhles S, Dagnell M, et al. Interferon α-induced apoptosis in tumor cells is mediated through the phosphoinositide 3-kinase/mammalian target of rapamycin signaling pathway. J Biol Chem. 2004;279:24152–24162. doi: 10.1074/jbc.M312219200. [DOI] [PubMed] [Google Scholar]
- 72.Sarkar SN, Peters KL, Elco CP, Sakamoto S, Pal S, Sen GC. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat Struct Mol Biol. 2004;11:1060–1067. doi: 10.1038/nsmb847. [DOI] [PubMed] [Google Scholar]
- 73.Matskevich AA, Moelling K. Stimuli-dependent cleavage of Dicer during apoptosis. Biochem J. 2008;412:527–534. doi: 10.1042/BJ20071461. [DOI] [PubMed] [Google Scholar]