Abstract
The biclustering problem has been extensively studied in many areas, including e-commerce, data mining, machine learning, pattern recognition, statistics, and, more recently, computational biology. Given an n × m matrix A (n ≥ m), the main goal of biclustering is to identify a subset of rows (called objects) and a subset of columns (called properties) such that some objective function that specifies the quality of the found bicluster (formed by the subsets of rows and of columns of A) is optimized. The problem has been proved or conjectured to be NP-hard for various objective functions. In this article, we study a probabilistic model for the implanted additive bicluster problem, where each element in the n × m background matrix is a random integer from [0, L − 1] for some integer L, and a k × k implanted additive bicluster is obtained from an error-free additive bicluster by randomly changing each element to a number in [0, L − 1] with probability θ. We propose an O (n2m) time algorithm based on voting to solve the problem. We show that when  , the voting algorithm can correctly find the implanted bicluster with probability at least
, the voting algorithm can correctly find the implanted bicluster with probability at least  . We also implement our algorithm as a C++ program named VOTE. The implementation incorporates several ideas for estimating the size of an implanted bicluster, adjusting the threshold in voting, dealing with small biclusters, and dealing with overlapping implanted biclusters. Our experimental results on both simulated and real datasets show that VOTE can find biclusters with a high accuracy and speed.
. We also implement our algorithm as a C++ program named VOTE. The implementation incorporates several ideas for estimating the size of an implanted bicluster, adjusting the threshold in voting, dealing with small biclusters, and dealing with overlapping implanted biclusters. Our experimental results on both simulated and real datasets show that VOTE can find biclusters with a high accuracy and speed.
Key words: additive bicluster, computational biology, gene expression data analysis, polynomial-time algorithm, probability model
1. Introduction
Biclustering has proved extremely useful for exploratory data analysis. It has important applications in many fields, for example, e-commerce, data mining, machine learning, pattern recognition, statistics, and computational biology (Yang et al., 2002). Data arising from, for example, text analysis, market-basket data analysis, web logs, and microarray experiments are usually arranged in a co-occurrence table or matrix, such as a word-document table, product-user table, cpu-job table, or webpage-user table. Discovering a large bicluster in a product-user matrix indicates, for example, which users share the same preferences. Biclustering has therefore applications in recommender systems and collaborative filtering, identifying web communities, load balancing, discovering association rules, etc.
Recently, biclustering has become an important approach to microarray gene expression data analysis (Cheng and Church, 2000). The underlying bases for using biclustering in the analysis of gene expression data are as follows: (i) similar genes may exhibit similar behaviors only under a subset of conditions instead of all conditions, and (ii) genes may participate in more than one function, resulting in a regulation pattern in one context and a different pattern in another. Using biclustering algorithms, one may obtain subsets of genes that are co-regulated under certain subsets of conditions.
Given an n × m matrix A, the main goal of biclustering is to identify a subset of rows (called objects) and a subset of columns (called properties) such that a pre-determined objective function which specifies the quality of the bicluster (consisting of the found subsets of rows and columns) is optimized. Biclustering is also known under several different names, for example, “co-clustering,” “two-way clustering,” and “direct clustering.” The problem was first introduced by Hartigan in the 1970s (Hartigan, 1972). Since then, it has been extensively studied in many areas and many approaches have been introduced. Several objective functions have also been proposed for measuring the quality of a bicluster; almost all of them have been proved or conjectured to be NP-hard (Lonardi et al., 2004; Peeters, 2003).
Let A(I, J) be an n × m (n ≥ m) matrix, where  is the set of rows representing the genes and
is the set of rows representing the genes and  is the set of columns representing conditions. In practice, the number of genes is much bigger than the number of conditions. Each element ai,j of A(I, J) is an integer in [0, L − 1] indicating the weight of the relationship between object i and property j. A bicluster of A (I, J) is a submatrix of A(I, J). For any given subset I′ ⊆ I and subset J′ ⊆ J, A(I′, J′) denotes the bicluster of A(I, J) that contains only the elements ai,j satisfying
is the set of columns representing conditions. In practice, the number of genes is much bigger than the number of conditions. Each element ai,j of A(I, J) is an integer in [0, L − 1] indicating the weight of the relationship between object i and property j. A bicluster of A (I, J) is a submatrix of A(I, J). For any given subset I′ ⊆ I and subset J′ ⊆ J, A(I′, J′) denotes the bicluster of A(I, J) that contains only the elements ai,j satisfying  and
and  . When a bicluster contains only a single row i and a column set J′, we simply use A(i, J′) to represent it. Similarly, we use A(I′, j) to represent the bicluster consisting of a row set I′ and a single column j.
. When a bicluster contains only a single row i and a column set J′, we simply use A(i, J′) to represent it. Similarly, we use A(I′, j) to represent the bicluster consisting of a row set I′ and a single column j.
The following are two popular models of biclusters that assume different relationships between objectives (or genes) (Yang et al., 2002).
Constant model: A bicluster A(I′, J′) is an error-free constant bicluster if for each column  , ai,j = cj for all
, ai,j = cj for all  , where cj is a constant for any column j.
, where cj is a constant for any column j.
Additive model: A bicluster A(I′, J′) is an error-free additive bicluster if for any pair of rows i1 and i2 in A(I′, J′),  for every column j, where
for every column j, where  is a constant for any pair of rows i1 and i2.
is a constant for any pair of rows i1 and i2.
Clearly, the additive model is more general than the constant model. In microarray gene expression analysis, the additive model can be used to capture groups of genes whose expression levels change in the same/simlar way under the same set of conditions (Madeira and Oliveira, 2004). The additive model also covers several other popular models in the literature as its special cases. For example, a multiplicative bicluster B is a submatrix that looks like:
 |
where each row can be obtained from another by multiplying a number (Madeira and Oliveira, 2004). If we replace each element b1,j (or cib1,j) with log b1,j (or log(cib1,j), respectively), we get an additive bicluster. For a detailed discussion on various models of biclusters, see Madeira and Oliveira (2004). The additive model has many applications and has been extensively studied in the literature (Barkow et al., 2006; Kluger et al., 2003; Li et al., 2006; Liu and Wang, 2007; Lonardi et al., 2004; Madeira and Oliveira, 2004; Peeters, 2003; Prelić et al., 2006). It was first implicitly applied to microarray gene expression analysis by Cheng and Church (2000), who proposed the mean squared residue score H to measure the coherence of the rows and columns in a bicluster. It is easy to show that for an error-free additive bicluster, its score H is 0. Several efficient heuristic algorithms for solving the additive model were in Prelić et al. (2006) and Yang et al. (2002). Recently, Liu and Wang (2007) proposed the maximum similarity score to measure the quality of an additive bicluster. They designed an algorithm that runs in O (nm(n + m)2) time to find an optimal solution under such a score. To our best knowledge, this is the first score admitting a polynomial time algorithm for additive biclusters.
In this article, we will focus on the additive model for biclusters. In particular, we study a probabilistic model, in which the background matrix and a size k × k additive bicluster are generated based on certain probability methods, and then we implant the additive bicluster by replacing a size k × k submatrix of the background matrix with the k × k additive bicluster. This probabilistic model has recently been used in the literature for evaluating biclustering algorithms (Liu and Wang, 2007; Prelić et al., 2006).
The probabilistic additive model: More precisely, our probabilistic model for generating the implanted bicluster and background matrix is as follows. Let A(I, J) be an n × m matrix, where each element ai,j is a random number in [0, L − 1] generated independently. Let B be an error-free k × k additive bicluster. The additive bicluster B′ with noise is generated from B by changing each element bi,j, with probability θ, into a random number in [0, L − 1]. We then implant B′ into the background matrix A′(I, J) and randomly shuffle its rows and columns to obtain a new matrix A′(I, J). For convenience, we will still denote the elements of A′(I, J) as ai,j's.
From now on, we will consider the matrix A′(I, J) as the input matrix. Let IB ⊆ I and JB ⊆ J be the row and column sets of the implanted bicluster in A′. The implanted bicluster is denoted as A′(IB, JB).
The implanted additive bicluster problem: Given the n × m matrix A′(I, J) with an additive bicluster implanted as described above, find the implanted additive bicluster B′.
Our results: We propose an O(n2m) time algorithm for finding an implanted bicluster based on a simple voting technique. We show that when  , the voting algorithm can correctly find the implanted bicluster with probability at least 1 − 9n−2. We also implement our algorithm as a C+ + program named VOTE. In order to make the program applicable in a real setting, the implementation has to incorporate several ideas for estimating the size of an implanted bicluster, adjusting the threshold in voting, dealing with small biclusters, and dealing with overlapping biclusters. Our experiments on both simulated and real datasets show that VOTE can find implanted additive biclusters with high accuracy and efficiency. More specifically, VOTE has a performance/accuracy comparable to the best programs that were recently compared in the literature (Prelić et al., 2006; Liu and Wang, 2007), but with a much faster speed.
, the voting algorithm can correctly find the implanted bicluster with probability at least 1 − 9n−2. We also implement our algorithm as a C+ + program named VOTE. In order to make the program applicable in a real setting, the implementation has to incorporate several ideas for estimating the size of an implanted bicluster, adjusting the threshold in voting, dealing with small biclusters, and dealing with overlapping biclusters. Our experiments on both simulated and real datasets show that VOTE can find implanted additive biclusters with high accuracy and efficiency. More specifically, VOTE has a performance/accuracy comparable to the best programs that were recently compared in the literature (Prelić et al., 2006; Liu and Wang, 2007), but with a much faster speed.
To our knowledge, the work in bioinformatics that is the most related to our above result is the work of Ben-Dor et al. (1999) concerning the clustering of gene expression patterns. In the article, they studied a probabilistic graph model, where each gene is a vertex in a clique graph H and each group of related genes form a clique in H. The (error-free) clique graph consists of d disjoint cliques, and the input graph is obtained from the clique graph H by (1) removing each edge in H with probability α < 0.5 and (2) adding each edge not in H with probability α < 0.5. They designed an algorithm that can successfully recover the original clique graph H with a high probability. Due to the difference in the models, our voting algorithm is totally different from their algorithm.
We note in passing that the problem of finding an implanted clique/distribution in a random graph has also been studied in the graph theory community (Alon et al., 1998; Feige and Krauthgamer, 2000; Kucera, 1995). Kucera Kucera (1995) claimed that when the size of the implanted clique is at least  , where n is the number of vertices in the input random graph, a simple approach based on counting the degrees of vertices can find the clique with a high probability. Alon et al. (1998) gave an improved algorithm that can find an implanted clique of size at least
, where n is the number of vertices in the input random graph, a simple approach based on counting the degrees of vertices can find the clique with a high probability. Alon et al. (1998) gave an improved algorithm that can find an implanted clique of size at least  with a high probability. Feige and Krauthgamer (2000) gave an algorithm that can find implanted cliques of similar sizes in semi-random graphs. It is easy to see that this problem of finding implanted cliques is a special case of our implanted bicluster problem, where the input matrix is binary and all the elements in the bicluster matrix are 1's. We observe that while it may be easy to modify Kucera's simple degree-based method to work for implanted constant biclusters under our probabilistic model, it is not obvious that the above results would directly imply our results on implanted additive biclusters. Moreover, these methods cannot easily be extended to discover multiple cliques/biclusters as often required in practice.
with a high probability. Feige and Krauthgamer (2000) gave an algorithm that can find implanted cliques of similar sizes in semi-random graphs. It is easy to see that this problem of finding implanted cliques is a special case of our implanted bicluster problem, where the input matrix is binary and all the elements in the bicluster matrix are 1's. We observe that while it may be easy to modify Kucera's simple degree-based method to work for implanted constant biclusters under our probabilistic model, it is not obvious that the above results would directly imply our results on implanted additive biclusters. Moreover, these methods cannot easily be extended to discover multiple cliques/biclusters as often required in practice.
In the rest of the article, we first present the voting algorithm and analyze its theoretical performance on the above probabilistic model. We then describe the implementation of the C++ program VOTE, and present the experimental results. Some concluding remarks are given at the end. For the convenience of the reader, the proofs of some of the technical lemmas in the theoretical analysis will be deferred to the Appendix.
2. The Three-Phase Voting Algorithm
We start the construction of the algorithm with some interesting observations. Recall that B is an error-free k × k additive bicluster and A′ is the random input matrix with a noisy additive bicluster B′ implanted.
Observation 1
Consider the k rows in B. There are at least  rows that are identical. This implies that there exists a row set IC ⊆ IB with
rows that are identical. This implies that there exists a row set IC ⊆ IB with  such that A′(IC, JB) is a constant bicluster with noise.
such that A′(IC, JB) is a constant bicluster with noise.
Consider a row  and a column
and a column  . For each row
. For each row  ,
,  is an integer in
is an integer in  . Based on the value
. Based on the value  , we can partition IB into L different row sets
, we can partition IB into L different row sets  ,
,  . Let IC be one of the row sets with the maximum cardinality,
. Let IC be one of the row sets with the maximum cardinality,  . Then, A′(IC, JB) is a constant bicluster (since
. Then, A′(IC, JB) is a constant bicluster (since  for any {i, i′} ⊆ IC and
for any {i, i′} ⊆ IC and  ) and
) and  Let |IC| = l.
Let |IC| = l.
Our algorithm has three phases. In the first phase of the algorithm, we want to find the row set IC in A′(I, J). In order to vote, we first convert the matrix A′(I, J) into a distance matrix D(I, J) containing the same sets of rows and columns, and then focus on D(I, J).
Distance matrix. Given an n × m matrix A′(I, J), we can convert it into a distance matrix based on a row in the matrix. Let  be any row in the matrix A. We refer to row i* as the reference row. Define
be any row in the matrix A. We refer to row i* as the reference row. Define  . In the transformation, we subtract the reference row i* from every row in A′(I, J). We use D(I, J) to denote the n × m distance matrix containing the set of rows I and the set of columns J with every element di,j. For a row
. In the transformation, we subtract the reference row i* from every row in A′(I, J). We use D(I, J) to denote the n × m distance matrix containing the set of rows I and the set of columns J with every element di,j. For a row  and a column set J′ ⊆ J, the number of occurrences of u,
and a column set J′ ⊆ J, the number of occurrences of u,  , in D(i, J′) is the number of elements with value u in D(i, J′), denoted by
, in D(i, J′) is the number of elements with value u in D(i, J′), denoted by  . The number of occurrences of the element that appears the most in D(i, J′) is f*(i, J′) = maxu f(i, J′, u). Similarly, for a row set I′ ⊆ I and a column
. The number of occurrences of the element that appears the most in D(i, J′) is f*(i, J′) = maxu f(i, J′, u). Similarly, for a row set I′ ⊆ I and a column  , the number of occurrences of u in D(I′, j) is the number of elements with value u in D(I′, j), denoted by f(I′, j, u). The number of occurrences of the element that appears the most in D(I′, j) is f*(I′,j) = maxu f(I′, j, u).
, the number of occurrences of u in D(I′, j) is the number of elements with value u in D(I′, j), denoted by f(I′, j, u). The number of occurrences of the element that appears the most in D(I′, j) is f*(I′,j) = maxu f(I′, j, u).
Observation 2
Suppose that we use a row  as the reference row. For each row i in IC, the expected number of 0's in row i of D(I, J) is at least
as the reference row. For each row i in IC, the expected number of 0's in row i of D(I, J) is at least  . For each row i in IB − IC, the expected number of 0's in row i of D(I, J) is at most
. For each row i in IB − IC, the expected number of 0's in row i of D(I, J) is at most  . For each row i in I − IB, the expected number of 0's in row i of D(I, J) is at most
. For each row i in I − IB, the expected number of 0's in row i of D(I, J) is at most  .
.
Proof. For each row i in IC, there are k elements from B′ and m − k elements from A. Based on the model, among the k elements in B′, the expected number of 0's in row i of D(I, J) is at least (1 − θ)2k (since in both row i and row i*, there are at least (1 − θ)k of the k elements remaining unchanged) and the expected number of 0's among the m − k elements in A is  . Similarly, for each row
. Similarly, for each row  , there are k elements from B′ and m − k elements from A. Among the k elements in B′, if the element di,j is 0 in D(I, J), then it must be one of the three cases: (1) ai,j is changed to
, there are k elements from B′ and m − k elements from A. Among the k elements in B′, if the element di,j is 0 in D(I, J), then it must be one of the three cases: (1) ai,j is changed to  and
and  remains the same, (2)
remains the same, (2)  is changed to ai,j and
is changed to ai,j and  remains the same, and (3) both ai,j and
remains the same, and (3) both ai,j and  are changed to an identical number in [0, L − 1]. The expected numbers for the three cases are
are changed to an identical number in [0, L − 1]. The expected numbers for the three cases are  and
and  , respectively. The total expected number of 0's for the three cases is at most
, respectively. The total expected number of 0's for the three cases is at most  . For the m − k elements in A, the expected number of 0's is
. For the m − k elements in A, the expected number of 0's is  . Therefore, for each row i in IB − IC, the expected number of 0's in row i of D(I, J) is at most
. Therefore, for each row i in IB − IC, the expected number of 0's in row i of D(I, J) is at most  . For each row i in I − IB, the expected number of 0's in row i of D(I, J) is at most
. For each row i in I − IB, the expected number of 0's in row i of D(I, J) is at most  since the probability that each element ai,j in row i is identical to
since the probability that each element ai,j in row i is identical to  is
is  . ▪
. ▪
Based on the observation, the expected number of 0's in each row of IC, which is at least  , is much more than that in the other rows. Therefore, with a high probability, the rows with the most 0's are in IC as long as the reference row i* is in IC. Note that,
, is much more than that in the other rows. Therefore, with a high probability, the rows with the most 0's are in IC as long as the reference row i* is in IC. Note that,  and the voting algorithm works when
and the voting algorithm works when  . Thus, we will show that
. Thus, we will show that  is a good threshold on the number of 0's to differentiate the rows in IC from the rows that are not in IC. More specifically, we can use this threshold to find a row set I0 by applying the following voting method.
is a good threshold on the number of 0's to differentiate the rows in IC from the rows that are not in IC. More specifically, we can use this threshold to find a row set I0 by applying the following voting method.
The first phase voting:
for i = 1 to n do
compute f(i, J, 0).
select rows i such that
 to form I0.
to form I0.
When m and k are sufficiently large and θ is sufficiently small, we can prove that, with high probability, the row set I0 is equal to IC. The proof will be given in the next section (in Lemma 4 and the discussion following its proof). If we cannot find any row i such that  , then the whole algorithm will not output any bicluster. However, this has never happened in our experiments.
, then the whole algorithm will not output any bicluster. However, this has never happened in our experiments.
In the second phase voting of the algorithm, we attempt to find the column set JB of the implanted bicluster. It is based on the following observation.
Observation 3
For a column j in JB, the expected number of occurrences of the element that appears the most in D(IC, j) is at least (1 − θ)|IC|. For a column j in J − JB, the expected number of occurrences of an element u in D(IC, j) is at most  .
.
Proof. In the error free matrix B, all rows in IC are identical. For a column  , the corresponding column in B has the same element, say u, in all rows IC. After adding noise with probability θ, the expected number of unchanged u's is (1 − θ)|IC|. Therefore, in the column D(IC, j), the expected number of occurrences of
, the corresponding column in B has the same element, say u, in all rows IC. After adding noise with probability θ, the expected number of unchanged u's is (1 − θ)|IC|. Therefore, in the column D(IC, j), the expected number of occurrences of  is at least (1 − θ)|IC|.
is at least (1 − θ)|IC|.
For a column  and a row
and a row  , if
, if  , the probability that
, the probability that  is 1/L, otherwise, the probability is 0. Therefore, the expected number of occurrences of u in D(IC, j2) is at most
is 1/L, otherwise, the probability is 0. Therefore, the expected number of occurrences of u in D(IC, j2) is at most  . ▪
. ▪
With high probability (and again assuming that θ is sufficiently small), the number of occurrences of the element that appears the most in the columns of JB is greater than the number of occurrences of the element that appears the most in the columns of J − JB. That is, for two columns  and
and  , with high probability,
, with high probability,  . Based on the property, we can use voting to find a column set J1.
. Based on the property, we can use voting to find a column set J1.
The second phase voting:
for j = 1 to m do
compute f*(I0, j).
select columns j such that
 to form J1.
to form J1.
We can prove (in the next section) that, with high probability, J1 is equal to the implanted column set JB.
Similarly, the third phase voting of the algorithm is designed to locate the row set IB of the implanted bicluster. But, before the voting, we need to correct corrupted columns of the distance matrix D(I, J) caused by the elements of the reference row i* that were changed during the generation of B′. Recall that f*(I0, j) = maxu f(I0, j, u). Let f(I0, j, uj) = f*(I0, j). For every  , if uj ≠ 0, then the element ai*,j was changed when B′ was generated (assuming J1 = JB), and we can thus correct each element di,j in the j th column of the matrix D(I, J) by subtracting uj from it.
, if uj ≠ 0, then the element ai*,j was changed when B′ was generated (assuming J1 = JB), and we can thus correct each element di,j in the j th column of the matrix D(I, J) by subtracting uj from it.
In the following, let us assume that the entries in the submatrix D(I, JB) have been adjusted according to the correct reference row i* as described above. The following observation holds.
Observation 4
For a row i in IB, the expected number of occurrences of the element that appears the most in D(i, JB) is at least (1 − θ)k. For a row i in I − IB, the expected number of occurrences of the element that appears the most in D(i, jB) is  .
.
We can thus find a row set I1 in A′(I, J1) as follows.
The third phase voting:
for i = 1 to n do
compute f*(i, J1).
select rows i such that
 to form I1.
to form I1.
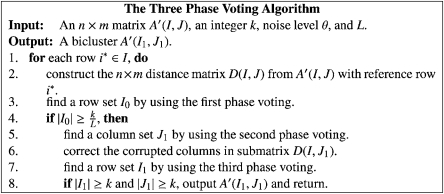
We can prove (in the next section) that, if |I1| ≥ k, with high probability, I1 is equal to the implanted row set IB. Therefore, a voting algorithm based on the above procedures, as given in Figure 1, can be used to find the implanted bicluster with high probability. Since the time complexity of the steps 2–7 of the algorithm is O(nm) and these steps are repeated n times, the time complexity of the algorithm is O(n2m). Note that, if for any phase, we cannot find any row or column that satisfies the bounds, then the algorithm will not output any bicluster. However, this has never happened in our experiments.
FIG. 1.
The three-phase voting algorithm.
3. Analysis of the Algorithm
In this section, we will prove that, with high probability, the above voting algorithm correctly outputs the implanted bicluster.
Recall that in the submatrix A′(IB, JB), each element was changed with probability θ to generate B′ from B. We will show that, with high probability, there exists a row  such that row i has at least (1 − δ) (1 − θ)k unchanged elements in A′(i, JB) for any 0 < δ < 1.
such that row i has at least (1 − δ) (1 − θ)k unchanged elements in A′(i, JB) for any 0 < δ < 1.
In the analysis, we need the following two lemmas from Li et al. (2002) and Motwani and Raghavan (1995).
Lemma 1 (Motwani and Raghavan, 1995)
Let  be n independent random binary (0 or 1) variables, where Xi takes on the value of 1 with probability pi, 0 < pi, < 1. Let
be n independent random binary (0 or 1) variables, where Xi takes on the value of 1 with probability pi, 0 < pi, < 1. Let  and μ = E[X]. Then for any 0 < δ < 1,
and μ = E[X]. Then for any 0 < δ < 1,
 |
Lemma 2 (Li et al., 2002)
Let Xi, 1 ≤ i ≤ n, X and μ be defined as in Lemma 1. Then for any 0 < ε < 1,
 |
Lemma 3
For any 0 < δ < 1, with probability at least  , there exists a row
, there exists a row  that has at least (1 − δ) (1 − θ)k unchanged elements in A′(i, JB).
that has at least (1 − δ) (1 − θ)k unchanged elements in A′(i, JB).
Proof. Let xi,j be a 0/1 random variable, where xi,j = 0 if ai,j is changed in generating B′, and xi,j = 1 otherwise. Based on the probabilistic model, the expected value of  is (1 − θ)kl.
is (1 − θ)kl.
By Lemma 1 and  ,
,
 |
Note that, if  for all rows
for all rows  , then we have
, then we have  . Thus,
. Thus,
 |
(1) |
The inequality (1) implies the lemma. ▪
Suppose that there is a row  with (1 − δ) (1 − θ)k unchanged elements in A′(i*, JB). Now, let us consider the distance matrix D(I, J) with the reference row i*. We now show that, with high probability, the rows in IC have more 0's than those in I − IC in matrix D(I, J). That is, with high probability, our algorithm will find the row set IC in the first phase voting.
with (1 − δ) (1 − θ)k unchanged elements in A′(i*, JB). Now, let us consider the distance matrix D(I, J) with the reference row i*. We now show that, with high probability, the rows in IC have more 0's than those in I − IC in matrix D(I, J). That is, with high probability, our algorithm will find the row set IC in the first phase voting.
Lemma 4
Let  IC be the reference row with (1 − δ) (1 − θ)k unchanged elements in A′(i*, JB), and D(I, J) the distance matrix as described in Section 2. When
IC be the reference row with (1 − δ) (1 − θ)k unchanged elements in A′(i*, JB), and D(I, J) the distance matrix as described in Section 2. When  and
and  , with probability at least 1 − m−7 − nm−5,
, with probability at least 1 − m−7 − nm−5,  for all
for all  , and
, and  for all
for all  ,
,
Proof. Let JC be a subset of JB such that |JC| = (1 − δ) (1 − θ)k, and for all  ,
,  is unchanged. Consider a row
is unchanged. Consider a row  , i ≠ i*.
, i ≠ i*.  be m random variables. For each random variable Xj, Xj = 1 if di,j = 0, otherwise, Xj = 0. Then,
be m random variables. For each random variable Xj, Xj = 1 if di,j = 0, otherwise, Xj = 0. Then,  . We consider two different column sets: JC and J − JB. (1) For
. We consider two different column sets: JC and J − JB. (1) For  , we have Pr(Xj = 1) = 1 − θ) and Pr(Xj = 0) = θ. The expectation of Xj is μj = 1 − θ. (2) For
, we have Pr(Xj = 1) = 1 − θ) and Pr(Xj = 0) = θ. The expectation of Xj is μj = 1 − θ. (2) For  ,
,  and
and  . The expectation of
. The expectation of  . Let JD = JC ∪ (J − JB). From the above analysis,
. Let JD = JC ∪ (J − JB). From the above analysis,
 |
(2) |
By the definition, the random variables  in row i are independent. By Lemma 2,
in row i are independent. By Lemma 2,
 |
(3) |
Since |IC| ≤ k, the probability that there exists a row in IC with no more than  0's in D(I, J) is at most km−8 ≤ m−7.
0's in D(I, J) is at most km−8 ≤ m−7.
Now, we consider a row  . Let
. Let  be m random variables. For each random variable Yj, Yj = 1 if di,j = 0, otherwise, Yj = 0. We have
be m random variables. For each random variable Yj, Yj = 1 if di,j = 0, otherwise, Yj = 0. We have  . The expectation of Yj is
. The expectation of Yj is  . From the analysis, we have
. From the analysis, we have  . The random variable
. The random variable  in row i are also independent. By Lemma 2,
in row i are also independent. By Lemma 2,
 |
Now let  , we get
, we get
 |
Since |I − IB| ≤ n, the probability that there exists a row in I − IB with at least  0's in D(I, J) is at most nm−5. Therefore, the lemma holds. ▪
0's in D(I, J) is at most nm−5. Therefore, the lemma holds. ▪
The above lemma shows that, when a row i* with (1 − δ)(1 − θ)k unchanged elements in A′(i, JB) is selected as the reference row, and m and k are large enough, I0 = IC with high probability. Next, we prove that, with high probability, our algorithm will find the implanted column set JB.
Lemma 5
Suppose that the row set I0 found in the first phase voting of Algorithm 1 is indeed equal to IC. With probability at least  , the column set J1 found in the second phase voting of Algorithm 1 is equal to JB.
, the column set J1 found in the second phase voting of Algorithm 1 is equal to JB.
Proof. The ideas of the proof is the same as those in the above lemma. For the benefit of readability, we defer the proof to the Appendix. ▪
Similarly, we can prove that, with high probability, our algorithm will find the implanted row set IB.
Lemma 6
Suppose that the column set J1 found in the second phase voting of Algorithm 1 is indeed equal to JB. With probability at least  , the row set I1 found in the third phase voting of Algorithm 1 is equal to IB.
, the row set I1 found in the third phase voting of Algorithm 1 is equal to IB.
Proof. Again, for the sake of readability, we defer the proof to the Appendix. ▪
Finally, we can prove that, with high probability, no column or row other than those in the implanted bicluster will be output by the voting algorithm.
Lemma 7
With probability at least  , no column or row of A′(I, J) other than those in A′(IB, JB) will be output by the Algorithm 1.
, no column or row of A′(I, J) other than those in A′(IB, JB) will be output by the Algorithm 1.
Proof. See the Appendix. ▪
Based on Lemmas 3, 4, 5, 6, and 7, we can show that, when m and k are large enough, the three-phase voting algorithm can find the implanted bicluster with high probability. Let c be a constant such that  . In most applications, we may assume that n < m3. Then, we have the following theorem.
. In most applications, we may assume that n < m3. Then, we have the following theorem.
Theorem 8
When  and
and  , the voting algorithm correctly outputs the implanted bicluster with probability at least 1 − 9m−2.
, the voting algorithm correctly outputs the implanted bicluster with probability at least 1 − 9m−2.
Proof. It follows from Lemma 3 that, when  , we can find a row with at least (1 − δ)(1 − θ)k unchanged elements in A′(i, JB) with probability 1 − m−2. Suppose that such a row is selected as the reference row. Lemma 4 shows that, when
, we can find a row with at least (1 − δ)(1 − θ)k unchanged elements in A′(i, JB) with probability 1 − m−2. Suppose that such a row is selected as the reference row. Lemma 4 shows that, when  and
and  , the row set IC will be correctly found in the first phase voting with probability 1 − 2m−2. If the row set IC is found, Lemma 5 shows that, when
, the row set IC will be correctly found in the first phase voting with probability 1 − 2m−2. If the row set IC is found, Lemma 5 shows that, when  , the implanted column set will be correctly found in the second phase voting with probability 1 − 2m−2. Similarly, if the implanted column set is found, in the third phase voting, when
, the implanted column set will be correctly found in the second phase voting with probability 1 − 2m−2. Similarly, if the implanted column set is found, in the third phase voting, when  , the implanted row set will be found with probability 1 − 2m−2. Therefore, when all the required conditions hold, all the rows and columns in the implanted bicluster will be found by our algorithm with probability 1 − 7m−2.
, the implanted row set will be found with probability 1 − 2m−2. Therefore, when all the required conditions hold, all the rows and columns in the implanted bicluster will be found by our algorithm with probability 1 − 7m−2.
It also follows from Lemma 7 that, when n < m3 and  , with probability 1 − 2m−2, no other row or column will be output by our algorithm. Therefore, our algorithm will correctly output the implanted bicluster with probability 1 − 9m−2. ▪
, with probability 1 − 2m−2, no other row or column will be output by our algorithm. Therefore, our algorithm will correctly output the implanted bicluster with probability 1 − 9m−2. ▪
If we replace m by n in the above analysis, the same proof shows that
Corollary 9
When  and
and  , the voting algorithm correctly outputs the implanted bicluster with probability at least 1 − 9n−2.
, the voting algorithm correctly outputs the implanted bicluster with probability at least 1 − 9n−2.
In the practice of microarray data analysis, the number of conditions m is much smaller than the number of genes n. Thus, Theorem 8 allows the parameter k to be smaller (i.e., it works for smaller implanted biclusters) than Corollary 9, although it assumes an upper bound on n (n < m3) and has a slightly worse success probability.
4. The Implementation of the Voting Algorithm
The voting algorithm described in Section 2 is originally based on the probabilistic model for generating the implanted additive bicluster. Many assumptions have been used to prove its correctness. To deal with real data, we have to carefully resolve the following issues.
Estimation of the bicluster size. In the voting algorithm, we assume that the size k of the implanted bicluster is part of the input. However, in practice, the size of the implanted bicluster is unknown. Here we develop a method to estimate the size of the bicluster. We first set k to be a large number such that k ≥ |JB|. Let q be the maximum number of rows such that f(i, J, u) > (m − k)Pr(di,j = u) + k among all  . Our key observation here is that if k is greater than |JB|, then q will be smaller than |IB|. If k is smaller than |JB|, then q will be greater than |IB|. Thus, we can gradually decrease the value of k while observing that the value of q increases accordingly. The process stops when q ≥ 2k.
. Our key observation here is that if k is greater than |JB|, then q will be smaller than |IB|. If k is smaller than |JB|, then q will be greater than |IB|. Thus, we can gradually decrease the value of k while observing that the value of q increases accordingly. The process stops when q ≥ 2k.
To set the initial value of k such that k ≥ |JB|, we set k = 3 · maxu(Pr(di,j = u)) · m. This worked very well in our experiments.
Dealing with rectangular biclusters. Many interesting biclusters in the practice of microarray gene expression data are non-square. Without loss of generality, assuming |IB|≥ |JB|. To obtain rectangular biclusters, we estimate the size of the bicluster as a k × k square, where k = |JB| in this case. We then use the first phase voting and the second phase voting normally. The third phase voting may automatically generate a rectangular bicluster by selecting all the rows i such that  .
.
Discretization of real data. We need to do discretization before we can apply our algorithm. After we obtain matrix D(I, J), we will transform D(i, J) into a discrete matrix, where each element is an integer in [0, L − 1]. We will do that row by row. Let min and max be the smallest number and the biggest number in the row, respectively. We divide the range [min, max] into L disjoint ranges of the same size. All the numbers in the i -th range will be mapped to integer i − 1. Note that, in our probability model, we did not consider small deviations. Based on the discretization method, if small deviations happen in the middle of the L ranges, we can still get the correct discrete value. However, if the small deviations happen at the ends of the L ranges, then they may lead to wrong discrete value. This is a disadvantage of our method. However, the voting algorithm can still find the rows and columns as long as most of the values in the rows and columns are correctly discretized.
Adjusting the threshold used in the first phase voting for a real input matrix. In Step 3 of the first phase voting, we use the threshold  to select rows to form I0. This is based on the assumption that in the random background matrix, di,j = 0 with probability
to select rows to form I0. This is based on the assumption that in the random background matrix, di,j = 0 with probability  . In order for the algorithm to work for any input data, we consider the distribution of numbers in the whole input matrix. We calculate the probability Pr(di,j = l) for each
. In order for the algorithm to work for any input data, we consider the distribution of numbers in the whole input matrix. We calculate the probability Pr(di,j = l) for each  in the discrete matrix. Here
in the discrete matrix. Here  , where p is the number of l's in the input discrete matrix. In Step 3 of the first phase voting, we choose all the rows such that f (i, J, u) > (m − k) P r (di,j = u) + k. In this way, we were able to make our algorithm to work well for real microarray data where the background did not seem to follow some simple uniform/normal distribution.
, where p is the number of l's in the input discrete matrix. In Step 3 of the first phase voting, we choose all the rows such that f (i, J, u) > (m − k) P r (di,j = u) + k. In this way, we were able to make our algorithm to work well for real microarray data where the background did not seem to follow some simple uniform/normal distribution.
When |Ic| is too small for voting. Recall that IC is the set of the rows identical to the reference row I* in the implanted bicluster. In other words, the set IC contains all the rows i with di,j = 0 for  . The expectation of |IC| is
. The expectation of |IC| is  . When k is small and L is large, |IC| (and thus I0) could be too small for the voting in the second phase to be effective. To enhance the performance of the algorithm, we consider the set
. When k is small and L is large, |IC| (and thus I0) could be too small for the voting in the second phase to be effective. To enhance the performance of the algorithm, we consider the set  for each
for each  as defined in the beginning of Section 2, and approximate it using a set
as defined in the beginning of Section 2, and approximate it using a set  in the algorithm just like how we approximated the set
in the algorithm just like how we approximated the set  by the set I0 in the first phase voting. Thus, the second phase voting becomes:
by the set I0 in the first phase voting. Thus, the second phase voting becomes:
The second phase voting:
for j = 1 to m do
compute
 for each
for each  .
.select columns j such that
 to form J1.
to form J1.
Dealing with multiple and overlapping biclusters. In microarray gene expression analysis, a real input matrix may contain multiple biclusters of interest, some of which could overlap. We could modify the voting algorithm to find multiple implanted biclusters by forcing it to go through all the n rounds (i.e., considering each of the n rows as the reference row) and recording all the biclusters found. If the two biclusters found in two different rounds overlap (in terms of the area) by more than 25% of the area of the smaller biclcuster, then we consider them as the same bicluster and eliminate the smaller one. Eventually, the biclusters found in all n rounds (that were not eliminated) would be output, in the decreasing order of sizes.
5. Experimental Results
We have implemented the above voting algorithm in C++ and produced a software, named VOTE. In this section, we will compare VOTE with some well-known biclustering algorithms in the literature on both simulated and real microarray datasets. The tests were performed on a desktop PC with P4 3.0-G CPU and 512-M memory running Windows operating system.
To evaluate the performance of different methods, we use a measure (called match score) similar to the score introduced by Prelić et al. (2006). Let M1, M2 be two sets of biclusters. The match score of M1 with respect to M2 is given by
 |
Let Mopt denote the set of implanted biclusters and M the set of the output biclusters of a biclustering algorithm. S (Mopt, M) represents how well each of the true biclusters is discovered by a biclustering algorithm.
5.1. Simulated datasets
Following the method used by Prelić et al. (2006) and Liu and Wang (2007), we consider an n × m background matrix A. Let L = 30. We generate the elements in the background matrix A such that the data fits the standard normal distribution with the mean of 0 and the standard deviation of 1. To generate an additive b × c bicluster, we first randomly generate the expression values in a reference row ( ) according to the standard normal distribution. To obtain a row (
) according to the standard normal distribution. To obtain a row ( ) in the additive bicluster, we randomly generate a distance di (based on the standard normal distribution) and set ai,j = aj + di for
) in the additive bicluster, we randomly generate a distance di (based on the standard normal distribution) and set ai,j = aj + di for  . After we obtain the b × c additive bicluster, we add some noise by randomly selecting θ · b · c elements in the bicluster and changing their values to a random number (according to the standard normal distribution). Finally, we insert the obtained bicluster into the background matrix A and shuffle the rows and columns. We compare our program, VOTE, with several well-known programs for biclustering from the literature, including ISA (Ihmels et al., 2004), CC (Cheng and Church, 2000), OPSM (Ben-Dor et al., 2002), and RMSBE (Liu and Wang, 2007). The program OPSM is originally designed for order preserving biclusters. (A bicluster is order preserving if its columns can be permuted so that every row is monotonically increasing.) Obviously, an error-free additive bicluster is also an order preserving bicluster. However, when errors are added into a additive bicluster, only part of the bicluster is still order preserving biclusters. Here we also include OPSM in our comparison in various cases though it is not fair to OPSM in some cases. The parameter settings of different methods are listed in Table 1.
. After we obtain the b × c additive bicluster, we add some noise by randomly selecting θ · b · c elements in the bicluster and changing their values to a random number (according to the standard normal distribution). Finally, we insert the obtained bicluster into the background matrix A and shuffle the rows and columns. We compare our program, VOTE, with several well-known programs for biclustering from the literature, including ISA (Ihmels et al., 2004), CC (Cheng and Church, 2000), OPSM (Ben-Dor et al., 2002), and RMSBE (Liu and Wang, 2007). The program OPSM is originally designed for order preserving biclusters. (A bicluster is order preserving if its columns can be permuted so that every row is monotonically increasing.) Obviously, an error-free additive bicluster is also an order preserving bicluster. However, when errors are added into a additive bicluster, only part of the bicluster is still order preserving biclusters. Here we also include OPSM in our comparison in various cases though it is not fair to OPSM in some cases. The parameter settings of different methods are listed in Table 1.
Table 1.
Parameter Settings for Different Biclustering Methods
| Method | Type of bicluster | Parameter setting |
|---|---|---|
| BiMax (Prelić et al., 2006) | Constant | Minimum number of genes and chips: 4 |
| ISA (Ihmels et al., 2004) | Constant/additive | tg = 2.0, tc = 2.0, seeds = 500 |
| CC (Cheng and Church, 2000) | Constant | δ = 0.5, α = 1.2 |
| CC (Cheng and Church, 2000) | Additive | δ = 0.002, α = 1.2 |
| RMSBE (Liu and Wang, 2007) | Constant/additive | α = 0.4, β = 0.5, γ = γe = 1.2 |
| OPSM (Ben-Dor et al., 2002) | Order preserving | l = 100 |
| SAMBA (Tanay et al., 2002) | Constant | D = 40, N1 = 4, N2 = 4, k = 20, L = 10 |
Testing the performance on small biclusters. First, we test the ability of finding small implanted additive biclusters. Let n = m = 100 and b = c = 15 × 15, and consider implanted biclusters generated with different noise levels θ in the range of [0, 0.25]. For each case, we run 100 instances and calculate the average match score. As illustrated in Table 2, the variances of the match scores of the biclusters found by the programs RMSBE and VOTE are very small when the noise level is small, but they increase quickly as the noise gets larger. Figure 2 shows that VOTE and RMSBE perform very well at all noise levels.
Table 2.
Match Score Variances of RMSBE and VOTE in the Test of Performance on Small Biclusters at Different Noise Levels
| |
Noise level |
|||||
|---|---|---|---|---|---|---|
| 0 | 0.05 | 0.10 | 0.15 | 0.20 | 0.25 | |
| Variance (RMSBE) | 0.0 | 0.0002 | 0.0004 | 0.0005 | 0.0009 | 0.0008 |
| Variance (VOTE) | 0.0 | 0.0 | 0.0001 | 0.0005 | 0.008 | 0.03 |
FIG. 2.
Performance on small additive biclusters.
Testing the performance on biclusters of different sizes. Since RMSBE has the best performance among the existing programs considered here, we compare VOTE with RMSBE on different bicluster sizes. In this test, the noise level is set as θ = 0.15. The sizes of the implanted (square) biclusters vary from 25 × 25 to 100 × 100 and the background matrix is of size 500 × 500. For each case, we run 100 instances and calculate the average match score. Table 3 shows the variances of the match scores of the biclusters found by the two programs, which are small except when the size of the implanted bicluster reaches below 32. As illustrated in Figure 3, VOTE outperforms RMSBE when the size of the implanted bicluster is greater than 32, while RMSBE is better at finding small biclusters.
Table 3.
Match Score Variances of RMSBE and VOTE in the Test of Performance on Small Biclusters of Different Sizes
| |
Size |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 27 | 29 | 30 | 32 | 34 | 36 | 38 | 40 | 50 | 75 | 100 | |
| Variance (RMSBE) | 0.012 | 0.012 | 0.018 | 0.021 | 0.028 | 0.019 | 0.006 | 0.002 | 0.0005 | 0.0003 | 0.00008 | 0.00006 |
| Variance (VOTE) | 0 | 0.019 | 0.15 | 0.13 | 0.0001 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
FIG. 3.
Performance on biclusters of different sizes.
Testing the performance on matrices of different k/n ratios. In Figure 3, the accuracy of VOTE drops quickly when the ratio k/n decreases below 30/500. In real applications, the ratio may be quite small. To use VOTE in practice, it is important to find the minimum ratio k/n that can guarantee a good performance. Here, we test VOTE and RMSBE on matrices of various sizes to find such a minimum k/n ratio. The noise level is set as θ = 0.15 and the size of the background matrix is in the range  . For each fixed n and k, we run 50 instances and calculate the average match score. For each fixed n, we try to find the smallest k such that the average match score of the obtained bicluster with respect to the implanted k × k bicluster is at least 80%. The values of k attained by VOTE and RMSBE for each n with match score ≥ 80% are listed in the upper half of Table 4. As shown in the table, the variances of the match scores of the biclusters found by the two programs are generally pretty high here since k is at its minimum value. Note that the match score of RMSBE depends on the quality of the reference row and column. When the reference row and column contain noise, RMSBE may miss some rows and columns in the resulting bicluster. However, VOTE is able to correct some noise in the reference row (Step 6 in Figure 1).Moreover, since VOTE does not require a reference column, it does not suffer from the impact of a noisy reference column. In general, VOTE outperforms RMSBE when n is large (i.e., it allows for a smaller k value). It should be pointed out that when the implanted bicluster size is smaller than the k value listed in Table 4, the match score of VOTE drops more quickly than that of RMSBE as illustrated in Figure 3. We also observe that (the results are not shown in this article), when the average match score is bigger than 80%, say, for example, 99%, VOTE is much better than RMSBE, but when the average match score is smaller than 69%, RMSBE performs better. To illustrate the latter point, we compare the two programs for match score 60%. The values of k attained by VOTE and RMSBE for each n with match score ≤ 60% are listed in the lower half of Table 4.
. For each fixed n and k, we run 50 instances and calculate the average match score. For each fixed n, we try to find the smallest k such that the average match score of the obtained bicluster with respect to the implanted k × k bicluster is at least 80%. The values of k attained by VOTE and RMSBE for each n with match score ≥ 80% are listed in the upper half of Table 4. As shown in the table, the variances of the match scores of the biclusters found by the two programs are generally pretty high here since k is at its minimum value. Note that the match score of RMSBE depends on the quality of the reference row and column. When the reference row and column contain noise, RMSBE may miss some rows and columns in the resulting bicluster. However, VOTE is able to correct some noise in the reference row (Step 6 in Figure 1).Moreover, since VOTE does not require a reference column, it does not suffer from the impact of a noisy reference column. In general, VOTE outperforms RMSBE when n is large (i.e., it allows for a smaller k value). It should be pointed out that when the implanted bicluster size is smaller than the k value listed in Table 4, the match score of VOTE drops more quickly than that of RMSBE as illustrated in Figure 3. We also observe that (the results are not shown in this article), when the average match score is bigger than 80%, say, for example, 99%, VOTE is much better than RMSBE, but when the average match score is smaller than 69%, RMSBE performs better. To illustrate the latter point, we compare the two programs for match score 60%. The values of k attained by VOTE and RMSBE for each n with match score ≤ 60% are listed in the lower half of Table 4.
Table 4.
Minimum k of Different Matrix Sizes
| n | 100 | 200 | 300 | 400 | 500 | 600 | 700 | 800 | 900 | 1000 |
|---|---|---|---|---|---|---|---|---|---|---|
| k (VOTE) | 13 | 19 | 24 | 27 | 31 | 34 | 37 | 40 | 43 | 45 |
| Variance (VOTE) | 0.048 | 0.056 | 0.030 | 0.028 | 0.070 | 0.083 | 0.069 | 0.031 | 0.047 | 0.098 |
| k (RMSBE) | 13 | 19 | 24 | 29 | 35 | 37 | 41 | 46 | 49 | 55 |
| Variance (RMSBE) | 0.016 | 0.023 | 0.026 | 0.022 | 0.010 | 0.025 | 0.024 | 0.016 | 0.021 | 0.013 |
| n | 100 | 200 | 300 | 400 | 500 | 600 | 700 | 800 | 900 | 1000 |
|---|---|---|---|---|---|---|---|---|---|---|
| k (VOTE) | 12 | 19 | 23 | 27 | 30 | 34 | 37 | 39 | 42 | 44 |
| Variance (VOTE) | 0.064 | 0.070 | 0.096 | 0.099 | 0.12 | 0.080 | 0.070 | 0.15 | 0.10 | 0.15 |
| k (RMSBE) | 10 | 15 | 19 | 25 | 27 | 31 | 34 | 38 | 42 | 48 |
| Variance (RMSBE) | 0.029 | 0.017 | 0.013 | 0.016 | 0.011 | 0.014 | 0.010 | 0.013 | 0.016 | 0.015 |
The upper half of the table corresponds to the case match score ≥80%, and the lower half corresponds to match score ≤60%.
We observe that the above k values attained by VOTE are much smaller than the theoretical bound given in Corollary 9. For example, using α = 0.8, θ = 0.09 and L = 30, when n = 1000, k would have to be at least 831 in order for the success probability of VOTE to be close to 1. This shows that the theoretical bound could be very conservative in practice.
Identification of overlapping biclusters. To test the ability of finding overlapping biclusters, we first generate two b × b additive biclusters with o overlapped rows and columns. The parameter o is called the overlap degree. The background matrix size is fixed as 100 × 100. Both the background matrix and the biclusters are generated as before. To find multiple biclusters in a given matrix, some methods, for example, CC, need to mask the previously discovered biclusters with random values. One of the advantages of the approaches based on a reference row, for example, VOTE and RMSBE, is that it is unnecessary to mask previously discovered biclusters. We test the performance of VOTE, RMSBE, CC and OPSM on overlapping biclusters by using 20 × 20 additive biclusters with noise level θ = 0.1 and overlap degree o ranging from 0 to 10. The results are shown in Figure 4. We can see that both VOTE and RMSBE are only marginally affected by the overlap degree of the implanted biclusters. VOTE is slightly better than RMSBE, especially when o increases.
FIG. 4.
Performance on overlapping biclusters.
Finding rectangular biclusters. We generate rectangular additive biclusters with different sizes and noise levels. The row and column sizes of the implanted biclusters range from 20 to 50. The noise level θ is from the range [0, 0.25]. The background matrix is of size 100 × 100. The results are shown in Figure 5. We can see that the performance of VOTE is not affected by the shapes of the rectangular biclusters. On the other hand, RMSBE can only find near square biclusters (Liu and Wang, 2007), and it has to be extended to work for general rectangular biclusters. By comparing Figure 5 with the test results in Liu and Wang (2007), we can see that VOTE is better in finding rectangular biclusters than the extended RMSBE.
FIG. 5.
Performance on rectangular biclusters.
Running time. To compare the speeds of VOTE and RMSBE, we consider background matrices of 200 columns. The number of rows ranges from 1000 to 6000. The size of the implanted bicluster is 50 × 50. The running time of VOTE and RMSBE is shown in Figure 6. In the test, we let RMSBE randomly select 10% rows as the reference row and 50 columns as the reference column. We can see that VOTE is much faster than RMSBE. Moreover, for the real gene expression data of S. cerevisiae provided by Gasch et al. (2000), our algorithm runs in 66 seconds and RMSBE (randomly selecting 300 genes as the reference row and 40 conditions as the reference column) runs in 1230 seconds.
FIG. 6.
Speeds of the programs.
5.2. Real dataset
Gene ontology. Similar to the method used by Tanay et al. (2002) and Prelić et al. (2006), we investigate whether the set of genes discovered by a biclustering method shows significant enrichment with respect to a specific GO annotation provided by the Gene Ontology Consortium (Gasch et al., 2000). We use the web tool funcAssociate of Berriz et al. (2003) to evaluate the discovered biclusters. FuncAssociate first uses Fisher's exact test to compute the hypergeometric functional score of a gene set. Then, it uses the Westfall and Young (1993) procedure to compute the adjusted significance score of the gene set. The analysis is performed on the gene expression data of S. cerevisiae provided by Gasch et al. (2000). The dataset contains 2993 genes and 173 conditions. We set L = 30 to discretize the data for VOTE. (Here, the value of L is chosen empirically.) For all the programs, we output the best 100 biclusters according to their own criteria. For VOTE, we output the largest 100 biclusters since our algorithm is based on counting. The running time of VOTE on this dataset is 66 seconds. The adjusted significance scores (adjusted p-values) of the 100 best biclusters are computed by using FuncAssociate. Here, we compare the significance scores for RMSBE (Liu and Wang, 2007), OPSM (Ben-Dor et al., 2002), BiMax (Prelić et al., 2006), ISA (Ihmels et al., 2004), Samba (Tanay et al., 2002), and CC (Cheng and Church, 2000). The result is summarized in Figure 7. We can see that 92% of discovered biclusters by VOTE are statistically significant, i.e., with α ≤ 5%. Moreover, the performance of VOTE in this regard is comparable to (although slightly worse than) that of RMSBE and is better than those of the other programs compared by Liu and Wang (2007). However, VOTE is much faster than RMSBE since VOTE runs in O(n2m) time, while RMSBE runs in O(nm(n + m)2) time in the worse case.
FIG. 7.
Proportion of biclusters significantly enriched in a GO category. Here, α is the adjusted significance score of a bicluster.
Colon cancer dataset. Murali and Kasif (2003) used a colon cancer dataset introduced by Alon et al. (1999) to test their biclustering algorithm XMOTIF. The matrix contains 40 colon tumor samples and 22 normal colon samples over about 6500 genes. The dataset is available at www.weizmann.ac.il/physics/complex/compphys (Getz et al., 2000). The two best biclusters found by Murali and Kasif (2003) using XMOTIF are B1 and B2 as shown in Table 5. B1 contains 11 genes and 15 samples. Among the 15 samples, 14 of them are tumor samples and 1 is a normal sample. B2 contains 13 genes and 18 samples. Among the 18 samples, 16 of them are normal and 2 are tumor. We use L = 472 to run our program VOTE. The two best biclusters that we find are B3 and B4 in Table 2. B3 contains 35 genes and 27 samples, where all of the 27 samples are tumor samples. B4 contains 91 genes and 11 samples. Among the 11 samples, 10 of them are normal and 1 is tumor. Clearly, the bicluster B3 characterizes tumor samples and B4 normal samples. To evaluate the chance of observing such phenotype (tumor or normal) enrichments at random, we compare the output sample subsets to those obtained by randomized selection. Since the number of phenotypes in a random sample subset fits the hypergeometric distribution, the p value can be computed based on the hypergeometric distribution., All the four biclusters found by both XMOTIF and VOTE are statistically significant as shown by their hypergeometric p-values in Table 5. This result shows that VOTE is able to find high quality biclusters on the dataset.
Table 5.
Biclusters Found in the Colon Cancer Dataset
| Bi-cluster | Method | No. of. genes | No. of samples | No. of tumors | No. of normal | p-value |
|---|---|---|---|---|---|---|
| B1 | XMOTIF | 11 | 15 | 14 | 1 | 3.6e–2 |
| B2 | XMOTIF | 13 | 18 | 2 | 16 | 1.0e–5 |
| B3 | VOTE | 35 | 27 | 27 | 0 | 7.3e–6 |
| B4 | VOTE | 91 | 11 | 1 | 10 | 3.7e–4 |
Carcinoma dataset. The dataset is available at http://microarray.princeton.edu/oncology/. The matrix contains 18 tumor samples and 18 normal samples over about 7464 genes. We use L = 400 to run VOTE. The most tumor-related bicluster contains 152 genes and 14 samples, where all of the 14 samples are tumor samples. The largest normal bicluster contains 1266 genes and 12 samples, where all of the 12 samples are normal samples. This result shows that VOTE can classify the tumor and normal samples well.
6. Conclusion
Based on a simple probabilistic model, we have designed a three-phase voting algorithm to find implanted additive biclusters. We proved that when the size of the implanted bicluster is  , the voting algorithm can correctly find the implanted bicluster with high probability. We have also implemented the voting algorithm as a software tool, VOTE, for finding novel biclsuters in real microarray gene expression data. Our extensive experiments on simulated datasets demonstrate that VOTE performs very well in terms of both accuracy and speed. Future work includes testing VOTE on more real datasets, which could be a bit challenging since true biclusters for most gene expression datasets are unknown. Another direction is to extend the probability model to include small deviations of the values in the input matrix. Perhaps, this will lead to algorithms that work much better in practice. Finally, we note that the automatic selection of parameters in our algorithm is a nontrivial issue and requires further research.
, the voting algorithm can correctly find the implanted bicluster with high probability. We have also implemented the voting algorithm as a software tool, VOTE, for finding novel biclsuters in real microarray gene expression data. Our extensive experiments on simulated datasets demonstrate that VOTE performs very well in terms of both accuracy and speed. Future work includes testing VOTE on more real datasets, which could be a bit challenging since true biclusters for most gene expression datasets are unknown. Another direction is to extend the probability model to include small deviations of the values in the input matrix. Perhaps, this will lead to algorithms that work much better in practice. Finally, we note that the automatic selection of parameters in our algorithm is a nontrivial issue and requires further research.
7. Appendix: The Missing Proofs
Proof of Lemma 5
Let xi,j be a 0/1 random variable, where xi,j = 1 if ai,j is unchanged in generating B′, and xi,j = 0, otherwise. Consider a column  . Let |I0| = l. The expectation for
. Let |I0| = l. The expectation for  is (1 − θ)l. By Lemma 2 and the fact that
is (1 − θ)l. By Lemma 2 and the fact that  ,
,
 |
(4) |
Note that for all  ,
,  . Therefore, the probability that
. Therefore, the probability that  is also at most
is also at most  . The probability that column
. The probability that column  is added into J1 is at least
is added into J1 is at least  . The probability that all columns in JB are added into J1 is at least
. The probability that all columns in JB are added into J1 is at least  .
.
For a column  and an integer
and an integer  , the expectation for
, the expectation for  .
.
By Lemma 2 and  ,
,
 |
(5) |
For  , the probability that there exists an integer
, the probability that there exists an integer  such that
such that  is at most
is at most  . The probability that column j is not added into J1 is at least
. The probability that column j is not added into J1 is at least  . The probability that no column in J − JB is added into J1 is at least
. The probability that no column in J − JB is added into J1 is at least  . Therefore, the probability that J1 = JB is at least
. Therefore, the probability that J1 = JB is at least  . ▪
. ▪
Proof of Lemma 6
Let xi,j be a 0/1 random variable, where xi,j = 1 if ai,j is unchanged in generating B′, and xi,j = 0 otherwise. Consider a row  . The expectation for
. The expectation for  .
.
By Lemma 2,
 |
(6) |
Note that, the distance between row i and i* in A (IB, JB) is a constant  . Since the submatrix D(I, J1) has been corrected,
. Since the submatrix D(I, J1) has been corrected,  . Thus, the probability that
. Thus, the probability that  is also at most
is also at most  . That is, the probability that row i is added into I1 is at least
. That is, the probability that row i is added into I1 is at least  . By considering all the k rows in IB, the probability that all the rows in IB are added into I1 is at least
. By considering all the k rows in IB, the probability that all the rows in IB are added into I1 is at least  .
.
For a row  and an integer
and an integer  , the expectation for f(i, J1, u) is no more than
, the expectation for f(i, J1, u) is no more than  .
.
By Lemma 2,
 |
(7) |
In the algorithm, the probability that there exists an integer  such that
such that  is at most
is at most  .
.
Therefore, the probability that row  is not added into I1 is at least
is not added into I1 is at least  . The probability that no row in I − IB is added into I1 is at least
. The probability that no row in I − IB is added into I1 is at least  . With the above analysis, with probability at least
. With the above analysis, with probability at least  , we have I1 = IB. ▪
, we have I1 = IB. ▪
Proof of Lemma 7
For any column  , similar to Lemma 5, we can prove that with probability
, similar to Lemma 5, we can prove that with probability  , the column j is not added into J1 in any of the n rounds of Algorithm 1. Since |J − JB| = m − k, with probability at least
, the column j is not added into J1 in any of the n rounds of Algorithm 1. Since |J − JB| = m − k, with probability at least  , no column in J − JB are added into J1. In other words, no column other than those of JB are output by the algorithm.
, no column in J − JB are added into J1. In other words, no column other than those of JB are output by the algorithm.
For any row  , similar to Lemma 6, we can prove that with probability
, similar to Lemma 6, we can prove that with probability  , the row i is not added into I1 in any of the n rounds of Algorithm 1. Since |I − IB| = n − k, with probability at least
, the row i is not added into I1 in any of the n rounds of Algorithm 1. Since |I − IB| = n − k, with probability at least  , no row in I − IB are added into I1. In other words, no row other than those of IB are output by the algorithm.
, no row in I − IB are added into I1. In other words, no row other than those of IB are output by the algorithm.
The above analysis shows that, with probability at least  , no row or column other than those in A′(IB, JB) will be output by Algorithm 1. ▪
, no row or column other than those in A′(IB, JB) will be output by Algorithm 1. ▪
Acknowledgments
We wish to thank the anonymous referees for their many helpful suggestions and constructive comments. The research of JX was supported in part by the National Natural Science Foundation of China (grant 60553001) and the National Basic Research Program of China (grant 2007CB807900). LW was supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (project no. CityU 121207). The research of TJ was supported by the NSF (grant IIS-0711129), the NIH (grant LM008991), the National Natural Science Foundation of China (grant 60528001), and a Changjiang Visiting Professorship at Tsinghua University.
Disclosure Statement
No conflicting financial interests exist.
References
- Alon N. Krivelevich M. Sudakov B. Finding a large hidden clique in a random graph. Random Struct. Algorithms. 1998;13:457–466. [Google Scholar]
- Alon U. Barkai N. Notterman D.A., et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkow S. Bleuler S. Prelić A., et al. BicAT: a biclustering analysis toolbox. Bioinformatics. 2006;22:1282–1283. doi: 10.1093/bioinformatics/btl099. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A. Chor B. Karp R., et al. Discovering local structure in gene expression data: the order-preserving submatrix problem. Proc. RECOMB; 2002. pp. 45–55. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A. Shamir R. Yakhini Z. Clustering gene expression patterns. J. Comput. Biol. 1999;6:281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- Berriz G.F. King O.D. Bryant B., et al. Charactering gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Cheng Y. Church G.M. Biclustering of expression data. Proc. ISMB-00; 2000. pp. 93–103. [PubMed] [Google Scholar]
- Feige U. Krauthgamer R. Finding and certifying a large hidden clique in a semirandom graph. Random Struct. Algorithms. 2000;16:195–208. [Google Scholar]
- Gasch A.P. Spellman P.T. Kao C.M., et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz G. Levine E. Domany E., et al. Super-paramagnetic clustering of yeast gene expression profiles. Physica A. 2000;279:457–464. [Google Scholar]
- Hartigan J.A. Direct clustering of a data matrix. J. Am. Statist. Assoc. 1972;67:123–129. [Google Scholar]
- Ihmels J. Bergmann S. Barkai N. Defining transcription modules using large-scale gene expression data. Bioinformatics. 2004;20:1993–2003. doi: 10.1093/bioinformatics/bth166. [DOI] [PubMed] [Google Scholar]
- Kluger Y. Basri R. Chang J., et al. Spectral biclustering of microarray data: coclustering genes and conditions. Genome Res. 2003;13:703–716. doi: 10.1101/gr.648603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera L. Expected complexity of graph partitioning problems. Discrete Appl. Math. 1995;57:193–212. [Google Scholar]
- Li H. Chen X. Zhang K., et al. A general framework for biclustering gene expression data. J. Bioinform. Comput. Biol. 2006;4:911–933. doi: 10.1142/s021972000600217x. [DOI] [PubMed] [Google Scholar]
- Li M. Ma B. Wang L. On the closest string and substring problems. J. ACM. 2002;49:157–171. [Google Scholar]
- Liu X. Wang L. Computing the maximum similarity biclusters of gene expression data. Bioinformatics. 2007;23:50–56. doi: 10.1093/bioinformatics/btl560. [DOI] [PubMed] [Google Scholar]
- Lonardi S. Szpankowski W. Yang Q. Finding biclusters by random projections. Proc. 15th Annu. Symp. Combin. Pattern Matching; 2004. pp. 102–116. [Google Scholar]
- Madeira S.C. Oliveira A.L. Biclustering algorithms for biological data analysis: a survey. IEEE/ACM Trans. Comput. Biol. Bioinform. 2004;1:24–45. doi: 10.1109/TCBB.2004.2. [DOI] [PubMed] [Google Scholar]
- Motwani R. Raghavan P. Randomized Algorithms. Cambridge University Press; Cambridge, UK: 1995. [Google Scholar]
- Murali T.M. Kasif S. Extracting conserved gene expression motifs from gene expression data. Pac. Symp. Biocomput. 2003:8. [PubMed] [Google Scholar]
- Peeters R. The maximum edge biclique problem is NP-complete. Discrete Appl. Math. 2003;131:651–654. [Google Scholar]
- Prelić A. Bleuler S. Zimmermann P., et al. A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics. 2006;22:1122–1129. doi: 10.1093/bioinformatics/btl060. [DOI] [PubMed] [Google Scholar]
- Tanay A. Sharan R. Shamir R. Discovering statistically significant biclusters in gene expression data. Bioinformatics. 2002;18(Suppl 1):136–144. doi: 10.1093/bioinformatics/18.suppl_1.s136. [DOI] [PubMed] [Google Scholar]
- Westfall P.H. Young S.S. Resampling-Based Multiple Testing. Wiley; New York: 1993. [Google Scholar]
- Yang J. Wang W. Wang H., et al. δ-clusters: capturing subspace correlation in a large data set. Proc. 18th Int. Conf. Data Eng.; 2002. pp. 517–528. [Google Scholar]