Abstract
Genomes are vulnerable to selfish genetic elements (SGEs), which enhance their own transmission relative to the rest of an individual's genome but are neutral or harmful to the individual as a whole. As a result, genetic conflict occurs between SGEs and other genetic elements in the genome. There is growing evidence that SGEs, and the resulting genetic conflict, are an important motor for evolutionary change and innovation. In this review, the kinds of SGEs and their evolutionary consequences are described, including how these elements shape basic biological features, such as genome structure and gene regulation, evolution of new genes, origin of new species, and mechanisms of sex determination and development. The dynamics of SGEs are also considered, including possible “evolutionary functions” of SGEs.
The idea that some components of the genome can be “selfish” or “parasitic” has a long and controversial history. The first recognition that a gene could increase in frequency by imparting a drive relative to its homolog came with the description of X-chromosome meiotic drive dynamics in Drosophila obscura by Gershenson (1). Later, Oestergren (2) investigated accumulation of supernumerary (extra nonvital) “B” chromosomes in plants and made the first explicit argument that some genetic material in an organism can be “parasitic.” These observations and their evolutionary implications were not widely known among biologists, however, in part because meiotic drive and supernumerary chromosomes were perceived as genetic peculiarities rather than important general phenomena. Three parallel threads then set the stage for more serious considerations of selfish genetic elements (SGEs) and genetic conflict ideas. First, empirical and conceptual developments in genetics and evolutionary biology led to wider acceptance of a gene-centric view of evolution (3, 4). Noteworthy in this regard was Dawkin's influential book (4) entitled The Selfish Gene, which described genes as “selfish replicators” encoding phenotypes that increase their transmission to future generations and organisms fundamentally as “vehicles” for the transmission of genes. Second, rapid advances in molecular biology began to reveal that many eukaryotic genomes contain large amounts of repetitive DNA without any clear function, although their potential role within the genome was the subject of much speculation (5). Seminal papers by Doolittle and Sapienza (6) and Orgel and Crick (7) first proposed that repetitive DNA could be considered parasitic or selfish replicators. In 1981, Cosmides and Tooby (8) explicitly introduced the concept of genetic conflict between nuclear and cytoplasmic (e.g., mitochondria) elements over sex determination. The idea of SGEs and genetic conflict remained highly controversial, however, and a counterview was that such elements exist because they play important regulatory roles in cells and in evolution. Third, an increasing number of genetic studies began to uncover non-Mendelian and other elements within diverse organisms that appeared to have “self-promoting” features that cannot simply be explained as adaptations for the organism. These included discoveries of meiotic drive in diverse organisms; heritable elements, such as killer plasmids; and a genome-eliminating supernumerary chromosome that was an unequivocal example of a nonadaptive self-promoting replicator (9).
Werren et al. (10) published the first general review of selfish or parasitic genes and defined an SGE as an element that has characteristics enhancing its own transmission relative to the rest of an individual's genome but neutral or detrimental to the organism as a whole. Examples include transposable elements (TEs), meiotic drivers, supernumerary B chromosomes, postsegregation killers, and heritable microbes and organelles that distort sex determination. In 1988, the idea that elements in the genome could be parasitic was still contrary to prevailing opinions of many molecular biologists, who viewed the cell and organism as a highly integrated machine, and therefore considered the idea that components of the cell could be maintained because of their selfish replication as a bizarre and foreign concept. In contrast, the SGE model is a more “ecological” view that considers the genome as a set of genetic elements with potentially different kinds of interactions, ranging from cooperative (mutualistic), to neutral (commensal), to selfish (parasitic) (11). According to this paradigm, genetic conflict can arise among components of the genome that have different transmission patterns (e.g., transposons, nuclear genes, cytoplasmic genes), and therefore conflicting genetic interests. The basic idea is as follows: When components of the genome have different transmission patterns, selection can act on an element to increase its transmission even if that is detrimental to the organism and/or other heritable components of the genome. Genetic conflict within the genome will then result, because enhanced transmission of an SGE decreases transmission of other genetic elements. An evolutionary “arms race” can then occur among different components of the genome over basic biological processes.
Werren et al. (10) raised three basic questions about SGEs that are still the subject of study today: (i) What are their origins, (ii) how are SGEs maintained, and (iii) are SGEs important in evolution? Regarding this last question, they concluded that “selfish elements, and the ‘intragenomic conflict’ they create, may be an important force promoting evolutionary change. However, this possibility has not been demonstrated conclusively in any system” (10). They further observed that the pace of understanding of SGEs “is expected to accelerate with the application of molecular cloning techniques” (10). Subsequent advances have eclipsed this expectation. What has occurred in the intervening years is the genomics revolution, a veritable explosion of information and techniques that have begun to open the “black box” of genome structure, function, and evolution. Today, there are over 1,000 bacterial genomes and over 100 eukaryote genomes sequenced, with the numbers growing almost daily (http://www.ebi.ac.uk/genomes/). These data and advances in genetic techniques have helped reveal how genomes evolve and function. The story that is emerging increasingly supports a central role of SGEs in shaping structure and function of genomes and in playing an important role in such fundamental biological processes as gene regulation, development, evolution of genetic novelty, and evolution of new species.
Here, I describe the conceptual framework for SGEs and genetic conflict as well as their types, and I then discuss developments that reveal the role of SGEs in important biological processes. There are several common themes to the topic that are briefly listed here and elaborated on below:
i) Antagonistic selection occurs between SGEs and other genome components, and this can lead to evolutionary change and novelty.
ii) Sexual recombination and lateral movement between lineages is important to SGE maintenance and evolution.
iii) Many genetic elements have mixed phenotypes with a combination of parasitic and beneficial features.
iv) SGEs can also lead to an “evolved dependency” by the host, which has the appearance of mutualism but is not.
v) SGEs can occupy “safe havens” within the genome, where their negative effects are mitigated or they are less likely to be excised or repressed.
vi) SGEs can be “domesticated” or “co-opted” by genomes, resulting in the evolution of novel genes and functions.
I will end the paper with a discussion of “why” SGEs persist in nature and contrast the evidence and predictions for the view that SGEs persist because of their ability to replicate within genomes (the parasitic hypothesis) vs. the view that they persist because they promote the ability of populations to adapt and evolve (the “evolvability” hypothesis).
Types of Genetic Conflict
Genetic conflict occurs when different genetic elements (either within an individual or between individuals) have influence over the same phenotype, and an increase in transmission of one element by its phenotypic effects causes a decrease in transmission of the other. Not included in this definition are population changes in allele frequency at a locus, unless they result from antagonistic selection acting on the alternative alleles for the shared phenotype (12, 13). An example will illustrate the point. A meiotic drive allele reduces the nondriving allele among the gametes of heterozygous individuals; therefore, these alternative alleles (and tightly linked loci) experience antagonistic selective pressures over the phenotype (one is selected to drive and the other to suppress the drive).
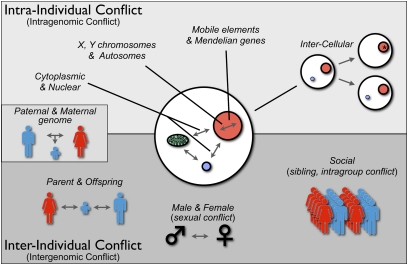
Genetic conflicts historically have been divided into “intragenomic” conflict, which occurs within the genome of an individual, and “intergenomic” conflict, which occurs between individuals (e.g., male-female or sexual conflict, parent-offspring, social conflict) (8, 10, 12, 14–16) (Fig. 1). Because the term “genome” is often used to include the sum of DNA across individuals within a species, however, less confusing terms to distinguish these levels may be “intraindividual” conflict and “interindividual” conflict, because these terms distinguish genetic conflicts within individual organisms (e.g., for transmission through gametes) as opposed to between individuals (e.g., male-female or parent-offspring conflict over reproductive effort). Fig. 1 shows several kinds of conflict that can occur within and between individuals. At one end of the spectrum are genetic conflicts between mobile elements (e.g., transposons), which are generally selected to transpose within a genome, and Mendelian components of the nuclear genome, which are selected to suppress transposition because of fitness costs to the individual. Evidence of this conflict includes diverse mechanisms that have evolved to restrain transposition (17). Similarly, cytoplasmically inherited and nuclear inherited elements experience genetic conflict, primarily over sex determination, because of their differences in transmission through male sperm and female eggs (8, 18). As indicated in Fig. 1, intercellular conflict can occur when heritable differences arise within the cell lineages of an organism by de novo mutation or unequal transmission of heritable organelles or microbes into daughter cells. It has been argued that development has been molded to minimize such conflicts by uniparental inheritance of organelles, metazoan development from single cells, and germline sequestering (8, 19). At the other end of the spectrum are conflicts between individuals, such as parents, offspring, mates, or members of social groups, over phenotypes they jointly influence, such as resource use. Nature is not always as clean as our paradigms, and there is at least one category of genetic conflict that has features of both intraindividual and interindividual conflict, that is, paternal-maternal genome conflicts over resource allocation. This form of conflict is manifested as genomic imprinting of alleles during male and female gametogenesis, which differentially affects their expression in offspring (20).
Fig. 1.
Types of genetic conflicts. Genetic conflicts can be categorized as intraindividual (or intragenomic) and interindividual (or intergenomic). Intraindividual conflicts occur among genetic elements with different inheritance patterns (e.g., cytoplasmic genes; nuclear genes; X, Y, and autosomally located genes; mobile elements). Intraindividual conflict also arises among cells within an organism that are genetically different because of de novo mutations or transpositions (*) or heteroplasmy attributable to unequal segregation (blue circle). Genetic conflicts also occur between individuals, including parent-offspring, sexual, or social conflict. Paternal-maternal genome interactions within offspring have features of both intraindividual and interindividual conflict.
Types of SGEs and Their Consequences
Here, we have a rogue's gallery of the genome. SGEs can be placed into the following broad categories (10, 15, 16): TEs, biased gene converters, meiotic drivers, postsegregation drivers, and cytoplasmic drivers. These elements act to increase their own transmission to the detriment of other components of an individual's genome. This does not mean that such elements cannot have positive long-term evolutionary consequences, and some of the elements in this list can have both selfish and beneficial components.
Transposons and Other Mobile Elements.
Mobile elements include plasmids, endogenous viruses, and TEs. TEs have the ability to copy and move to new locations within the genome; as a consequence, they can accumulate. Doolittle and Sapienza (6) and Orgel and Crick (7) first proposed that they can be considered SGEs, and this view is now widely accepted (although see below). TEs fall into two main categories: DNA transposons move via DNA copies, and retrotransposons use an RNA intermediate (21). TEs can also be autonomous (encoding proteins that promote their transposition) or nonautonomous (not encoding proteins needed for transposition but using the cellular machinery or proteins provided by other TEs). An interesting category of mobile elements is group I and II self-splicing introns (22), which can be tolerated in the typically streamlined genomes of prokaryotes and organelles because self-splicing restores functional open reading frames in genes with the inserts, thus reducing negative fitness costs. Although group II introns are not found in eukaryotes, shared features with the spliceosome of eukaryotes have led to the proposal that the spliceosome machinery and eukaryotic introns evolved from group II introns (22). If correct, this hypothesis implies that acquisition of the spliceosome machinery to remove mobile group II introns set the stage for evolutionary expansion of introns in the genomes of higher eukaryotes.
In bacteria, the amount of mobile DNA ranges from 0–21% and varies with bacterial ecology rather than phylogeny (23)—bacteria with greater exposure to other bacterial lineages show higher levels of mobile DNA. This suggests that opportunities for lateral acquisition, rather than benefits to the host, explain relative abundances of these elements. It is also clear, however, that mobile elements in bacteria (e.g., plasmids) can encode proteins that increase survival of their bacterial hosts, such as antibiotic resistance (24). Evidence suggests that mobile elements in bacteria can be maintained by a combination of selfish features that promote their acquisition and retention in bacterial genomes and (in some cases) beneficial effects on their bacterial hosts. Very large plasmids tend to become immobile and carry important bacterial functions, indicating their evolution to mutualism (24).
In eukaryote genomes, the abundance of TEs can vary widely (25). For example, ∼40% of the human genome is composed of TEs, whereas only 3% of the pufferfish genome is (26). Plants vary similarly. As a result, TEs and other repetitive DNA can be major determinants of genome size within taxa (27). TEs also vary considerably in the taxonomic breadth of their distribution (28, 29). Around 10 different DNA TE superfamilies are currently recognized (28), and many show a broad host taxonomic distribution and signature of lateral transfer between taxa. There is evidence that poxviruses have vectored retroposons between reptiles and mammals, and members of four DNA TE families are found in both vertebrates and blood-sucking triatomid bugs, suggesting possible mechanisms for intertaxon transfers. A study in Drosophila genomes finds that approximately one-third of TE families originated from recent interspecies lateral transfers, with an estimated transfer rate of 0.04 events per family per million years (30). In fact, lateral movement across host taxa is believed to be an important mechanism for long-term maintenance of TE families. The rationale is that evolutionary suppression of TEs by the host will lead to their eventual mutational degradation and loss, except for TEs that move laterally to “infect” and invade new hosts.
Although many active TEs have relatively short evolutionary associations with particular hosts, some can be maintained for long evolutionary time frames in a lineage because they occupy (or target) a safe haven within the genome. Safe havens are genome locations with either reduced fitness costs to the host or where hosts cannot readily remove the insert or evolve countermeasures. R1 and R2 retroelements appear to use a safe haven. They insert into highly conserved segments of the ribosomal RNA genes subject to strong selective constraint (31). Ribosomal RNA genes typically occur in large tandem arrays in eukaryotes; therefore, an insertion into any single copy has relative low fitness costs and new uninserted ribosomal RNA copies are continually produced by unequal chromatid exchange. These features probably explain why R1 and R2 elements have been maintained within lineages over long evolutionary time scales despite little evidence of fitness benefits or lateral element transfer (31).
TEs are known to induce harmful effects through various mechanisms, including insertions that disrupt coding sequences or cis-regulation regions, ectopic recombination between TE copies resulting in deletions and rearrangements, and the costs of transcription and translation of large numbers of TEs (21, 32, 33). As a result, there has been strong selection on plant and animal genomes to evolve machinery to suppress TE activity, including DNA methylation suppression, repeat-induced point mutation in fungi, RNAi, and small RNA suppression pathways (17, 26). These mechanisms could have originally evolved for suppression of TEs and other exogenous DNA, and have subsequently acquired gene regulatory functions. In Drosophila melanogaster, the flamenco locus is a large genomic region containing TE insertions that is used by the piwi-interacting RNA (piRNA) pathway to suppress active TEs dispersed elsewhere in the genome. In plants, suppression of TEs can involve small noncoding RNAs that assist in transcriptional and posttranscriptional silencing and guide targeting of TEs for DNA methylation inactivation (34). By increasing rates of CG-to-TA mutation, DNA methylation of TEs also accelerates their mutational degradation. Neurospora shows targeted repeat TE degradation by this mechanism. One side effect of their very efficient repeat elimination is that the maintenance of gene duplications is difficult in Neurospora, thus affecting its evolutionary trajectory (17). DNA methylation suppression of TEs occurs in both animals and plants and has been invoked as a likely preadaptation for evolution of the placenta and genomic imprinting in mammals (35, 36).
Given the ubiquity and abundance of TEs, it is inevitable that some will be recruited by genomes for new cellular functions (reviewed in refs. 28, 37, and 38). This is variously referred to as “domestication,” “co-option,” or “exaptation.” Classic examples include the utilization of TART and HetA TEs for telomeres in drosophilid flies and the likely origin of V[D]J recombination (used in vertebrates to generate immunoglobin diversity) from mariner-TC1 family TEs. New and exciting discoveries further indicate that domestication of TEs is important in the evolution of genomes, such as the evolution of new protein-coding genes (including regulatory DNA binding factors), cis-regulatory sequences, and regulatory small RNAs from TEs (28, 37, 38). Both the DNA binding and catalytic domains from the transposase genes of DNA TEs have been involved in domestication events in animals, plants, and fungi. The SETMAR gene in primates is a chimera derived from fusion of a mariner-like element with the SET domain from a histone methyltransferase gene 40–58 million years ago (37, 38). Its function is unknown but possibly involved in DNA repair. Widely distributed TEs have been involved in independent domestication events in diverse taxa, such as Pogo elements in both mammals and fission yeast (37). Intriguingly, a number of regulatory DNA binding proteins appear to have evolved from the DNA binding domains of TEs, such as PAX6 (sensory development in metazoans), CENP-B (centromere function in vertebrates), and Bric-a-Brac (tissue development in insects). Feschotte (38) reports that at least seven key DNA binding proteins probably evolved from TEs in taxa ranging from plants and fungi to metazoans. Retrogenes occur when host mRNA is reverse-transcribed and inserted into the genome. The process is dependent on RT proteins from retroelements. Retrogenes have been stripped of introns and usually degenerate as pseudogenes. They have also evolved into new functional genes [e.g., ∼109 examples in the genome of Populus], however, and have acquired introns in some cases (39, 40).
Although the vast majority of TEs that insert near or in protein-coding regions are deleterious, mounting evidence indicates that fragments of inserted TE DNA have also evolved cis-regulatory or posttranscriptional regulatory functions (38). Evidence for this includes conservation of TE-derived fragments in ∼25% of human promoters and deeply conserved fragments in cis-regulatory modules of mammals, as well as evidence of a regulatory role in some cases. These regulatory elements have evolved from ancient insertions of TEs that are no longer active in the mammalian lineage. A basic interpretation is that TE insertions provide abundant sequence variation in regulatory regions on which selection can act.
In Leishmania, 3′ UTRs are important in posttranscriptional gene regulation. Bringaud et al. (41) found evidence of accumulation of fragments from a now inactive family of retrotransposons in the UTRs of some predicted mRNAs, and further found that these genes showed lower than average mRNA levels. The pattern is suggestive of a possible evolution of a regulatory function. Caution is recommended in interpreting apparent overrepresentation of TEs near coding genes as an indicator of function, however. Elements inserted near protein-coding genes are less likely to be deleted because their removal increases the chance of harmful consequences to the adjacent gene. Therefore, inserts in these safe havens will persist longer, even if they are mildly deleterious. Studies that look for possible functional TE insertions based on distributions require null hypotheses that consider the mutational spectra (e.g., deletions) tolerated in regions of different distance from functional genes. Safe havens adjacent to genes also mean that such inserts have more time before deletion to evolve into functional cis-regulators, through mutational amelioration of their deleterious side effects and refinement of cis-regulatory effects.
Do TEs increase the rate of speciation in their hosts? Nonhomologous recombination among TEs can lead to chromosomal rearrangements that contribute to chromosomal-based speciation (21). TE differences between related species may also contribute to reproductive isolation, however; arguing against this scenario is the rapidity by which TEs can jump species boundaries, as observed by P-elements in Drosophila (21). Regarding extinction rates, a comparative study suggests that high TE loads increase the probability of extinction in plants, birds, and reptiles but not mammals (42). Theoretical treatments indicate that TE activity can play a significant role in the extinction of parthenogenetic species through mutational load accumulation, particularly in small populations (43, 44). In larger populations, clonal selection will lead to loss of active TEs, assuming their effects are mostly deleterious (43). Zeh et al. (45) propose an “epi-transposon” hypothesis that changing environments can lead to stress-induced breakdown of epigenetic suppression of TEs (e.g., methylation, piRNAs), with resulting extensive transposition providing new material for rapid adaptive shifts. They also note that such transposon release could lead to increased extinction rates. Alternatively, a changing environment could simply provide transient advantages for elevated mutation rates, which TEs can readily provide. Although intriguing, there is currently little direct support for these ideas.
Biased Gene Converters.
Gene converters are a special class of SGEs that preferentially insert themselves into homologous uninserted sites in the genome. The most famous of these are the homing endonucleases (HEs) found in bacteria and eukaryotes (46). HEs are self-splicing group I introns coding for highly specific endonucleases that cut rare specific sites within the genome, typically the uninserted homologous sequence (hence the name “homing”). Cellular mechanisms use the inserted sequence and homologous flanking DNA as templates for repair, resulting in insertion of the HE into the previously unoccupied site. Other HEs actually splice out of proteins following translation (“inteins”). HEs are known to spread laterally between distant bacterial lineages and appear to maintain strong site fidelity. Their self-splicing ability ensures that the protein function is not disrupted. Using functionally important regions as insertion sites reduces the ability of hosts to evolve resistance to HEs, thus providing a “safe harbor.”
Other forms of biased gene conversion have been found in recombination hotspots in humans through detailed analysis of the products of recombination (47). The extent to which such biased gene conversion can be considered selfish depends on whether conversion bias is dependent on the sequence of the putative SGE. There is growing evidence in eukaryotes of a general GC gene conversion bias in DNA repair of double-strand breaks, resulting in AT/GC heterozygotes producing more GC than AT gametes (48). Can we therefore consider G and C to be our smallest selfish elements? Probably not, because conversion is likely attributable to a general bias in using G and C during double-stranded break repair rather than to a biased conversion attributable to a specific sequence motif. In any case, GC-biased conversion clearly has major consequences for genome composition and evolution (48).
Meiotic Drivers.
Meiosis results in a reduction of the diploid germ cells to haploid gametes. In general, meiosis is “fair,” meaning that the two homologous chromosomes have an equal probability of ending up in functional gametes (sperm or eggs). Meiosis creates opportunities for SGEs that can increase their transmission relative to a nondriving homolog, however. Meiotic drivers are widespread in nature and include such examples as segregation distorter (SD) in D. melanogaster, X-chromosome drive in many animals, X and Y drive in plants, knob-containing chromosomes in maize, the t-locus in mice, supernumerary or B chromosomes in animals and plants, and centromere drive in different organisms (49–53). Meiotic drive can take three basic forms. “True” meiotic drive (e.g., many B chromosomes, centromere drive) is accomplished by preferential segregation to the functional (egg or ovule) pole during gametogenesis. Germline overreplication occurs in some B chromosomes and results in their increased transmission to gametes. “Gamete killer” drive acts by selective elimination or functional disruption of gametes that do not carry the meiotic driving element. This latter form is typically found in males because they produce an excess of gametes that effectively compete for fertilization of eggs. Gamete killing results in increased fertilization of eggs by sperm with the driving chromosome from heterozygous males. SD illustrates the basic mechanism (52). SD is composed of two tightly linked loci near the centromere of chromosome 2 of D. melanogaster. The distorter locus Sd encodes a partial duplication of the gene RanGAP, and the Rsp responder locus contains variable numbers of a tandem repeat. WT chromosomes contain a normal copy of RanGAP and higher copy numbers of Rsp. In heterozygotes, WT sperm fail to develop properly because of interactions between the variant RanGAP (which mislocalizes to the nucleus) and Rsp repeats. A third linked locus enhances drive and a number of unlinked loci reduce drive, as predicted by conflict theory.
Centromere drive has been proposed as a mechanism for evolution of centromeric DNA (51). The basic idea is that competition for spindle binding favors expansion of centromere binding sequences to promote segregation to the function (egg) pole in meiosis. This process could have played an important role in the evolution of chromosome structure and will result in meiotic drive. B chromosomes are “extra” chromosomes that are not essential for viability. They are widespread in animals and plants, and many have mechanisms for increasing their transmission during gametogenesis, including overreplication in germ cells and/or preferential segregation to the egg nucleus rather than the polar body (49). Most B chromosomes appear to be mildly parasitic, and their maintenance can be readily explained by drive mechanisms. Recent studies reveal that B chromosomes in some organisms can code for beneficial effects as well, however (49). At the other end of the spectrum, the most extreme examples of SGEs are the B chromosomes found in haplodiploid insects that persist by destroying other chromosomes after fertilization of the egg (54).
Sex chromosome drive is widespread (50). As a result of evolution of drive repression, however, X drive is often cryptic and only revealed in crosses between populations or species. For example, there are at least three cryptic X-drive systems in Drosophila simulans alone (50). Drive can also be difficult to detect if there is not an associated phenotype or linked genetic marker to detect deviation from Mendelian ratios. Genomic techniques are now opening new avenues for detecting drive. For example, a recent study in chickens using genome-wide approaches revealed previously undetected drive around the centromere and telomeres of chromosome 1 (55), as predicted by the centric drive model. Such genome-wide approaches are likely to uncover many more examples of drive in the near future.
The possible role of meiotic drive in speciation has a controversial and interesting history. Frank (56) and Hurst and Pomiankowski (57) first proposed that divergence in X and Y meiotic drive and suppression of drive could lead to abnormal gametogenesis and sterility in hybrids. They were, in part, attempting to explain Haldane's rule: the observation that when hybrid incompatibilities are asymmetrical, it is usually the heterogametic sex (XY males or ZW females) that show hybrid sterility. The drive model was vigorously dismissed by leading speciation researchers at the time (58, 59) for two primary reasons: Meiotic drive was considered to be uncommon, and there was lack of direct empirical evidence for an association of drive and hybrid sterility. Today, there is mounting evidence in support of a significant role of meiotic drive in speciation (60–62). The change in landscape is attributable to the discovery that meiotic drive is often cryptic and much more common than previously thought and to detailed molecular and genetic studies of hybrid incompatibility genes that have revealed or implicated meiotic drive. Presgraves (61) concludes that some form of genetic conflict is implicated in ∼6 of the 14 hybrid incompatibility genes that have been relatively well characterized.
Post Segregation Distorters (PSDs).
A diverse array of PSDs exist, which, when present in an organism, act after progeny are produced to reduce the survival/fitness of progeny that have lost the driver. Although PSDs have arisen independently in many different organisms, a key feature of all PSDs is the involvement of a modification-rescue system, also called a toxin-antidote. I use the more general modification-rescue terminology because it does not assume a particular biochemical mechanism (cell toxicity) as the mode of action. A modification occurs in the “parent,” and this modification must be rescued in the offspring. If the PSD element is not transmitted to the offspring, rescue cannot occur and “harm” will come to the progeny (often death).
Dramatic examples of PSDs are the killer plasmids of bacteria and yeast (63). In killer plasmids, the longer persistence of the encoded toxin relative to the antidote protein ensures that daughter cells die if the plasmid is lost. This acts to prevent both segregation loss of the plasmid and its displacement by a competitor plasmid. Restriction-modification (R-M) systems also have PSD properties (64). They are widespread in bacteria and typically involve an enzyme that modifies DNA (e.g., by methylation) and a restriction enzyme that will cut DNA of a specific sequence that lacks the modification. These were originally believed to have evolved to provide protection to the cell against foreign DNA (e.g., bacteriophages). Kobayashi (64) has convincingly shown that R-M systems also behave as postsegregation killers. R-M systems located on plasmids are like other PSDs in having a modification and rescue. The restriction enzyme protein persists longer in progeny cells than does the modification enzyme; therefore, if the plasmid is lost in a bacterial daughter cell, the result will be restriction of its DNA and death. It has been shown that R-M systems do indeed kill cells that lose them, either through segregation or displacement by a different plasmid, thus maintaining the selfish R-M system.
The dynamics of PSDs, such as killer and R-M system plasmids, can be complex and dependent on population structure (64). In general, daughter cell killing only imparts an indirect and weak advantage to the progenitor cell lineage unless daughter cells compete with each other for resources, in which case the benefits are more direct. Therefore, the advantages occur most strongly in structured populations where related microbes co-occur. A second advantage of PSDs is prevention of displacement by competing plasmids. Some R-M systems are integrated into bacterial chromosomes, and this would seem to limit their advantage as postsegregation killers. Recent work shows that some integrated R-M systems are associated with TE-like structures that could facilitate their lateral movement between bacterial clones, however (65). PSDs may have been co-opted by bacterial and yeast genomes for defense against competitors, viral protection, or other phenotypes. For example, killer plasmids share some features with bacteriocins in bacteria and diffusible killer toxins in yeast, which can act to kill competitor cells lacking the linked modification-rescue mechanism (63). Bacteriocins typically produce diffusible toxins, however, and therefore target different cell lineages. Nevertheless, some bacteriocin systems may have PSD features and may have originated from PSD plasmids in some cases. Frank and Wolfe (63) have found the evolutionary addition of killer plasmid and killer virus DNA into yeast chromosomes, contributing to “killer chromosome” genotypes that produce diffusible substances targeting competitor cell lineages.
PSDs are also found in complex multicellular eukaryotes. A most intriguing example is the maternal effect dominant embryonic arrest (Medea) system in Tribolium beetles (66), which is chromosomally integrated and was discovered in crosses between populations with and without the driver. When females carry the Medea element, their zygotes must also receive the element (either maternally or paternally) or the offspring perish. The maternal modification factor and zygotic responder are tightly linked. Recent work suggests that Medea may be caused by a TE insertion just upstream of a gene with maternal and zygotic function. It is a quandary why Medea-like elements have not been found more widely in eukaryotes, but one explanation could be that they can quickly go to fixation in populations and become hidden. Medea elements have the potential to drive desirable traits into host populations (e.g., vector resistance to pathogens). To explore this idea, an artificial Medea element was successfully constructed in D. melanogaster by coupling a gene for micro-RNA silencing of a maternally expressed gene required for embryogenesis with a zygotically expressing rescue (67).
An usual example of B chromosome-induced PSD occurs in Nasonia and Trichogramma wasps, which have haplodiploid sex determination; haploid males develop from unfertilized eggs and females from fertilized eggs (54). The paternal sex ratio (psr) chromosome occurs in some males of these species. These males produce functional sperm, but psr induces improper condensation of the paternal chromosomes (except itself) in the fertilized egg, resulting in total loss of the normal paternal set. This converts the embryo into a haploid male that carries the supernumerary psr chromosome. That male's genome will be destroyed in the next generation, and psr will associate itself with yet another set of chromosomes destined for destruction. Because psr totally destroys the genome with which it becomes associated in each generation, it represents the most extreme example of an SGE in any organism. Haplodiploid sex determination promotes these extreme SGEs under certain population structures, and they have evolved independently in different wasp species.
Heritable Organelles and Microbes.
It may seem odd to include vital organelles, such as mitochondria and chloroplasts, in a treatise on SGEs. These organelles can have genetic interests that diverge from that of the nuclear genome, however, resulting in genetic conflict (Fig. 1). Mitochondria and chloroplasts evolved from ancient bacterial symbionts, and each retains its own DNA despite extensive transfer of genetic material to the nucleus. Such symbioses are not simply events of the ancient past, because heritable microorganisms (those that are inherited during the reproduction of their hosts, often through the egg cytoplasm) are widespread in plants and animals and involve a diverse array of microbial taxa. Once it was assumed that these microbes must always be beneficial to their hosts because they are dependent on host reproduction for their transmission. Although many heritable microbes are mutualistic, others manipulate host reproduction in ways that enhance the microbes’ transmission, and hence can be considered “reproductive parasites” (68).
The difference in inheritance patterns between heritable cytoplasmic elements and nuclear genes causes genetic conflict (8, 18, 69). With a few exceptions, inherited cytoplasmic elements are passed through the cytoplasm of the egg but not through sperm (in part, because of little cytoplasm in sperm). As a consequence, females transmit these elements, whereas males do not. In contrast, (autosomal) nuclear genes are typically inherited through both sexes. The result is cytonuclear conflict over sex determination and sex ratios, which, along with other forms of sex determination conflict, has likely played a role in sex determination evolution (18). For example, mitochondrial variants induce pollen sterility in many plants, resulting in evolution of nuclear suppressor genotypes. Some of these systems are cryptic, as a result of evolutionary suppression, but are revealed in crosses between populations or related species.
Many different inherited microbes have evolved mechanisms to manipulate host reproduction because of their preferential transmission through females (14, 18, 68). Among the kinds of manipulations are conversion of males to functional females, induction of parthenogenetic reproduction in females, male-killing, and a form of sperm-egg incompatibility termed “cytoplasmic incompatibility” (CI). Noteworthy by its abundance in invertebrates and ability to perform all these manipulations is the α-proteobacterium Wolbachia (68). This bacterium is transmitted through eggs but also moves laterally between taxa. As a result, it is found in ∼70% of terrestrial arthropods (70). Many strains of Wolbachia induce a form of sperm-egg incompatibility called CI. These Wolbachia strains modify the sperm (by unknown biochemical mechanisms), such that the same strain of Wolbachia must be present in the egg to rescue the modification. If not, the sperm chromatin condenses improperly in the embryo, usually killing the offspring. By reducing the fitness of uninfected females, the infection can spread very rapidly in populations. Different strains of Wolbachia can be reciprocally incompatible because of differences in their modification-rescue system or host genetic interactions. An interesting possible case of evolved dependency occurs in the wasp Asobara tabida (71). Removal of Wolbachia results in sterility as a result of elevated apoptosis in the developing female reproductive tract. Because closely related species do not require Wolbachia for ovarian development, this is likely either a case of PSD (killing of stem cells that have lost the bacteria) or an evolved host dependency on the presence of the parasite in reproductive tissues, which results in abnormal apoptosis in their absence. Such evolved dependencies to SGEs are likely to be common and are distinct from mutualisms, even though both will result in reduced fitness when the element is removed or inhibited.
Do inherited symbionts promote speciation in their hosts? Because CI can induce partial or complete reproductive incompatibility between diverging populations with different infections, it may promote reproductive isolation and speciation (reviewed in refs. 72 and 73). For example, in the Nasonia species complex, reciprocal CI between the species is a major contributor to hybrid reproductive incompatibilities and evolved early in the speciation process. A general role for Wolbachia in speciation has been criticized for several reasons, however, including the beliefs that (i) Wolbachia differences will not be stable between populations, (ii) CI is insufficient to maintain genetic divergence between populations, and (iii) Wolbachia infections are not common enough to be a major player. Nevertheless, supporting data continue to grow, including theoretical studies indicating that CI differences can be stable, maintain divergence, and select for premating isolation; empirical studies showing CI as contributing to reproductive isolation and reinforcement of mate discrimination between species (74); and the finding that Wolbachia infections are much more common than previously recognized (in ∼70% of species rather than original estimates of 20%). Parthenogenetic species have also arisen courtesy of Wolbachia (75). In haplodiploids, Wolbachia causes parthenogenesis by inducing diploidization of unfertilized haploid eggs, which leads to female development. Parthenogenesis causing Wolbachia often occurs as a polymorphism in sexual species, but host genetic changes favoring females that do not mate can lead to fully parthenogenetic species (75). Subsequent loss of genes needed for sexual reproduction makes the process irreversible. Over 20 examples of Wolbachia-induced parthenogenetic species have been described in haplodiploids.
Wolbachia occurs within the germline in intimate proximity to the nucleus. An exciting recent discovery is that lateral gene transfers from Wolbachia to animals are common (76). Approximately one-third of invertebrate genomes show such transfers, sometimes involving large amounts of DNA (e.g., nearly the entire 1.2-MB genome of Wolbachia in Drosophila anannassae). Such transfers can either degrade over evolutionary time as a result of mutation accumulation or evolve novel functional genes, and there is evidence of the latter in some species (76, 77).
Other Evolutionary Consequences of SGEs.
SGEs have been invoked to play a role in many important biological phenomena, with variable levels of empirical and theoretical support. Examples include the evolution of sex, recombination, anisogamy, germline sequestration and uniparental inheritance of plastids, and alteration of mating systems (14–16). Here, I will mention a few recent studies of interest. The scarcity of males caused by meiotic drivers and cytoplasmic sex ratio distorters has selected for changes in mating systems (78). The germ granule, a key constituent in germ cell determination, contains proteins important in TE suppression (79), and SGEs may have promoted the evolution of a sequestered germline (80). Suzuki et al. (35) propose that DNA methylation suppression of TEs was a precursor for the evolution of genomic imprinting and the placenta in mammals. Such studies suggest that SGEs have an enormous potential range of evolutionary consequences.
Evolutionary Dynamics, Evolvability, and “Function” of SGEs.
A long-standing debate concerns the evolutionary function of TEs, and this debate has been reinvigorated with recent discoveries of their evolutionary domestication into functional genes. Are TEs maintained over long evolutionary time scales because they induce beneficial mutations and innovations, thus allowing species to adapt (the evolvability hypothesis) or because they are self-replicating elements that are maintained by replicating at faster rates than they are lost (the parasite hypothesis)? Note that the parasite hypothesis does not preclude TE insertions evolving beneficial functions in the host (i.e., domestication) but argues that this is a consequence of TEs rather than the “reason” for the existence of TEs. There has been a recent resurgence of articles either implicitly or explicitly stating that an important evolutionary “function” of TEs is to promote genetic innovation and evolvability (81–84). For example, Aziz et al. (83) argue that the ubiquity of TEs is proof that they must exist to provide benefits in evolution. They state that “ubiquity is one of the indicators of essentiality” and further claim that these elements are “indispensible in every genome (elements of core genome) or every ecosystem (eco-essential genes)” (83). The near-ubiquity of TEs can be readily explained by their ability to replicate within genomes and to move laterally between species, however. There is ample evidence that TEs can accumulate in genomes as a result of transposition and induce harmful mutations, that organisms have evolved many mechanisms to suppress TEs, that genomes are littered with fossil suppressed and degraded elements, and that TEs move laterally between species, often across large evolutionary distances. These observations are consistent with the parasitic hypothesis, and no additional conditions are needed to explain the persistence of TEs over evolutionary time or their widespread occurrence. In contrast, there is much less direct evidence supporting the argument that TEs persist in evolution because they enhance the evolvability of organisms or play some vital role in ecosystems (81, 83).
To evaluate the concept of evolvability as it applies to SGEs, the questions need to be framed clearly. The evolvability concept can be applied to short-term adaptive evolution or long-term adaptation and innovation. Implicit to the short-term concept is that active TEs are maintained because of the beneficial mutations they induce (i.e., those insertions contribute substantially to the pool of active elements). If this is correct, there should be evidence of recent active TE insertions associated with adaptive evolutionary changes within species. TEs can be a significant source of standing genetic variation (33), but the question remains of how often they induce beneficial as opposed to deleterious mutations. There is ample evidence of mutational costs of TE insertion events (21, 32, 33). There has been relatively little evidence that young insertions lead to adaptive mutations, although some possible cases have emerged recently (85). Examples include inserts associated with pesticide resistance in D. melanogaster and D. simulans as well as a genome-wide study implicating TE insertions with temperature adaptation in two separate latitudinal clines in D. melanogaster.
González and Petrov (85) also note that if adaptive TE insertions are frequent, we should observe fixation of TE insertions in species more often in high-recombination regions than are observed. They offer one possible explanation that beneficial insertions quickly evolve through mutation, and therefore are not readily recognizable as TEs. If true, this would suggest that they then contribute little to the generation of new active TEs. It is also possible that they are maintained as ancient polymorphisms in related species; however, again, we would expect them to degrade as active TEs. A third possible explanation is that TE insertions provide short-term adaptive mutations (e.g., attributable to habitat differences) but that they are eventually replaced by point mutations with fewer negative pleiotropic consequences. Although the data for frequent adaptive insertions are tantalizing, they are still largely indirect. Nevertheless, the findings suggest that a more nuanced evaluation of the mechanisms that maintain active TEs is needed, which includes models that incorporate deleterious, neutral, and beneficial insertions as well as mutational changes in insertions (86). The key empirical and theoretical question becomes “What portion of TE transposition comes from elements associated with beneficial vs. neutral or deleterious effects?” This is crucial for determining what maintains active TEs in a species.
A key observation argues against beneficial insertions being important in the maintenance of active TEs. If TEs are beneficial, we would expect to see specific TE types being maintained within lineages over evolutionary time (i.e., the phylogenies of TE elements should parallel that of the organisms in which they occur). With a few exceptions, this is not the case. Instead, the patterns of TE variation in most species indicate that they tend to invade a species, rapidly proliferate, and then are suppressed (28, 38).
The long-term evolvability argument is that TEs exist because of their long-term contributions to adaptation and innovation (e.g., new genes and gene regulation networks). In its naive form, the long-term evolvability argument is teleological and confuses cause and consequence. Simply, evolution is not anticipatory, and the observation that TE inserts can evolve into functional genes is not proof that they are maintained because they provide genetic material for long-term evolutionary innovation. The idea also suffers from the irreversibility problem. Once a TE insertion has evolved into a function gene or regulatory element, it is unlikely to evolve back into a TE, and thus does not contribute to the pool of active TEs supposedly being maintained for their long-term benefits. In other words, there is no evolutionary feedback to maintain active TEs for long-term benefits.
A more reasonable hypothesis for maintenance of TEs attributable to long-term benefits is based on clade selection (i.e., competition among lineages of species) (81, 84). The clade selection hypothesis is that those clades (e.g., species, genera) with active TE elements are more likely to persist and radiate because of their ability to evolve to changing environments or to evolve innovations. This hypothesis is difficult to test because of the near-ubiquity of TEs. The parasitic vs. evolvability hypotheses do make clear contrasting predictions about what should happen in asexual vs. sexual species, however. The parasitic hypothesis predicts that active TEs will decline in asexual species because of the deleterious effects of transposition, whereas the evolvability hypothesis predicts that they will be maintained (or increase) because of their beneficial effects. Indeed, the benefits of TEs might be expected to be even greater in asexual species as a source of beneficial mutations to resist mutational decline and for adaptation to new environments. What few data currently exist support the parasitic hypothesis. Asexual Bdelloid rotifers have significantly reduced TE numbers compared with sexual relatives, including loss of retroelements (87), and asexual Daphnia species show reduced numbers of active TEs compared with sexual Daphnia species (88). More comparisons are needed between related asexual and sexual species to test the predictions of these alternative hypotheses.
A theme in some recent papers is that although TEs were “dismissed” over the past several decades as mere selfish DNA, new evidence now shows that they have evolved functions within the genome (81, 83). Although perhaps a useful literary foil, these statements are not very accurate. Even the original paper by Doolittle and Sapienza (6), which asserted that repetitive elements can be maintained because of their self-replicating properties, goes on to state that “we do not deny that [such elements] may have roles of immediate phenotypic benefit to the organism. Nor do we deny roles for these elements in the evolutionary process.” This same theme has been maintained by advocates of the parasitic hypothesis for the past 2 decades (10, 14–16). Discoveries that TE insertions can evolve into function sequences and induce favorable mutations are not contrary to the SGE hypothesis for TE maintenance. What has been criticized by Doolittle and Sapienza (6) onward are uncritical evolvability claims that TEs exist because they provide evolutionary benefits to organisms. As outlined above, evolvability arguments need to be precisely framed (and in nonteleological terms) to provide testable predictions, such as that beneficial mutations are required for maintenance of active TEs or that clade selection favors lineages with active TEs. This can then lead to more rigorous tests of the evolvability idea. To date, the data continue to support the view that TEs have important evolutionary consequences but are maintained because of their selfish replicative features.
Conclusions
Rapidly growing evidence emerging from genomics and advances in genetics indicate that SGEs are important motors for evolutionary change and innovation. Several general principles have reoccurred in the discussion above, and I will briefly revisit these themes here. The first is that SGEs lead to antagonistic coevolution with other components of the genome. Important features of eukaryotic genomes (e.g., DNA methylation, RNAi, small RNA regulatory pathways, R-M systems) have evolved, at least in part, as defense mechanisms against SGEs. Many genetic elements have mixed phenotypes, with both selfish (parasitic) and “beneficial” (“mutualistic”) features. The classic example is the mitochondrion, which is clearly beneficial but also shows selfish features (e.g., cytoplasmic male sterility) that reduce nuclear gene fitness, thus leading to genetic conflict. Evolutionary dependency can also evolve in hosts with ubiquitous SGEs, which can lead to irreversible dependence. Growing evidence supports a significant role of SGEs in eukaryotic development and speciation, and possibly also in extinction of species. Genome domestication of SGEs leads to evolutionary innovations, including acquisition of new genes and gene regulation from TEs, heritable microbes (e.g., Wolbachia), and selfish plasmids. Safe havens can promote longer associations of SGEs with host lineages and also may facilitate their domestication. Finally, distinctions are made between the evolutionary consequences of SGEs and the factors that maintain them over evolutionary time. Clear formulations of the idea of evolvability as a means for evolutionary maintenance of SGEs will facilitate rigorous testing of this idea. Nevertheless, current evidence strongly supports the view that SGEs are maintained by their transmission-enhancing phenotypes and that evolutionary innovations emerging from them are a consequence of their existence rather than the cause.
Acknowledgments
I thank J. Strassmann, D. Queller, J. Avise, and F. Ayala for organizing In the Light of Evolution V: Cooperation; and A. Avery, R. Edwards, T. Eickbush, C. Feschotte, D. Loehlin, M. Clark, J. Jaenike, M. Lenoe, C. Landry, R. Minckley, P. Nagarajan, and D. Wheeler for assistance and comments on the manuscript. Because of space limitations, topic reviews are sometimes cited in lieu of the original research paper and the reader is encouraged to access the reviews for primary research sources. Support for this work came from National Science Foundation Grants DEB0821936 and NIH GM084917.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution V: Cooperation and Conflict,” held January 7–8, 2011, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/SACKLER_cooperation.
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gershenson S. A new sex-ratio abnormality in Drosophila obscura. Genetics. 1928;13:488–507. doi: 10.1093/genetics/13.6.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Östergren G. Parasitic nature of extra fragment chromosomes. Bot Not. 1945;2:157–163. [Google Scholar]
- 3.Williams GC. Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought. Princeton: Princeton Univ Press; 1996. [Google Scholar]
- 4.Dawkins R. The Selfish Gene. London: Paladin; 1976. [Google Scholar]
- 5.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 7.Orgel LE, Crick FH. Selfish DNA: The ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 8.Cosmides LM, Tooby J. Cytoplasmic inheritance and intragenomic conflict. J Theor Biol. 1981;89:83–129. doi: 10.1016/0022-5193(81)90181-8. [DOI] [PubMed] [Google Scholar]
- 9.Nur U, Werren JH, Eickbush DG, Burke WD, Eickbush TH. A “selfish” B chromosome that enhances its transmission by eliminating the paternal genome. Science. 1988;240:512–514. doi: 10.1126/science.3358129. [DOI] [PubMed] [Google Scholar]
- 10.Werren JH, Nur U, Wu CI. Selfish genetic elements. Trends Ecol Evol. 1988;3:297–302. doi: 10.1016/0169-5347(88)90105-X. [DOI] [PubMed] [Google Scholar]
- 11.Avise JC. Evolving genomic metaphors: A new look at the language of DNA. Science. 2001;294:86–87. doi: 10.1126/science.294.5540.86. [DOI] [PubMed] [Google Scholar]
- 12.Rice WR, Holland B. The enemies within: Intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 13.Frank SA, Crespi BJ. Pathology from evolutionary conflict, with a theory of X chromosome versus autosome conflict over sexually antagonistic traits. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10886–10893. doi: 10.1073/pnas.1100921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurst GDD, Hurst LD, Johnstone RA. Intranuclear conflict and its role in evolution. Trends Ecol Evol. 1992;7:373–378. doi: 10.1016/0169-5347(92)90007-X. [DOI] [PubMed] [Google Scholar]
- 15.Hurst GDD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 16.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Belknap; 2006. [Google Scholar]
- 17.Johnson LJ. The genome strikes back: The evolutionary importance of defence against mobile elements. Evol Biol. 2007;34:121–129. [Google Scholar]
- 18.Werren JH, Beukeboom LW. Sex determination, sex ratios, and genetic conflict. Annu Rev Ecol Syst. 1998;29:233–261. [Google Scholar]
- 19.Maynard Smith J, Szathmary E. The Major Transitions in Evolution. Oxford: Oxford Univ Press; 1995. p. 360. [Google Scholar]
- 20.Haig D. The kinship theory of genomic imprinting. Annu Rev Ecol Syst. 2000;31:9–32. [Google Scholar]
- 21.Kidwell MG, Lisch DR. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 22.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 23.Newton ILG, Bordenstein SR. Correlations between bacterial ecology and mobile DNA. Curr Microbiol. 2011;62:198–208. doi: 10.1007/s00284-010-9693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biémont C, Vieira C. Genetics: Junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 26.Blumenstiel JP. Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet. 2011;27:23–31. doi: 10.1016/j.tig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Bennetzen JL. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev. 2005;15:621–627. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends in Ecology & Evolution. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartolomé C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eickbush TH, Eickbush DG. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371 doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell KA, Burns KH. Mobilizing diversity: Transposable element insertions in genetic variation and disease. Mob DNA. 2010;1:21. doi: 10.1186/1759-8753-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantu D, et al. Small RNAs, DNA methylation and transposable elements in wheat. BMC Genomics. 2010;11:408–422. doi: 10.1186/1471-2164-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki S, et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekita Y, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243–248. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- 37.Sinzelle L, Izsvák Z, Ivics Z. Molecular domestication of transposable elements: From detrimental parasites to useful host genes. Cell Mol Life Sci. 2009;66:1073–1093. doi: 10.1007/s00018-009-8376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Zhang Y, Long M. Extensive structural renovation of retrogenes in the evolution of the Populus genome. Plant Physiol. 2009;151:1943–1951. doi: 10.1104/pp.109.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fablet M, Bueno M, Potrzebowski L, Kaessmann H. Evolutionary origin and functions of retrogene introns. Mol Biol Evol. 2009;26:2147–2156. doi: 10.1093/molbev/msp125. [DOI] [PubMed] [Google Scholar]
- 41.Bringaud F, et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vinogradov AE. Genome size and extinction risk in vertebrates. Proc Biol Sci. 2004;271:1701–1705. doi: 10.1098/rspb.2004.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolgin ES, Charlesworth B. The fate of transposable elements in asexual populations. Genetics. 2006;174:817–827. doi: 10.1534/genetics.106.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuzhdin SV, Petrov DA. Transposable elements in clonal lineages: Lethal hangover from sex. Biol J Linn Soc Lond. 2003;79:33–41. [Google Scholar]
- 45.Zeh DW, Zeh JA, Ishida Y. Transposable elements and an epigenetic basis for punctuated equilibria. Bioessays. 2009;31:715–726. doi: 10.1002/bies.200900026. [DOI] [PubMed] [Google Scholar]
- 46.Stoddard BL. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 48.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 49.Camacho JP, Sharbel TF, Beukeboom LW. B-chromosome evolution. Philos Trans R Soc Lond B Biol Sci. 2000;355:163–178. doi: 10.1098/rstb.2000.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jaenike J. Sex chromosome meiotic drive. Annu Rev Ecol Syst. 2001;32:25–49. [Google Scholar]
- 51.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 52.Presgraves D. Drive and sperm: Evolution and genetics of male meiotic drive. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Burlington, MA: Elsevier; 2009. pp. 471–506. [Google Scholar]
- 53.Lyttle TW. Segregation distorters. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- 54.Werren JH, Stouthamer R. PSR (paternal sex ratio) chromosomes: The ultimate selfish genetic elements. Genetica. 2003;117:85–101. doi: 10.1023/a:1022368700752. [DOI] [PubMed] [Google Scholar]
- 55.Axelsson E, et al. Segregation distortion in chicken and the evolutionary consequences of female meiotic drive in birds. Heredity. 2010;105:290–298. doi: 10.1038/hdy.2009.193. [DOI] [PubMed] [Google Scholar]
- 56.Frank SA. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 57.Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: A new explanation of Haldane's rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coyne JA. Genetics and speciation. Nature. 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 59.Coyne JA, Orr HA. Further evidence against meiotic-drive models of hybrid sterility. Evolution. 1993;47:685–687. doi: 10.1111/j.1558-5646.1993.tb02123.x. [DOI] [PubMed] [Google Scholar]
- 60.McDermott SR, Noor MAF. The role of meiotic drive in hybrid male sterility. Philos Trans R Soc Lond B Biol Sci. 2010;365:1265–1272. doi: 10.1098/rstb.2009.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 62.Johnson NA. Hybrid incompatibility genes: Remnants of a genomic battlefield? Trends Genet. 2010;26:317–325. doi: 10.1016/j.tig.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Frank AC, Wolfe KH. Evolutionary capture of viral and plasmid DNA by yeast nuclear chromosomes. Eukaryot Cell. 2009;8:1521–1531. doi: 10.1128/EC.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furuta Y, Abe K, Kobayashi I. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 2010;38:2428–2443. doi: 10.1093/nar/gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorenzen MD, et al. The maternal-effect, selfish genetic element Medea is associated with a composite Tc1 transposon. Proc Natl Acad Sci USA. 2008;105:10085–10089. doi: 10.1073/pnas.0800444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen CH, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 68.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 69.Eberhard WG. Evolutionary consequences of intracellular organelle competition. Q Rev Biol. 1980;55:231–249. doi: 10.1086/411855. [DOI] [PubMed] [Google Scholar]
- 70.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kremer N, et al. Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog. 2009;5:e1000630. doi: 10.1371/journal.ppat.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Werren JH. Endless Forms: Species and Speciation. New York: Oxford University Press; 1998. Wolbachia and speciation; pp. 245–260. [Google Scholar]
- 73.Bordenstein SR. Insect Symbiosis. Boca Raton: CRC Press; 2003. Symbiosis and the origin of species; pp. 283–304. [Google Scholar]
- 74.Jaenike J, Dyer KA, Cornish C, Minhas MS. Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol. 2006;4:e325. doi: 10.1371/journal.pbio.0040325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stouthamer R, Russell JE, Vavre F, Nunney L. Intragenomic conflict in populations infected by Parthenogenesis Inducing Wolbachia ends with irreversible loss of sexual reproduction. BMC Evol Biol. 2010;10:229. doi: 10.1186/1471-2148-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunning Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 77.Werren JH, et al. Nasonia Genome Working Group Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Price TA, Hodgson DJ, Lewis Z, Hurst GD, Wedell N. Selfish genetic elements promote polyandry in a fly. Science. 2008;322:1241–1243. doi: 10.1126/science.1163766. [DOI] [PubMed] [Google Scholar]
- 79.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson LJ. Selfish genetic elements favor the evolution of a distinction between soma and germline. Evolution. 2008;62:2122–2124. doi: 10.1111/j.1558-5646.2008.00433.x. [DOI] [PubMed] [Google Scholar]
- 81.Biémont C. A brief history of the status of transposable elements: From junk DNA to major players in evolution. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci USA. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oliver KR, Greene WK. Transposable elements: Powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- 85.González J, Petrov DA. The adaptive role of transposable elements in the Drosophila genome. Gene. 2009;448:124–133. doi: 10.1016/j.gene.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Rouzic A, Boutin TS, Capy P. Long-term evolution of transposable elements. Proc Natl Acad Sci USA. 2007;104:19375–19380. doi: 10.1073/pnas.0705238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arkhipova I, Meselson M. Transposable elements in sexual and ancient asexual taxa. Proc Natl Acad Sci USA. 2000;97:14473–14477. doi: 10.1073/pnas.97.26.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schaack S, Pritham EJ, Wolf A, Lynch M. DNA transposon dynamics in populations of Daphnia pulex with and without sex. Proc Biol Sci. 2010;277:2381–2387. doi: 10.1098/rspb.2009.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]