Abstract
The critical challenge in virtually all cancer research is heterogeneity: “Breast cancer” and “lung cancer” are actually collections of disease with distinct molecular mechanisms and clinical characteristics. The challenge is evident in the complexity of most cancers with multiple mutations and alterations generating the cancer phenotype, requiring therapeutic strategies that can match the complexity with equally complex combination regimens. Substantial progress in treatment requires major advances in methods to define refined, “common mechanism” subgroups to allow development of combination therapeutics that target these individual mechanisms. Our work is on the use of genomic signatures of oncogenic signaling pathways that provide an opportunity to dissect the complexity of lung cancer and to serve as tools to direct the use of targeted therapeutic agents.
Keywords: lung cancer, genomics, pathway signatures
Lung cancer is not one disease but rather a multitude of disorders with distinct underlying mechanisms. Increasing evidence suggests that disruption of cellular pathways, rather than individual mutations, is largely responsible for directing tumorigenesis. Varied combinations of altered cellular pathways are responsible for patient–patient differences in tumor characteristics and may explain why some patients respond favorably to specific therapies, whereas others are resistant to these treatments. The focus of our work is squarely on these issues in lung cancer, aiming to develop and translate to clinical practice the use of multiple pathway signatures to dissect the complexity of non–small cell lung cancer (NSCLC), improve understanding of disease subtypes, and define and validate opportunities for novel therapeutic strategies.
The importance of dissecting the heterogeneity is no better illustrated than via Herceptin, a key breast cancer drug but relevant only in the small fraction of patients with Her2+ breast cancer. Given the complexity of most cancers with multiple mutations and alterations generating the cancer phenotype, developing therapeutic strategies that can match the complexity with equally complex combination regimens is a significant challenge. Substantial progress in treatment requires major advances in methods to define homogeneous subgroups with common mechanisms of disease to allow the development of combination therapeutics that target these individual mechanisms.
Single markers are often ineffective in identifying characteristics of tumors that define therapeutic responses. Only 30% of patients with Her2+ breast cancer respond to Herceptin, and similar results are seen in colon cancer, where response to Cetuximab is determined by K-Ras mutation status, but only a fraction of the wild-type Ras tumors respond to the drug. Our previous work focused, in part, on the development of genomic signatures of deregulation of oncogenic pathways important in NSCLC; these signatures can accurately predict responses to various therapeutics and chemotherapies in vitro, in vivo, and in human studies. The use of signatures to dissect the complexity of NSCLC can provide a detailed understanding of disease subtypes and opportunities for novel therapeutic strategies.
The overwhelming challenge in the understanding and treatment of NSCLC is to develop rational strategies for stratification and matching with therapeutics, including combination therapies. Our approach to this challenge is to use signatures of pathway activity as a basis for dissecting the heterogeneity of lung cancer samples, thereby vastly improving understanding and quantitative breakdown of the subtle distinctions in tumor phenotypes and reforming the ability to select drug combinations targeting these pathways. The clinical impact lies in this strategy as a basis for defining effective, individualized combinations of therapies.
Treatments with pathway-specific therapeutics have shown encouraging clinical success, but most patients with advanced solid tumors relapse and die of their disease. The challenge is to identify subpopulations of individuals who are most likely to respond to a given therapy. Traditional methods depend on the gross visual characterization of the tumor as well as a limited number of biochemical assays. Although recent work has shown the ability of specific biomarkers to classify patients for certain anticancer drugs, most therapies have no basis to guide their use. Furthermore, a single biomarker may be insufficient to predict response to a specific therapy, and these techniques are limited in their capacity to adequately define tumor subgroups with distinct biology, resulting in imprecisely defined, heterogeneous classes of tumors and patients. Because many recently developed therapeutic agents for cancer treatment target oncogenic cellular pathways for which the mutation or expression (altered or normal) of a single gene or protein is insufficient to predict pathway activity or drug efficacy, alternative tumor classification strategies are required to effectively target therapies to the proper groups of patients.
Perhaps the most striking limitation in the study of lung cancer is the absence of a robust framework for understanding the complexity and diversity of disease. Unlike the advances seen in breast cancer, as well as other cancers, that have made use of genomic data to identify molecular subtypes of disease with distinct biological properties, there has been relatively little done in lung cancer to achieve this goal beyond the work describing prognostic signatures. We believe there is an opportunity for substantially improved tumor classification, based on the activity of multiple cellular pathways, which will improve understanding of disease mechanisms and identify patient subgroups for targeted administration of pathway-specific chemotherapeutics. This will increase response rates to current adjuvant therapies, lead to more effective multidrug combination regimens, and reduce cytotoxic side effects and the development of chemotherapeutic resistance, thereby advancing the development of personalized approaches to the treatment of the disease.
CONCEPT OF AN EXPRESSION SIGNATURE
An overarching concept that characterizes our work is the use of gene expression data to develop “signatures” that represent cancer phenotypes. Expression signatures represent biological states, define surrogate representations of biological phenotypes, and are portable: A signature developed in a cell culture context can be used to measure pathway activity in a tumor sample, linking experimental states with in vivo states. Our past work, among others, defines the clinical predictive value of expression signatures reflecting phenomena, such as hormone receptor status, disease outcome, response to therapies, response to hypoxia, or the activation state of various signaling pathways, that contribute to oncogenic progression (1–4).
Our statistical models for classification and prediction of cancer phenotypes use “metagene” expression signatures (1) as markers of clinical outcomes, drug response, and predictors of modules of activation or deregulation of specific oncogenic and signaling pathways. Our past success in application of cancer-relevant expression phenotypes underlies our approach to developing methods that can translate to the improved use of targeted cancer therapeutics.
Our general approach is one that makes use of expression signatures developed to measure the activation state of various oncogenic signaling pathways. We use these signatures in a manner similar to the use of full-genome expression data as a means to identify subgroups of cancers. At the same time, these signatures have been shown to predict sensitivity to targeted therapeutics that can be matched with the individual pathways. As such, this provides an approach to identifying therapeutic opportunities that can be matched with the characteristics of individual tumors.
GENOMIC APPROACHES TO THE STUDY OF ONCOGENIC PATHWAYS
We have used gene expression profiles to identify signatures predictive of deregulated oncogenic pathways. These signatures provide a measure of the consequence of the oncogenic process, irrespective of how the pathway might have been altered. Thus, even if the known oncogene is not mutated, but rather another component of the pathway is altered, the expression profile detects the alteration. Recombinant adenoviruses containing various oncogenes were used to activate an otherwise quiescent cell, thereby isolating the subsequent events as defined by that single pathway activation and deregulation. Assays of various known pathway targets or activation events associated with the pathways provided confirmation that this approach led to pathway activation. Pathway gene expression signatures were identified using supervised classification methods of analysis as previously described (1–3). Metagene expression signatures represent groups of genes that together exhibit a consistent pattern of expression in the collection of samples and can be defined by genes most highly correlated with the classification of cell line samples into oncogene-activated/deregulated versus control. The dominant principal component from such a set of genes defines a phenotype-related metagene, and regression models assign the probability of pathway deregulation in tumor or cell line samples. Figure 2 illustrates examples of oncogenic signaling pathway signature development.
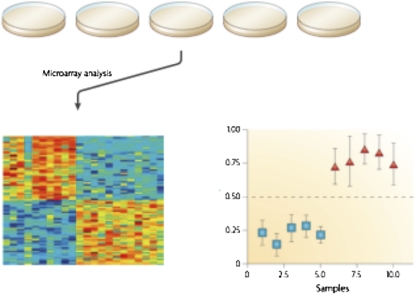
Figure 1.
Generation of an expression signature. A collection of cell cultures are assayed under specific conditions that define two states (pathway off/on). RNA is prepared from the cells and used for DNA microarray analysis. These data are then used for a supervised analysis in which a signature is derived that distinguishes the two cell states (bottom left). The signature is then evaluated in a test dataset to generate a probability score reflect the extent to which a sample exhibits the signature (bottom right).
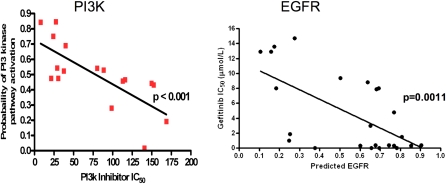
Figure 3.
Correlation of predicted pathway activity with drug sensitivity. RNA was prepared from a collection of cancer cell lines and used for microarray analysis and prediction of PI3K pathway activity (left) or epidermal growth factor receptor (EGFR) pathway activity (right) using relevant pathway signatures. Sensitivity to a PI3K inhibitor was also measured across the panel of cells (as IC50 values) as well as sensitivity to an EGFR tyrosine kinase inhibitor and then compared with the predicted pathway activity (as measured as a probability from 0 to 1).
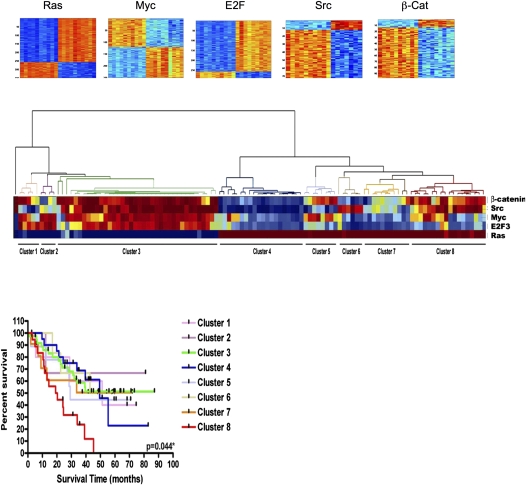
Figure 2.
Gene expression signatures of oncogenic pathway activity: patterns of pathway activity in non–small cell lung cancer (NSCLC) samples. Top: Expression images of genes in signatures of indicated pathways (red and blue: high and low expression, respectively; columns and rows: samples and genes, respectively) ordered by gene-signature regression coefficients. Bottom: Heatmap displaying prediction of pathway activation in NSCLC samples using pathway signatures (red and blue: high and low activation, respectively). Samples are clustered based on predicted pathway activation that relates clearly to survival of patients. (Right) Survival curves for patients within pathway-defined clusters.
A key use of expression signatures as predictors of pathway activity is the capacity to generate quantitative estimates, expressed as a probability that can be assessed in a collection of tumor samples. Moreover, these quantitative measures can be used as a basis for identifying patterns of overlapping pathway activity, displayed by hierarchical clustering. In short, the predicted pathway probabilities can be used in a manner similar to the use of raw gene expression data to identify structure within a tumor dataset. An example for profiling the status of various pathways in a series of lung cancer samples has been described (3), where clustering based on the oncogenic pathway signatures revealed distinct patterns in which subgroups of tumors were identified based on pathway patterns.
This analysis demonstrates the ability to identify patterns of pathway deregulation that coincide with clinical outcome because clusters identify patients with distinct survival characteristics. Also, the pathway analysis provides an opportunity within any selected group of patients to potentially match a targeted therapeutic agent with a patient based on the status of the relevant pathway. For instance, a group of patients exhibiting patterns of Src deregulation might be appropriate for a Src kinase inhibitor. Conversely, patients with activated Ras pathway would be treated with a drug that targets the Ras pathway.
USE OF PATHWAY SIGNATURES TO GUIDE THE USE OF TARGETED THERAPEUTICS
The potential to affect treatment decisions is shown in our work using prediction of pathway deregulation to predict drug sensitivity (3). We mapped the pathway deregulation to a series of lung cancer cell lines used for screening potential therapeutic drugs, using an initial limited set of available pathway. The probability of pathway activation in a given cell line is predicted from the experimentally developed signature using cell lines for assays with drugs that are known to target specific activities in oncogenic pathways. Assays consider growth inhibition measurements using standard colorimetric assays, as well as colony-forming assays in soft agar. It is evident from these studies that there is a strong predictive relationship between the probability of pathway activation and response to a pathway-specific inhibitor.
This pathway-centric systems view of cancer genome alterations provides a basis for understanding the complexity of cancer-associated gene mutations and advances in the ability to predict the functional and phenotypic consequences. Sequencing initiatives emphasize the interpretation of sequence data in the context of effects on distinct cell signaling pathways. Although mutations in any of several genes in a pathway can yield indistinguishable results in cellular proliferation, combinations of several altered cellular pathways define differences in tumors, and pathway assessment can predict variations in responses to therapies. Hence, characterizing tumors via multiple relevant pathways is practically mandated, and this underlies our goals of defining improved, quantified predictions of tumor status and subtypes. Our initial analyses of lung cancer indicates that gene expression patterns, as well as pathway patterns, can reveal variation beyond simple histology or smoking status, emphasizing the power to go beyond the limitations of current clinical information.
Predictive subgrouping based on pathway patterns also allows classification of cancer cell lines as experimental models of a given subgroup. We can evaluate the predicted pattern of pathway activity within a given cell line and use this information as the basis for prediction of membership in one of the subgroups, as has been described in the breast cancer work recently published (4). A similar approach can provide an opportunity to evaluate a large collection of lung cancer cell lines for pathway patterns that can be related to patient tumor samples. Again, this provides a robust and quantitative approach to determining the extent to which a given cell line is truly representative of a tumor subgroup. These predictions will provide the basis for cell line selection for xenograft studies that have the goal of exploring therapeutic strategies that can be matched with individual subgroups based on patterns of predicted pathway activity.
Supported by grants CA112952–05S1 and CA106520–06.
Author Disclosure: J.N. is on the Advisory Board for Millennium Pharmaceuticals and Qiagen. He owns Founders Shares of Expression Analysis and received grant support from the NIH and the V-Foundation.
References
- 1.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci USA 2001;98:11462–11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang E, Ishida S, Pittman J, Dressman H, West M, Nevins JR. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet 2003;34:226–230. [DOI] [PubMed] [Google Scholar]
- 3.Bild A, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi M-B, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353–357. [DOI] [PubMed] [Google Scholar]
- 4.Gatza ML, Lucas JE, Barry WT, Kim J-W, Wang Q, Crawford M, Datto M, Kelley M, Mathey-Prevot B, Potti A, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA 2010;107:6994–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]