Abstract
Glacier forefields are ideal ecosystems to study the development of nutrient cycles as well as single turnover processes during soil development. In this study, we examined the ecology of the microbial nitrogen (N) cycle in bulk soil samples from a chronosequence of the Damma glacier, Switzerland. Major processes of the N cycle were reconstructed on the genetic as well as the potential enzyme activity level at sites of the chronosequence that have been ice-free for 10, 50, 70, 120 and 2000 years. In our study, we focused on N fixation, mineralization (chitinolysis and proteolysis), nitrification and denitrification. Our results suggest that mineralization, mainly the decomposition of deposited organic material, was the main driver for N turnover in initial soils, that is, ice-free for 10 years. Transient soils being ice-free for 50 and 70 years were characterized by a high abundance of N fixing microorganisms. In developed soils, ice-free for 120 and 2000 years, significant rates of nitrification and denitrification were measured. Surprisingly, copy numbers of the respective functional genes encoding the corresponding enzymes were already high in the initial phase of soil development. This clearly indicates that the genetic potential is not the driver for certain functional traits in the initial phase of soil formation but rather a well-balanced expression of the respective genes coding for selected functions.
Keywords: real-time PCR, nitrogen cycle, functional genes, soil development, chronosequence
Introduction
Since the end of the Little Ice Age around 1850 many alpine glaciers have retreated (Maisch, 2000), exposing new terrain for soil formation. Consequently, glacier forefields represent chronosequences of different soil development stages offering an ideal system to study the development of functional microbial communities in soil. Initial stages of the glacier chronosequences are characterized by low plant diversity and abundance (Chapin et al., 1994; Tscherko et al., 2005; Hammerli et al., 2007), which increase over time and reach their maxima at sites being ice-free for more than 200 years. Consequently, the amount of available carbon (C) and nitrogen (N) is low during initial ecosystem development. Sigler and Zeyer (2002) reported highest bacterial richness at initial soils (ice-free for 2–10 years). This observation is in agreement with studies reporting highest diversity of functional groups, such as nitrate reducers and N-fixing microbes. (Deiglmayr et al., 2006; Duc et al., 2009; Schmalenberger and Noll, 2009). Moreover, initial sites are often dominated by microbes belonging to the r-strategists being able to respond quickly to changing environmental conditions, whereas with continuing succession a shift to k-strategists occurs that rather pursue the strategy of maintenance (Sigler and Zeyer, 2002). In contrast to these studies, some reports found no significant correlation between soil age and diversity (Edwards et al., 2006) or increasing diversity (Tscherko et al., 2003; Nemergut et al., 2007).
In the last decades, different studies at glacier forefields focused on the dynamic of selected processes and transformation steps of the N cycle (Deiglmayr et al., 2006; Kandeler et al., 2006; Duc et al., 2009; Towe et al., 2010a) because N as a macronutrient is essential for ecosystem development, but most bedrocks do not contain any N.
Although many authors have postulated the importance of N fixation at initial stages of soil development (Kohls et al., 1994; Schmidt et al., 2008), the role of mineralization is discussed controversially. For example, Tscherko et al. (2003) found a pronounced acceleration of N mineralization only after 50 years of soil development, paralleling plant succession as well as organic-matter assemblage. This is in agreement with the common assumption that autotrophic organisms like cyanobacteria, algae, mosses and lichens appear first (Walker and del Moral, 2003; Schmidt et al., 2008), followed by heterotrophic organisms. However, Bardgett et al. (2007) found evidence for a significant mineralization activity also at the initial phases of soil development, which was related to the degradation of ancient and recalcitrant soil organic matter.
Similarly, the development of denitrification over time remained vague. On the one hand, high gene abundance (copies per nanogram DNA) of nirS (nitrite reductase) and narG (nitrate reductase) were observed in pioneer soils (Kandeler et al., 2006); on the other hand, the nitrate reductase activity peaked in developed soils (Deiglmayr et al., 2006).
Besides some contradicting data, all these studies focused on a single N transformation step only, but did not consider consequences for other N cycle processes. A complete reconstruction of the microbial N cycle on the genetic as well as potential activity level at different soil development stages is still missing. Therefore, it was the aim of this study to reconstruct the microbial N cycle from bulk soil samples taken from sites that were ice-free for 10 to 2000a and thus represent different soil development stages. The study is restricted to bulk soil, because the initial sites of the Damma glacier forefield are dominated by bare substrate and are only sparsely vegetated.
We postulate that, although high gene copy numbers might occur at the initial sites, nitrification and denitrification activities parallel the succession of plants, whereas mineralization and N fixation are already important at the beginning of the ecosystem development. To test this hypothesis abundance pattern of genes encoding for subunits of enzymes involved in N fixation (nifH), proteolysis and chitinolysis (aprA and chiA) as proxy for N mineralization, nitrification (amoA of ammonia oxidizing bacteria and archaea) and denitrification (nirS, nirK and nosZ) were measured. Gene abundance data was linked to the corresponding potential enzymatic activities and available C and N in soil. In addition, microbial biomass was determined.
Materials and methods
Sampling site
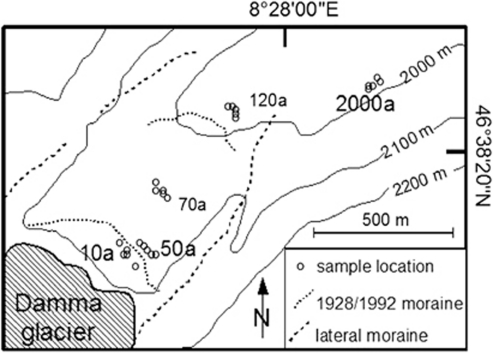
Sampling took place along the forefield of the Damma glacier (46°38′20′'N and 8°28′00′'E) in canton Uri, Switzerland (Figure 1; Edwards et al., 2006). The forefield is characterized by gneiss as bedrock and silty sandy soil texture (Lazzaro et al., 2009). It has a northeast exposition (Bernasconi, 2008) and an inclination of 25% (Sigler et al., 2002). Two terminal moraines dating from 1992 and 1928, as well as two lateral moraines dating from 1850, characterize the forefield (Figure 1). Mean annual air temperature at the forefield is 0–5 °C; average temperature during summer season is 6–10 °C (Climap, MeteoSchweiz, http://www.meteoswiss.admin.ch/). An annual precipitation of 2400 mm was measured at the forefield (Bernasconi, 2008).
Figure 1.
Site map illustrating the Damma glacier forefield. Circles mark the sampling locations. Five circles representing the soil samples of the same developmental stage are labeled with the corresponding sample code: 10, 50, 70, 120 and 2000a.
Chemical and physical soil parameters changed along the forefield. Soil pH values decreased from pH 5.1 in soils being ice-free for 10 years (a) to pH 4.6 in soils being ice-free for 120a and reached a minimum of pH 4.1 outside the forefield, where soils were ice-free for more than 2000a. On the contrary, water-holding capacity slightly increased from 26% (10a) to 33% (2000a). Plant coverage significantly changed from <10% at site 10 to >70% and 95% at sites 50 and 70a, respectively. Sites 120 and 2000a were densely covered with plants (Hans Goransson, ETH Zurich, personal communication). Dominant plant species were Leucanthemopsis alpina, Agrostis gigantea, Rumex scutatus, Salix sp. and Lotus alpinus (Hans Goransson, ETH Zurich, personal communication).
Nitrogen deposition
N depositions in alpine ecosystems of central Switzerland were modeled in different studies (Table 1). Input of total N is in the range between 10–15 kg N ha–1 a–1. To estimate the input of organically bound N and C to the forefield of the Damma glacier, we sampled snow patches at the end of the winter season (<1 m snow depth) in the forefields of the Damma glacier and two adjacent glaciers: Tiefen glacier and Stein glacier. At the end of May 2009, five replicates of surface snow per sampling site (0.04 m2) were collected, melted in the field and filtered through 2.7 μm glass fiber filters (Whatman GF/D, Opfikon, Switzerland). Filters were dried, ground in a bead mill and analyzed for C and N content on a microanalyzer (Leco CHNS-932, St Josephs, MI, USA). Average values were used to estimate the annual aerial input of particulate organic matter (Table 1).
Table 1. Nitrogen deposition at alpine ecosystems in central Switzerland.
| Type of deposition | Area, source of data |
Deposition (kg N ha−1 a−1) or (kg C ha−1 a−1) |
|||
|---|---|---|---|---|---|
| Dry N | Wet NH4+-N | Wet NO3−-N | Total N | ||
| Dry and wet deposition within a year (models) | Central Switzerlanda | — | — | — | 10–15 |
| Central Switzerlandb | 2.2 | 4.5 | 3.5 | — | |
| Central Switzerlandc | — | 5–7.5 | 2.5–3.5 | — | |
| Particulate deposition on snow (experimental) |
Particulate Nd |
Particulate Cd |
|||
| Forefield Damma glacier, this study | 0.63 (0.18) | 7.5 (1.5) | |||
| Forefield Stein glacier, this study | 0.77 (0.08) | 11.8 (0.4) | |||
| Forefield Tiefen glacier, this study | 0.24 (0.05) | 4.0 (0.7) | |||
Values indicate average of five replicates, s.e.m. in brackets.
Sampling procedure
Sampling took place at the Damma Glacier forefield during the growing season on 15th July 2008. Samples were taken in proximity to an already established and characterized sampling grid, representing sites that have been free of ice for 10–2000a (for details see Bernasconi, 2008 and http://www.cces.ethz.ch/projects/clench/BigLink/). In detail, four differently developed soils were sampled along the chronosequence. As the retreat of the glacier was monitored by the Swiss Glacier Monitoring Network (http://glaciology.ethz.ch/swiss-glaciers/), the sites chosen for sampling can be attributed to ice-free times of 10, 50, 70 and 120a (for details see Duc et al., 2009). The distance from the glacier terminus was 100 m for the 10a site and 650 m for the 120a site. In addition, a grassland soil, adjacent to the glacier forefield, was sampled as reference for a developed soil (Figure 1). This site is approximately 500 m away from the 120a site and was not glaciated for more than 2000a (Stefano Bernasconi, ETH Zurich, personal communication). Bulk soil samples from 0 to 2 cm depth were collected in five independent replicates from each site and treated separately. Each of the replicate samples consisted of five sub-samples taken within an area of 1 m2, avoiding plant roots. The distance between the sampling areas for each replicate was 5–10 m. In the field, the soil was sieved (2 mm) and kept on ice for chemical analyses and enzyme assays (Schutte et al., 2009); soil for molecular analysis was stored on dry ice.
Soil carbon and nitrogen content
Fresh soil and fumigated soil (see below) was extracted in 0.01 calcium chloride solution by 45 min horizontal shaking. Soil-to-liquid ratio was 1:2 for the 10–120a samples and 1:3 for the 2000a samples. The following parameters were measured for each soil filtrate (Whatman 595 ½, Dassel, Germany): dissolved organic carbon and nitrogen on DIMA-TOC 100 (Dima Tec, Langenhagen, Germany), nitrate (NO3−) using Spectroquant Nitrate-Test kit (Merck, Darmstadt, Germany) and ammonium (NH4+) using Nanocolor kit (Macherey-Nagel, Düren, Germany). Microbial carbon (Cmic) and microbial nitrogen (Nmic) were determined by the fumigation-extraction method (Vance et al., 1987; Joergensen, 1996). Total organic carbon of air-dried ground soil was determined on a CN-analyzer (Leco CNS2000, St Josephs, MI, USA). Total organic carbon was measured as total C, as the soil contains no carbonates. Total nitrogen was determined by the persulfate oxidation method as described for soil extracts (Cabrera and Beare, 1993). Approximately 200 mg (2000a) to 500 mg (10a) of ground soil was autoclaved (60 min at 121 °C) in 10 ml of oxidizing reagent. Total nitrogen was then measured as nitrate by ion chromatography (Dionex DX-320, IonPac AS11-HC column, Sunnyvale, CA, USA). All analyses for soil C and N content were performed in technical triplicates.
Enzyme assays
To measure the potential N fixation activity in soil, the protocol by Tsunogai et al. (2008) was modified. Briefly, 2 g of fresh soil was weighed into a 20 ml serum bottle. The headspace was flushed with helium for 15 min (Helium 5.0, Pangas, Switzerland) and 1 ml 15N2 gas (98%+, Cambridge Isotope Laboratories, Andover, MA, USA) was added. After 2 weeks of incubation at 12 °C in the dark, 10 ml of oxidizing reagent (Tsunogai et al., 2008) and 0.5 g (samples 10, 50 and 70a) or 1.0 g (samples 120 and 2000a) of low-N potassium persulfate (Fluka 60489, Buchs, Switzerland) was added to convert total N to nitrate. Then, the serum bottles were autoclaved (Tsunogai et al., 2008). After freeze-drying, 30–50 mg of the nitrate-containing sulfate salts were analyzed on a Thermo Fisher Scientific FlashEA (Waltham, MA, USA) coupled with a ConFlo IV (Thermo Fisher Scientific) interface to a Delta V IRMS system (Thermo Fisher Scientific). The instrument was calibrated with the international standards IAEA N1, IAEA N2 and IAEA N3. Samples only flushed with helium served as control. All samples were analyzed in technical quintuplicates.
As proxy for N mineralization activity, chitinase and protease potential activity were measured. Chitinase and protease catalyze the initial breakdown of the two most abundant natural macromolecules that contain N, chitin and protein. Chitinase and protease potential activity were determined using a slightly modified protocol by Hendel and Marxsen (2005). The modifications were: 100–200 mg fresh soil was incubated in 2 ml Eppendorf tubes, containing 50 m fluorescence substrate in 1.6 ml sterilized stream water from the forefield. As fluorogenic substrates, -Leucine 7-amido-4-methyl coumarin hydrochloride (Fluka 61888, Buchs, Switzerland) for protease activity and 4-Methylumbelliferyl N-acetyl-β--glucosaminide (Sigma M2133, Buchs, Switzerland) for chitinase activity were used. All samples were incubated at 12 °C for 6 h in the dark on a rotating shaker and analyzed in triplicate. Fluorescence was determined from 200 μl aliquots in 96-well microtiter plates and compared with 4-Methylumbelliferone (MUF) standards (Aldrich M1381, Buchs, Switzerland). An incubation temperature of 12 °C was chosen, because it is the approximately average soil temperature during summer season (unpublished results).
Potential nitrification activity was performed as a microtiter plate assay following the method described by Hoffmann et al. (2007). Briefly, 2.5 g soil was incubated in 10 ml 1 m ammonium sulfate solution amended with 50 μl of 1.5 sodium chlorate solution. Samples were incubated for 5 h in triplicates and reaction was stopped by adding 2.5 ml of a 2 KCl solution. Nitrite concentrations of non-incubated samples served as controls. Nitrite measurements were performed colorimetrically on a Spectramax 340 (Molecular Devices, Ismaning, Germany) (Hoffmann et al., 2007).
Potential denitrification activity was determined as reported by Luo et al. (1996). Three replicates of 10 g fresh soil were saturated with 0.1 m glucose and nitrate solution. The headspace of the serum bottle was flushed with helium and 10% acetylene was added. N2O was analyzed by gas chromatography on Shimadzu GC-14B (Düsseldorf, Germany). The soil was incubated at 20 °C and headspace samples were taken after 3 and 6 h. Preliminary experiments showed that in all soils considered, the N2O production was a linear function of time from 0 to 6 h (data not shown). It was also found that in these assays the denitrification rates were identical in the presence and absence of chloramphenicol (data not shown).
DNA Extraction and quantification
DNA was extracted from thawed soil using Fast DNA SPIN Kit for Soil (MP Biomedicals, Illkirch, France). In the final step, DNA was eluted two times from the DNA binding column, incubating the column with 30 μl water (supplied) for 5 min. To ensure quantitative DNA extraction, preceding tests stipulated to extract ∼500 mg of initial and transient soils (10, 50 and 70a) and ∼250 mg of developed soil (120 and 2000a). Then there was a linear relationship between DNA yield and amount of soil extracted. DNA yields were quantified using Sybr green I (Invitrogen, Basel, Switzerland) assay as described by Matsui et al. (2004).
Real-time PCR
Sybr Green-based quantification assays using Power Sybr Green (Applied Biosystems, Darmstadt, Germany) and Kapa Sybr Fast (Kapa Biosystems, LabGene, Chatel-St-Denis, Switzerland) PCR master mixes were run on a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Reaction volumes were 25 μl containing onefold PCR master mix. PCR conditions, efficiencies, primers and calibration standards used are summarized in Table 2. Preceding tests with dilution series of the extracted DNA showed no inhibitory effects at a 1:16 dilution. Each PCR run included triplicate sample templates, calibration standard series and no template controls.
Table 2. Reaction composition, thermal profiles, sources of calibration standards and primer references of qPCR reactions used in this study.
| Target gene | Reaction conditions | Source of calibration standard | Primer name and reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F- and R-primer (pmol μl−1) | BSA (μg μl−1) | DMSO (μg μl−1) | Denaturation time at 95 °C (s) | Annealing time and temperature | Elongation time at 72 °C (s) | Number of cycles | qPCR efficiency (%)a | |||
| nifHb | 0.2 | 0.2 | 0 | 15 | 25 s at 53 °Cc | 45 | 43 | 88–90 | Paenibacillus azotofixans DSM 5976 | nifH-F, nifH-R, Rosch et al., 2002 |
| ChiAb | 0.2 | 0.1 | 0 | 10 | 90 s at 65 °C | 30 | 40 | 95–96 | Streptomyces coelicolor DSM 41189 | chif2, chir, Xiao et al., 2005 |
| Aprb | 0.2 | 0.2 | 0 | 10 | 20 s at 54 °C | 30 | 42 | 72–74 | Pseudomonas fluorescens DSM 50090 | FR apr I, RP apr II, Bach et al., 2001 |
| nirKd | 0.2 | 0.6 | 25 | 15 | 30 s at 58 °Ce | 30 | 45 | 83–88 | Azospirillum irakense DSM 11586 | nirK 876, Henry et al., 2004, nirK 5R, Braker et al., 1998 |
| nirSd | 0.2 | 0.6 | 25 | 45 | 45 s at 57 °C | 45 | 40 | 88–95 | Pseudomonas stutzeri | nirS cd3af, Michotey et al., 2000, nirSR3cd Throback et al., 2004 |
| nosZd | 0.2 | 0.6 | 0 | 15 | 30 s at 60 °Cf | 30 | 45 | 92–95 | Pseudomonas fluorescens C7R12g | nosZ2F, nosZ2R, Henry et al., 2006 |
| amoAd (AOA) | 0.2 | 0.6 | 0 | 45 | 45 s at 55 °C | 45 | 40 | 85–87 | Fosmid clone 54d9h | 19F, Leininger et al., 2006, CrenamoA616r48x, Schauss et al., 2009 |
| amoAd (AOB) | 0.3 | 0.6 | 0 | 45 | 45 s at 60 °C | 45 | 40 | 78–79 | Nitrosomonas sp. | amoA 1F, amoa 2R, Rotthauwe et al., 1997 |
Abbreviation: AOB, ammonia-oxidizing bacteria.
Efficiency was calculated after Babic et al. (2008).
Amplified using Kapa SYBR Fast (Kapa Biosystems) master mix.
Touch down starting at 63 °C temperature decrease of 2 °C per cycle.
Amplified using Power SYBR Green (Applied Biosystems) master mix.
Touch down starting at 63 °C temperature decrease of 1 °C per cycle.
Touch down starting at 65 °C temperature decrease of 1 °C per cycle.
Statistical analysis
One-way analysis of varience (ANOVA) on ranks was performed using SigmaPlot 11.0 package (Systat Software Inc., San Jose, CA, USA). Before regression analysis, potential enzymatic activities and copy numbers were log-transformed according to log10(x+c), where c is the 2.5% quartile of the measured parameter.
Results
Soil parameters along the chronosequence
Measurements of the C and N pools indicate that soil-nutrient status changed substantially along the glacier chronosequence (Table 3). All C and N pools increased by one order of magnitude from 10 to 2000a. Total organic C increased from 0.7 to 40 mg C g−1 Similarly, microbial carbon increased from 58 to 902 μg C g−1 and was significantly correlated to the amount of DNA that was extracted from the soil (n=25, R2=0.53, P<0.001). Cell counts and abundance of rpoB gene copy numbers were less well-correlated to microbial carbon (n=25, R2=0.31, P=0.004 and n=25, R2=0.11, P=0.099; respectively) (unpublished data). Total N and microbial N contents were one order of magnitude lower than C contents. The resulting C/N ratios ranged from 12 to 18, whereby samples 50 and 70a displayed the highest C/N ratio. Nitrate concentration was five times higher in soils from the 10a site (0.13 μg N g−1) compared with ammonium; however, the ammonium content increased in proportion to the nitrate concentration, reaching one order of magnitude higher concentrations (12.8 μg N g−1) at site 2000a.
Table 3. Carbon, nitrogen and DNA content of the soil. Numbers give the average of five replicates, in brackets standard error of means.
|
Site |
|||||
|---|---|---|---|---|---|
| 10a | 50a | 70a | 120a | 2000a | |
| Carbon (μg C g−1) | |||||
| TOC ( × 103)a | 0.7 (0.2) | 4.7 (1.2) | 3.0 (0.5) | 12.0 (4.2) | 39.6 (12.4) |
| DOCb | 7.1 (2.3) | 18.2 (8.3) | 6.6 (1.2) | 42.4 (14.3) | 77.8 (16.7) |
| Cmicc | 58 (11) | 122 (26) | 73 (7) | 241 (71) | 902 (215) |
| Nitrogen (μg N g−1) | |||||
| TN ( × 103)d | 0.07 (0.02) | 0.25 (0.05) | 0.17 (0.03) | 0.73 (0.24) | 2.67 (0.83) |
| DONe | 0.3 (0.1) | 0.9 (0.2) | 0.6 (0.1) | 3.0 (2.3) | 12.2 (6.1) |
| Ammonium | 0.03 (0.01) | 0.18 (0.12) | 0.15 (0.08) | 6.69 (5.21) | 12.81 (6.32) |
| Nitrate | 0.13 (0.05) | 0.23 (0.08) | 0.18 (0.08) | 1.31 (0.32) | 0.81 (0.10) |
| Nmicf | 6 (1) | 19 (5) | 13 (2) | 29 (8) | 115 (28) |
| C/N ratiog | 12.8 (3.1) | 18.0 (1.2) | 18.5 (1.6) | 15.9 (1.0) | 15.0 (0.6) |
| Extracted DNA (μg g−1) | 2.3 (0.8) | 12.0 (2.9) | 7.8 (2.2) | 17.4 (4.6) | 28.0 (8.5) |
Total organic carbon.
Dissolved organic carbon.
Microbial carbon.
Total nitrogen.
Dissolved organic nitrogen.
Microbial nitrogen.
Ratio of TOC to TN.
Potential enzyme activities
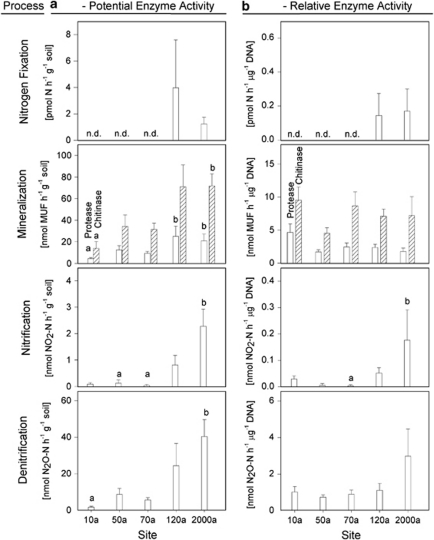
Potential enzyme activity was related to soil dry weight as well as the amount of extracted DNA to obtain a biomass-independent parameter (Figure 2). Generally, potential enzyme activities related to dry soil increased along the chronosequence. N-fixation activity was below the detection limit in samples 10–70a (detection limit 0.2 pmol N h−1 g−1) but ranged between 1 and 3 pmol N h−1 g−1 in samples 120 and 2000a. Relative N-fixation activity was similar in samples 120 and 2000a. As proxy for potential mineralization activity, protease and chitinase assays were performed. Significant increases of protease activity from 5 to 25 nmol MUF h−1 g−1 as well as of chitinase activity from 14 to 72 nmol MUF h−1 g−1 were observed along the chronosequence. At the same time, relative mineralization activity tended to decrease, however, the trend was not significant. Potential nitrification activity increased significantly from 0.1 (10a) to 2.3 nmol NO2−-N h−1 g−1 (2000a). The same pattern was observed for potential denitrification activity, where activities increased from 1.8 to 40 nmol N2O-N h−1 g−1 The relative nitrification and denitrification activities resulted in similar pattern but did not reach significant levels in most cases.
Figure 2.
Potential (a) and relative enzyme activities (b) for nitrogen fixation, mineralization, nitrification and denitrification are displayed (n=5, error bars represent standard error of means). Only significant differences as revealed by one way ANOVA on ranks (P<0.05) are indicated by different letters. n.d. indicates that all five replicates of potential N fixation were below the detection limit of 0.2 pmol N h−1 g−1 soil.
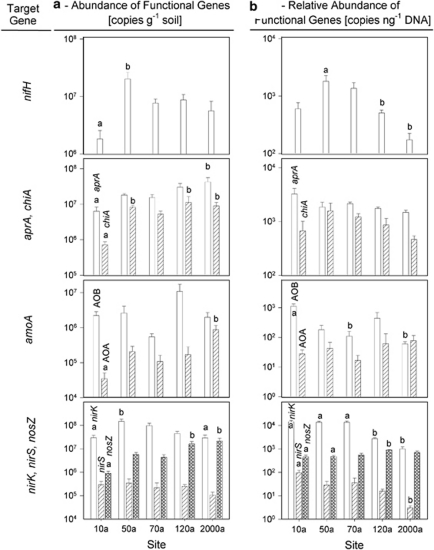
Abundance of functional genes
Gene copy numbers were related to gram soil as well as nanogram DNA (Figure 3). Abundance of nifH gene was lowest in sample 10a (2 × 106 copies per gram soil) and increased up to 2 × 107 copies per gram soil in soil samples from 50a. In the soil samples from sites that have been ice-free for a longer period nifH gene abundance decreased. Similar results were obtained when data were related to nanogram extracted DNA.
Figure 3.
Gene copy numbers of functional genes involved in major steps of the nitrogen cycle (nifH, aprA, chiA, amoA AOB, amoA AOA, nirK, nirS and nosZ) are displayed related to gram soil (a) and nanogram DNA (b) (n=5, error bars represent standard error of means). Only significant differences as revealed by one way ANOVA on ranks (P<0.05) are indicated by different letters.
Abundance of genes coding for enzymes involved in mineralization significantly increased (Figure 3): aprA genes from 6 × 106 to 4 × 107; chiA genes from 7 × 105 to 9 × 106 copies per gram soil. In contrast, relative abundances (related to nanogram of DNA) of both genes involved in mineralization were constant (Figure 3b) at all sites.
Abundance of the nitrification marker gene amoA was studied for ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB). Although amoA AOB gene abundances showed no significant differences related to gram of soil, AOA significantly increased from 3 × 104 to 8 × 105 copies per gram soil. Generally, AOB abundances were up to 80-times higher compared with AOA at the 10, 50, 70 and 120a site, whereas the AOB to AOA ratios were close to two in soil samples 2000a. Relative amoA AOA gene abundances remained stable, whereas relative amoA AOB gene abundances significantly decreased from 1.4 × 103 to 7.2 × 102 copies per nanogram DNA along the chronosequence.
To quantify the denitrifying bacteria, genes coding for the nitrite reductases (nirK and nirS) and nitrous oxide reductase (nosZ) were measured. Generally, nirK gene abundances were two orders of magnitude higher than nirS. Gene abundances of nirK decreased from 1.5 × 108 to 3 × 107 copies per gram soil along the chronosequence. The nosZ gene copy numbers were lowest in sample 10a with 9 × 105 copies per gram soil but equaled nirK gene abundance at site 2000a. Relative gene abundances showed comparable patterns for denitrification genes except nirS, which decreased along the chronosequence.
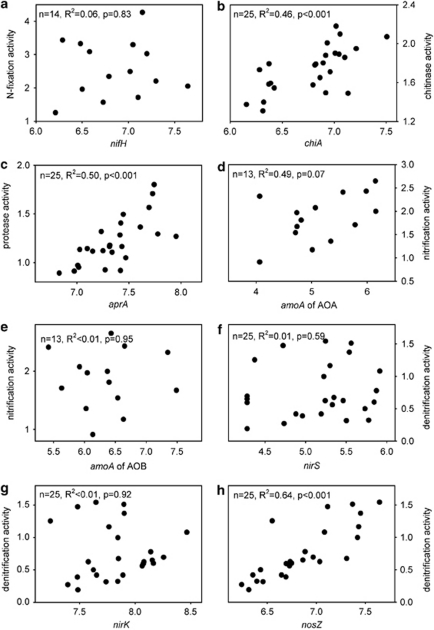
Correlation of abundances and activities
Correlations between gene abundance pattern (copies per gram soil) and potential enzyme activities (per gram soil) were assessed. In order to reduce variability and to include all available data points, both data sets were log-transformed before regression analysis (Figure 4). Although nifH, amoA AOB, nirS and nirK gene abundances did not correlate with the corresponding potential enzyme activities, significant correlations with the potential enzymatic activities were found for chiA, aprA, amoA AOA and nosZ gene copy numbers significantly correlated with the potential enzyme activities (α=0.1).
Figure 4.
Correlations of the gene copy numbers and the corresponding potential enzyme activities are displayed for nitrogen fixation (a), mineralization (b, c), nitrification (d, e) and denitrification (f, g and h). The graphs plot log-transformed copy numbers vs log-transformed enzyme activities. In the top left corner of each plot the parameters of the regression analysis, including P-values of t-tests, are provided.
Discussion
The initial state
Initial ecosystems are characterized by low nutrient content and scarce vegetation (Sigler and Zeyer, 2002; Tscherko et al., 2004; Yoshitake et al., 2007). For the Damma glacier it was shown that the initial sites (10a) have less than 10% vegetation cover, whereas the developed sites (120–2000a) have a closed plant cover (Hammerli et al., 2007; Hans Goransson, personal communication). Thus, the input of organic material by root exudates or plant litter might be low at the initial sites of the glacier forefield. Hence, N fixation and autotrophic CO2 incorporation may have a crucial role for ecosystem development (Kohls et al., 1994). Interestingly, our data revealed no potential N-fixation activity and the lowest nifH gene copy numbers per gram soil in the 10a soil. This is supported by the studies of Duc et al. (2009) and Nemergut et al. (2007) in which the lowest N fixation rate was measured in the bulk soil of the initial stage of the Damma glacier (Switzerland) and Puca glacier (Peru) forefield, respectively. In contrast, mineralization of organic material seemed to be the driver for N cycling at the initial stage of ecosystem development, indicated by the highest potential activities for proteases combined with highest relative aprA gene copy numbers (based on the amount of extracted DNA). Although other genes like nirK and amoA AOB showed similar or even higher gene copy numbers at the initial sites, the potential denitrification and nitrification rates were extremely low. This implies the presence of inactive nitrifying and denitrifying populations, attributed to the shortage of ammonium and nitrate. Obviously, at the initial stage of the glacier forefield, sufficient N and C are provided for mineralization: (i) allochthonous organic material (plant debris, insects and so on) is deposited in the forefield, as revealed by our and other measurements (Hodkinson et al., 2002; National Air Pollution Monitoring Network (NABEL), 2008; Nyiri et al., 2009), (Table 1), (ii) the cryoconite holes in the glacier ablation zone possibly inhabit small foodwebs of cyanobacteria and heterotrophs, which were relocated to the glacier forefield by the glacial stream after snowmelt (Schmalenberger and Noll, 2009; Xiang et al., 2009) and (iii) microbes might feed on ancient recalcitrant C (Hodkinson et al., 2002; Bardgett et al., 2007). In addition, significant inputs of nutrients may be due to atmospheric dry and wet deposition of N and C species (Table 1).
Plants as competitors for nutrients
With the first appearance of plant patches it could be expected that C will be provided in moderate quantities by rhizodeposition and exudation. However, it has rather been postulated for developing ecosystems that plants and microbes compete for nutrients, like N and phosphorus, as on the one hand nutrient availability is low and on the other hand plant performance and exudation is reduced (Chapin et al., 1994; Hammerli et al., 2007). Although ammonium and nitrate concentrations increased two- to sixfold from site 10a to the transient soils (50 and 70a), these values are still more than 10 times lower than values from pasture sites (Chronakova et al., 2009), confirming the assumption that strong competition for N still exists. Thus, it is likely that with the first occurrence of plant patches at site 50a, the nifH gene abundance reached its maximum even if the potential N-fixing activity was still negligible. However, it has to be taken into account that the used assay only covered the performance of free-living heterotrophic N fixers and did not consider symbiotic N fixation by Rhizobia, plant-associated N fixation or phototrophic N fixation. The observation that the activity of free-living N fixers is low at this stage of ecosystem development is not surprising, as the amounts of dissolved organic carbon in the bulk soil of the 50 and 70a sites were low and consequently, C sources were missing for the energy-consuming process of N fixation (Zehr et al., 2003). In this regard Duc et al. (2009) showed that once C-rich nutrients were delivered like exudates in the rhizosphere of plants, N-fixation activity significantly increased. Hence, it can be speculated that mainly symbiotic and plant-associated N fixing microbes might have a role for N input at these sites.
Along with the increase in plant abundance and diversity (Hammerli et al., 2007) also other organisms establish in the developing soils. Especially the abundance of fungi increased along glacial chronosequences (Ohtonen et al., 1999; Bardgett and Walker, 2004). Thus, the observed increase in chiA gene abundance and potential chitinase activity were not surprising. These findings are further in-line with data presented by Tscherko et al. (2004) who revealed highest N-acetyl-β-glucosaminidase activity in the 75a transient soils of the Rotmoosferner glacier. On the contrary, aprA gene abundances did not change, underlining the high relevance of chitin as alternative C and organic N source (Olander and Vitousek, 2000), which might result from the deposition of dead insects (Hodkinson et al., 2002).
Although potential mineralization was also prominent at the transient stages of ecosystem development (50 and 70a), potential nitrification and denitrification activity remained low. That is not surprising as nitrification causes the transformation of ammonium to highly mobile nitrate and in connection with denitrification, it would result in a substantial loss of N. Although amoA AOA, nirK and nosZ gene abundances increased with increasing biomass, their relative abundances in the microbial community were constant. Interestingly, that was not true for amoA AOB because their relative abundance significantly dropped towards site 70a, which might be a response to the decreasing pH values along the chronosequence with highest pH values of 5.1 at the initial sites. De Boer and Kowalchuk (2001) found that Nitrosospira species, which are the most common AOB in soil (Kowalchuk et al., 2000), were not able to oxidize ammonium at pH values below 5.5. In contrast, several studies indicate that AOA are able to oxidize ammonium in a broad pH range (Leininger et al., 2006; Nicol et al., 2008; Reigstad et al., 2008) and appear to be better adapted to ammonium-poor environments (Di et al., 2009, 2010) and soils with low pH (Nicol et al., 2008).
Plants as driver for nitrogen turnover
After 120a of soil development at the Damma glacier forefield the soil surface is densely covered with plants. The pronounced root penetration in the developed soils brings along an enhanced water retention potential resulting in less oxygen diffusion and reduced partial oxygen pressure (Rheinbaben and Trolldenier, 1984; Deiglmayr et al., 2006). In combination with the sevenfold increase of the nitrate concentrations, developed soils (120 and 2000a) provide good conditions for denitrification. That development is also reflected in our potential denitrification measurements, which showed highest activity in the 2000a soil. Our data showed a negligible role of nirS genes at the Damma glacier forefield for nitrite reduction. Obviously, nitrite reduction appeared to be driven by nirK-harboring bacteria. Similarly, Kandeler et al. (2006) found a dominance of nirK and nosZ genes along the Rotmoosferner glacier forefield, Austria. However, if comparing gene abundances of different genes, one should keep in mind that the quantitative PCR approach suffers from some biases such as unspecific primers or varying PCR amplification efficiencies. Thus, the displayed gene copy numbers did never reflect the actual in situ abundance. Nevertheless, the use of degenerated primers (Henry et al., 2004) and the accurate performance of the measurement (Towe et al., 2010b) allow the discussion of solid tendencies.
Beside highest potential enzyme activities, gene copy numbers (per gram of soil) of chiA, aprA and amoA AOA revealed significantly highest values in the 2000a soil. These data are congruent with observations from Frank et al. (2000) who found a positive correlation between nitrification, denitrification and N mineralization processes in Yellowstone Park grasslands. Additionally, associations between mineralizing and nitrifying organisms were described in previous studies (Schimel and Bennett, 2004).
Linking potential enzymatic activities to gene abundance
We found correlations between gene abundance and enzymatic activities for chitinolytic and proteolytic microbes, as well as for N2O reducers. This indicates that over the whole chronosequence no shifts in the physiology of the corresponding functional groups have occured, as turnover rates have not changed under optimal conditions (potential activities) in relation to the gene copy numbers. This is not surprising, as microbes that are able to mineralize N are restricted to a few specialized microbes that are often taxonomically closely related. For example, many proteolytic bacteria belong to different species of Bacillus (Fuka et al., 2008). For chitinases it has been postulated that horizontal gene transfer might have an important role for transfer of this functional trait, thus operon structures and induction pathways might be comparable (Cohen-Kupiec and Chet, 1998). In contrast to N-mineralizing microbes, N2O reducers occur in many different groups of microorganisms with different physiological backgrounds. However, induction of the N2O reductase is mainly linked to the presence of nitrate and thus similarly regulated for all groups of N2O reducers (Zumft, 1997; Hallin et al., 2009), which might explain the good correlation between gene abundance and enzymatic activities for this functional group.
Vice versa low or not existing correlations between gene abundance and enzymatic activities might indicate shifts in the diversity pattern and the physiology of the dominant functional groups along the chronosequence. This is obvious for N-fixing microbes: In the first phase of soil development N fixation is driven mainly by cyanobacteria, which have relatively low N-fixation activity. In contrast, N-fixing symbionts, who aquire carbon from the plant, fix N at higher rates. However, it must also be taken into account that the conditions of the enzymatic assays have been only optimal for a subgroup of each functional group. If environmental conditions change along the chronosequence (for example, pH values) these changing conditions have not been reflected in the assays performed. It might be speculated that this fact could explain the missing correlation between ammonia-oxidizing bacteria and potential nitrification, as some well adapted AOBs mainly at the developed sites (120 and 2000a) with lower soil pH (4.6–4.1), could respond differently to the conditions of the nitrification assay performed compared with AOBs from the initial and transient soils.
Overall it must be clearly stated that potential activities measured under laboratory conditions cannot be related to in situ fluxes and turnover rates (Roling, 2007). To address this type of question a different experimental setup is needed.
The chronosequence approach
Although a chronosequence approach might be biased by, for example, different microclimatic conditions along the slope of hill or the big spatial heterogeneity, it was shown in several studies that the influence of the soil age outcompetes minor influences of other site characteristics (Nemergut et al., 2007; Duc et al., 2009). Thus, it displayed a proper approach to compare and pursue N cycle processes during different soil development stages. The study did not intend to assess seasonal effects and therefore, all samples were taken at one single time-point.
Conclusion
In conclusion, our data revealed that initial (10a) and transient soils (50 and 70a) were dominated by processes, which were responsible for N input and release from organic matter, that is, N fixation and mineralization. Denitrification and nitrification that lead to N losses from the system were reduced at these stages. As soon as a closed plant cover developed and microbial associations are established, nitrification and denitrification potential activity increased. Moreover, we demonstrated that potential enzyme activities correlated with amoA AOA, nosZ, chiA and aprA gene abundance only. To corroborate these results further studies are needed with the aim to measure in situ enzyme activities and link these data to the corresponding gene transcript abundances. However, the enormous spatial heterogeneity and the low enzymatic activities mainly at the initial sites require sophisticated sampling strategies. In addition, future studies dealing with N fixation, should allow assessing the individual contributions of free-living heterotrophs, phototrophs, symbionts and plant-associated N-fixing microbes to the N budget.
Acknowledgments
The authors thank Stefano Bernasconi for stable isotope measurements and critical reviewers for their helpful comments on the manuscript. This study is part of the Transregional Collaborative Research Centre 38 (SFB/TRR 38), which is financially supported by the Deutsche Forschungsgemeinschaft (DFG, Bonn) and the Brandenburg Ministry of Science, Research and Culture (MWFK, Potsdam).
References
- Babic KH, Schauss K, Hai B, Sikora S, Redzepovic S, Radl V, et al. Influence of different Sinorhizobium meliloti inocula on abundance of genes involved in nitrogen transformations in the rhizosphere of alfalfa (Medicago sativa L.) Environ Microbiol. 2008;10:2922–2930. doi: 10.1111/j.1462-2920.2008.01762.x. [DOI] [PubMed] [Google Scholar]
- Bach HJ, Hartmann A, Schloter M, Munch JC. PCR primers and functional probes for amplification and detection of bacterial genes for extracellular peptidases in single strains and in soil. J Microbiol Meth. 2001;44:173–182. doi: 10.1016/s0167-7012(00)00239-6. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, Walker LR. Impact of coloniser plant species on the development of decomposer microbial communities following deglaciation. Soil Biol Biochem. 2004;36:555–559. [Google Scholar]
- Bardgett RD, Richter A, Bol R, Garnett MH, Baumler R, Xu XL, et al. Heterotrophic microbial communities use ancient carbon following glacial retreat. Biol Lett. 2007;3:487–490. doi: 10.1098/rsbl.2007.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi SM. Weathering, soil formation and initial ecosystem evolution on a glacier forefield: a case study from the Damma Glacier, Switzerland. Mineral Mag. 2008;72:19–22. [Google Scholar]
- Braker G, Fesefeldt A, Witzel K. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera ML, Beare MH. Alkaline persulfate oxidation for determining total nitrogen in microbial biomass extracts. Soil Sci Soc Am J. 1993;57:1007–1012. [Google Scholar]
- Chapin FS, Walker FS, Fastie CL, Sharman LC. Mechanisms of primary succession following deglaciation at Glacier Bay, Alaska. Ecol Monogr. 1994;64:149–175. [Google Scholar]
- Chronakova A, Radl V, Cuhel J, Simek M, Elhottova D, Engel M, et al. Overwintering management on upland pasture causes shifts in an abundance of denitrifying microbial communities, their activity and N2O-reducing ability. Soil Biol Biochem. 2009;41:1132–1138. [Google Scholar]
- Cohen-Kupiec R, Chet I. The molecular biology of chitin digestion. Curr Opin Biotechnol. 1998;9:270–277. doi: 10.1016/s0958-1669(98)80058-x. [DOI] [PubMed] [Google Scholar]
- De Boer W, Kowalchuk GA. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- Deiglmayr K, Philippot L, Tscherko D, Kandeler E. Microbial succession of nitrate-reducing bacteria in the rhizosphere of Poa alpina across a glacier foreland in the Central Alps. Environ Microbiol. 2006;8:1600–1612. doi: 10.1111/j.1462-2920.2006.01051.x. [DOI] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O′Callaghan M, Bowatte S, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O′Callaghan M, Bowatte S, et al. Ammonia oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. 2010;72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- Duc L, Noll M, Meier BE, Burgmann H, Zeyer J. High Diversity of diazotrophs in the forefield of a receding alpine glacier. Microb Ecol. 2009;57:179–190. doi: 10.1007/s00248-008-9408-5. [DOI] [PubMed] [Google Scholar]
- Edwards IP, Burgmann H, Miniaci C, Zeyer J. Variation in microbial community composition and culturability in the rhizosphere of Leucanthemopsis alpina L Heywood and adjacent bare soil along an alpine chronosequence. Microb Ecol. 2006;52:679–692. doi: 10.1007/s00248-006-9097-x. [DOI] [PubMed] [Google Scholar]
- Eparvier A, Lemanceau P, Alabouvette C. Population dynamics of non-pathogenic Fusarium and fluorescent Pseudomonas strains in rockwool, a substratum for soilless culture. FEMS Microbiol Lett. 1991;86:177–184. [Google Scholar]
- Ferderal Commission for Air Hygiene 2005Stickstoffhaltige Luftschadstoffe in der Schweiz, Schriftenreihe Umwelt Nr. 384Federal Office for the Environment: Bern.
- Frank DA, Groffman PM, Evans RD, Tracy BF. Ungulate stimulation of nitrogen cycling and retention in Yellowstone Park grasslands. Oecologia. 2000;123:116–121. doi: 10.1007/s004420050996. [DOI] [PubMed] [Google Scholar]
- Fuka MM, Engel M, Gattinger A, Bausenwein U, Sommer M, Munch JC, et al. Factors influencing variability of proteolytic genes and activities in arable soils. Soil Biol Biochem. 2008;40:1646–1653. [Google Scholar]
- Hallin S, Jones CM, Schloter M, Philippot L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 2009;3:597–605. doi: 10.1038/ismej.2008.128. [DOI] [PubMed] [Google Scholar]
- Hammerli A, Waldhuber S, Miniaci C, Zeyer J, Bunge M. Local expansion and selection of soil bacteria in a glacier forefield. Eur J Soil Sci. 2007;58:1437–1445. [Google Scholar]
- Hendel B, Marxsen J.2005Fluorometric determination of the activity of beta-glucosidase and other extracellular hydrolytic enzymesIn: Graça, MAS, Bärlocher F, Gessner MO (eds).Methods to study litter decomposition Springer: Dordrecht; 261–266. [Google Scholar]
- Henry S, Baudoin E, López-Gutiérrez JC, Martin-Laurent F, Brauman A, Philippot L. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Meth. 2004;59:327–335. doi: 10.1016/j.mimet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Henry S, Bru D, Stres B, Hallet S, Philippot L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol. 2006;72:5181–5189. doi: 10.1128/AEM.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodkinson ID, Webb NR, Coulson SJ. Primary community assembly on land – the missing stages: why are the heterotrophic organisms always there first. J Ecol. 2002;90:569–577. [Google Scholar]
- Hoffmann H, Schloter M, Wilke BM. Microscale-scale measurement of potential nitrification rates of soil aggregates. Bio Fert Soils. 2007;44:411–413. [Google Scholar]
- Joergensen RG. The fumigation-extraction method to estimate soil microbial biomass: Calibration of the k(EC) value. Soil Biol Biochem. 1996;28:25–31. [Google Scholar]
- Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl Environ Microbiol. 2006;72:5957–5962. doi: 10.1128/AEM.00439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohls SJ, Vankessel C, Baker DD, Gridal DF, Lawrence DB. Assessment of N2 fixation and N cycling by dryas along a chronosequence within the forelands of the Athabasca glacier, Canada. Soil Biol Biochem. 1994;26:623–632. [Google Scholar]
- Kowalchuk GA, Stienstra AW, Heilig GH, Stephen JR, Woldendorp JW. Changes in the community structure of ammonia-oxidizing bacteria during secondary succession of calcareous grasslands. Environ Microbiol. 2000;2:99–110. doi: 10.1046/j.1462-2920.2000.00080.x. [DOI] [PubMed] [Google Scholar]
- Lazzaro A, Abegg C, Zeyer J. Bacterial community structure of glacier forefields on siliceous and calcareous bedrock. Eur J Soil Sci. 2009;60:860–870. [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Luo J, White RE, Ball PR, Tillman RW. Measuring denitrification activity in soils under pasture: Optimizing conditions for the short-term denitrification enzyme assay and effects of soil storage on denitrification activity. Soil Biol Biochem. 1996;28:409–417. [Google Scholar]
- Maisch M. The longterm signal of climate change in the swiss alps: glacier retreat since the end of the little ice age and future ice decay scenarios. Geogr Fis Dinam Quat. 2000;23:139–151. [Google Scholar]
- Matsui K, Ishii N, Honjo M, Kawabata Z. Use of the SYBR Green I fluorescent dye and a centrifugal filter device for rapid determination of dissolved DNA concentration in fresh water. Aquat Microb Ecol. 2004;36:99–105. [Google Scholar]
- Michotey V, Méjean V, Bonin P. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl Environ Microbiol. 2000;66:1564–1571. doi: 10.1128/aem.66.4.1564-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Air Pollution Monitoring Network (NABEL) 2008NABEL-Luftbelastung 2007Umweltzustand Nr. 0823. Federal Office for the Environment: Bern [Google Scholar]
- Nemergut DR, Anderson SP, Cleveland CC, Martin AP, Miller AE, Seimon A, et al. Microbial community succession in an unvegetated, recently deglaciated soil. Microb Ecol. 2007;53:110–122. doi: 10.1007/s00248-006-9144-7. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Nyiri A, Gauss M, Klein H. Norwegian Meteorological Institute: Oslo; 2009. Transboundary air pollution by main pollutants (S, N, O3) and PM. [Google Scholar]
- Ohtonen R, Fritze H, Pennanen T, et al. Ecosystem properties and microbial community changes in primary succession on a glacier forefront. Oecologia. 1999;119:239–246. doi: 10.1007/s004420050782. [DOI] [PubMed] [Google Scholar]
- Olander LP, Vitousek PM. Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry. 2000;49:175–190. [Google Scholar]
- Reigstad LJ, Richter A, Daims H, Urich T, Schwark L, Schleper C. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol. 2008;64:167–174. doi: 10.1111/j.1574-6941.2008.00466.x. [DOI] [PubMed] [Google Scholar]
- Rheinbaben WV, Trolldenier G. Influence of plant growth on denitrification in relation to soil moisture and potassium nutrition. Z Pflanzenernähr Bodenkd. 1984;147:730–738. [Google Scholar]
- Roling WFM. Do microbial numbers count? Quantifying the regulation of biogeochemicalfluxes by population size and cellular activity. FEMS Microbiol Ecol. 2007;62:202–210. doi: 10.1111/j.1574-6941.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- Rosch C, Mergel A, Bothe H. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl Environ Microbiol. 2002;68:3818–3829. doi: 10.1128/AEM.68.8.3818-3829.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe JH, Witzel K, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, et al. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol. 2009;11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- Schimel JP, Bennett J. Nitrogen mineralization: Challenges of a changing paradigm. Ecology. 2004;85:591–602. [Google Scholar]
- Schmalenberger A, Noll M. Shifts in desulfonating bacterial communities along a soil chronosequence in the forefield of a receding glacier. FEMS Microbiol Ecol. 2009;71:208–217. doi: 10.1111/j.1574-6941.2009.00799.x. [DOI] [PubMed] [Google Scholar]
- Schmidt SK, Reed SC, Nemergut DR, Grandy AS, Cleveland CC, Weintraub MN, et al. The earliest stages of ecosystem succession in high-elevation (5000 metres above sea level), recently deglaciated soils. Proc R Soc B. 2008;275:2793–2802. doi: 10.1098/rspb.2008.0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte UME, Abdo Z, Bent SJ, William CJ, Schneider GM, Solhein B, et al. Bacterial succession in a glacier foreland of the High Arctic. ISME J. 2009;3:1258–1268. doi: 10.1038/ismej.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler WV, Crivii S, Zeyer J. Bacterial succession in glacial forefield soils characterized by community structure, activity and opportunistic growth dynamics. Microb Ecol. 2002;44:306–316. doi: 10.1007/s00248-002-2025-9. [DOI] [PubMed] [Google Scholar]
- Sigler WV, Zeyer J. Microbial diversity and activity along the forefields of two receding glaciers. Microb Ecol. 2002;43:397–407. doi: 10.1007/s00248-001-0045-5. [DOI] [PubMed] [Google Scholar]
- Throback IN, Enwall K, Jarvis A, Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol. 2004;49:401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Towe S, Albert A, Kleineidam K, Brankatschk R, Dumig A, Welzl G, et al. Abundance of microbes involved in nitrogen transformation in the rhizosphere of Leucanthemopsis alpina (L.) HEYWOOD grown in soils from different sites of the Damma glacier forefield. Microb Ecol. 2010a;60:762–770. doi: 10.1007/s00248-010-9695-5. [DOI] [PubMed] [Google Scholar]
- Towe S, Kleineidam K, Schloter M. Differences in amplification efficiency of standard curves in quantitative real-time PCR assays and consequences for gene quantification in environmental samples. J Microbiol Methods. 2010b;82:338–341. doi: 10.1016/j.mimet.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Tscherko D, Rustemeier J, Richter A, Wanek W, Kandeler E. Functional diversity of the soil microflora in primary succession across two glacier forelands in the Central Alps. Eur J Soil Sci. 2003;54:685–696. [Google Scholar]
- Tscherko D, Hammesfahr U, Marx MC, Kandeler E. Shifts in rhizosphere microbial communities and enzyme activity of Poa alpina across an alpine chronosequence. Soil Biol Biochem. 2004;36:1685–1698. [Google Scholar]
- Tscherko D, Hammesfahr U, Zeltner G, Kandeler E, Bocker R. Plant succession and rhizosphere microbial communities in a recently deglaciated alpine terrain. Basic Appl Ecol. 2005;6:367–383. [Google Scholar]
- Tsunogai U, Kido T, Hirota A, Ohkubo SB, Komatsu DD, Nakagawa F. Sensitive determinations of stable nitrogen isotopic composition of organic nitrogen through chemical conversion into N2O. Rapid Commun Mass Sp. 2008;22:345–354. doi: 10.1002/rcm.3368. [DOI] [PubMed] [Google Scholar]
- Vance ED, Brookes PC, Jenkinson DS. An extraction method for measuring soil microbial biomass-C. Soil Biol Biochem. 1987;19:703–707. [Google Scholar]
- Xiang SR, Shang TC, Chen Y, Yao TD. Deposition and postdeposition mechanisms as possible drivers of microbial population variability in glacier ice. FEMS Microbiol Ecol. 2009;70:9–20. doi: 10.1111/j.1574-6941.2009.00759.x. [DOI] [PubMed] [Google Scholar]
- Walker LR, del Moral R. Primary Succession and Ecosystem Rehabilitation. Cambridge University Press: Cambridge; 2003. [Google Scholar]
- Xiao X, Yin X, Lin J, Sun L, You Z, Wang P, et al. Chitinase genes in lake sediments of Ardley Island, Antarctica. Appl Environ Microbiol. 2005;71:7904–7909. doi: 10.1128/AEM.71.12.7904-7909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake S, Uchida M, Koizumi H, Nakatsubo T. Carbon and nitrogen limitation of soil microbial respiration in a High Arctic successional glacier foreland near Ny-angstrom lesund, Svalbard. Polar Res. 2007;26:22–30. [Google Scholar]
- Zehr JP, Jenkins BD, Short SM, Steward GF. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol. 2003;5:539–554. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:522–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]