Abstract
A novel high-light (HL)-adapted Prochlorococcus clade was discovered in high nutrient and low chlorophyll (HNLC) waters in the South Pacific Ocean by phylogenetic analyses of 16S ribosomal RNA (rRNA) and 16S–23S internal transcribed spacer (ITS) sequences. This clade, named HNLC fell within the HL-adapted Prochlorococcus clade with sequences above 99% similarity to one another, and was divided into two subclades, HNLC1 and HNLC2. The distribution of the whole HNLC clade in a northwest to southeast transect in the South Pacific (HNLC-to-gyre) and two 8°N to 8°S transects in the Equatorial Pacific was determined by quantitative PCR using specific primers targeting ITS regions. HNLC was the dominant HL Prochlorococcus clade (2–9% of bacterial 16S rRNA genes) at the three westernmost stations in the South Pacific but decreased to less than 0.1% at the other stations being replaced by the eMIT9312 ecotype in the hyperoligotrophic gyre. The highest contributions of HNLC Prochlorococcus in both Equatorial Pacific transects along the latitudinal lines of 170°W and 155°W were observed at the southernmost stations, reaching 16 and 6% of bacterial 16S rRNA genes, respectively, whereas eMIT9312 dominated near the Equator. Spearman Rank Order correlation analysis indicated that although both the HNLC clade and eMIT9312 were correlated with temperature, they showed different correlations with regard to nutrients. HNLC only showed significant correlations to ammonium uptake and regeneration rates, whereas eMIT9312 was negatively correlated with inorganic nutrients.
Keywords: 16S rRNA, Equatorial Pacific, HNLC, ITS, Prochlorococcus, qPCR
Introduction
Prochlorococcus is the most abundant photosynthetic organism in the World Ocean (Partensky et al., 1999), and hence has a key role in global biogeochemical cycles. Since its discovery over two decades ago, the study of strains in culture, as well as natural populations in situ on global scales, have rapidly increased our understanding of the success of this genus. It is now well-known that although Prochlorococcus strains show over 97% similarity of their 16S ribosomal RNA (rRNA) genes, they can be divided into at least six ecotypes on the basis of the phylogeny of the more variable 16S-23S rRNA internal transcribed spacer (ITS) sequences (Rocap et al., 2002). These ecotypes can be divided broadly into high-light- (HL) and low-light-adapted (LL) ecotypes that show distinct photophysiologies (Moore et al., 1998) and which are partitioned vertically in the water column allowing the colonization of the entire euphotic zone (Ferris and Palenik, 1998; West and Scanlan, 1999).
The HL-adapted clade can be divided into two subclades (Urbach et al., 1998; West and Scanlan, 1999) known as HLI and HLII, or alternatively eMED4 and eMIT9312 (Ahlgren et al., 2006). These HL ecotypes exhibit markedly different global distributions; whereas eMIT9312 is the most abundant and widespread ecotype, occurring in warmer, stratified, tropical and sub-tropical waters, eMED4 is usually found in colder and moderately stratified higher latitude waters (Bouman et al., 2006; Johnson et al., 2006; Zwirglmaier et al., 2007, 2008).
Although, light, temperature and physical forcing appear to be the major factors influencing the global distribution of the HL Prochlorococcus ecotypes (Bouman et al., 2006; Zwirglmaier et al., 2008), ambient nutrient concentrations may influence the composition of different populations within an ecotype (Martiny et al., 2009a, 2009b). Physiological studies with HL Prochlorococcus strains (Moore et al., 2002) and genome analyses indicated that strains isolated from nutrient-poor surface waters were not able to use nitrate and nitrite, relying on reduced forms of nitrogen (N) such as ammonium. However, this paradigm of N acquisition by HL Prochlorococcus has been recently challenged by a metagenomic study that revealed nitrate assimilation genes in naturally-occurring populations affiliated with the eMIT9312 ecotype (Martiny et al., 2009b).
The quantification of Prochlorococcus ecotypes by quantitative real-time PCR (qPCR) usually agrees with flow cytometric abundances for the HL-adapted ecotypes in surface waters (Zinser et al., 2006), suggesting that the existing primer sets or probes used, target the majority of HL-adapted ecotypes (Bouman et al., 2006; Johnson et al., 2006; Zwirglmaier et al., 2008). However, there is often a discrepancy between the molecular quantification and flow cytometry analysis for samples collected deeper in the water column (Zinser et al., 2006). A recent survey of Prochlorococcus diversity in the Atlantic and North Pacific Oceans showed that although 93% of 16S-ITS-23 sequences were affiliated to previously described clades, the remaining sequences belonged to several novel LL-adapted subclades (Martiny et al., 2009c).
Thus, it is becoming clear that the current picture of Prochlorococcus biogeography and ecology is not complete, and has been largely influenced by the strains that have been brought into culture, by the primer and probes designed so far, and by the oceanographic regions explored. The South Pacific Ocean is one of the underexplored regions of the World's Ocean and picoplankton diversity has only been studied during a few cruises such as those in the BIogeochemistry and Optics South Pacific Experiment (BIOSOPE) (October–December, 2004) and the Blue EArth Global Expedition (BEAGLE) studies (August 2003–February 2004). Quantification of the HL ecotypes along the BIOSOPE transect revealed that at the western part of the transect in high nutrient and low chlorophyll (HNLC) waters (Claustre et al., 2008), no HL Prochlorococcus were detected with the probes available despite the high concentrations of Prochlorococcus cells measured by flow cytometry (Zwirglmaier et al., 2008). Here, we confirm the presence of a novel, and environmentally abundant HL Prochlorococcus clade by 16S rRNA gene, and 16S-ITS-23S ITS cloning and sequencing. The abundance of the novel clade was quantified along the BIOSOPE transect and in other HNLC waters in the Equatorial Pacific by a newly developed and highly specific qPCR approach and the influence of a series of environmental factors on its distribution was assessed.
Materials and methods
Study sites
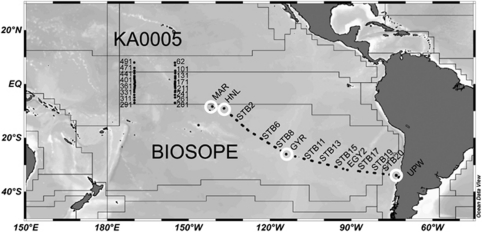
The BIOSOPE cruise track crossed the South Pacific Ocean from the west of the Marquesas Archipelago to the coastal waters of Chile (Claustre et al., 2008) between 26 October and 11 December 2004. The stations sampled are indicated in Figure 1 and consisted of six long-term (>2 days) stations (MAR, HNL, GYR, EGY, UPW and UPX) and 21 short-term (<5 h) stations. The samples analyzed in this study are indicated by a black dot and 16S rRNA gene-clone libraries were prepared at MAR, HNL, GYR and UPW. The NOAA KA0005 cruise was from Pearl Harbor to Kwajalein in the Equatorial Pacific from 13 June to 14 July 2000. Samples were taken between approximately 8°N and 8°S along two N–S lines at 170°W and 155°W (central equatorial Pacific) (Figure 1).
Figure 1.
Location of stations sampled during a NOAA cruise, KA0005, in 1996, in the Equatorial Pacific and during the BIOSOPE cruise in 2004 in the South Pacific Ocean. The BIOSOPE stations used for 16S rRNA gene-clone library construction are indicated with a white circle. The polygons represent the provinces defined by Longhurst (1998).
Sample collection
For the BIOSOPE cruise, seawater samples were collected at multiple depths at the long-term stations and at the surface and deep chlorophyll maximum depths at the short-term stations using a conductivity temperature depth (CTD)-rosette system equipped with 21 × 12 l Niskin bottles. The water samples were prefiltered through 0.8 μm filters, except for stations MAR and STB13 (3.0 μm in these stations). Prefiltered water was filtered onto 0.2 μm pore-size Sterivex cartridges (Millipore, Billerica, NY, USA) except for HNL, STB8, STB11, STB19 (47 mm, 0.2 μm pore-size Nuclepore polycarbonate filters (Whatman, Maidstone, UK) were used in these stations). Filtered volumes were noted and ranged between 4.5–8.1 l for the Sterivex collections and between 0.75–1.0 l for the polycarbonate filter collections. The filters and Sterivex cartridges were immediately stored in liquid nitrogen then at −80 °C until nucleic acid extraction.
For the Equatorial Pacific cruise, seawater was collected as above, prefiltered though GF/A filters (Whatman, 1.6 μm nominal pore size) and 100 ml volumes of seawater were filtered onto 13 mm, 0.2 μm pore-size Supor 200 filters (Pall Co., East Hills, NY, USA) frozen onboard at −20 °C and at −80 °C onshore before nucleic acid extraction.
Environmental and biological parameters
For the BIOSOPE cruise, data was obtained from: http://www.obs-vlfr.fr/proof/ vt/op/ec/biosope/bio.htm and had been measured as described previously for chlorophyll-a (Ras et al., 2008), inorganic nutrients (Raimbault et al., 2008), regenerated ammonium and ammonium-uptake rates (Raimbault and Garcia, 2008) and for picoplankton abundance (Grob et al., 2007a,2007b).
For the KA0005 cruise samples, total chlorophyll-a was measured by a standard fluorometric technique (Chavez et al., 1995). Inorganic nutrients were measured post cruise as described previously (Sakamoto et al., 1990) on 20 ml samples that had been preserved at −20 °C. The environmental and biological parameters are shown in Supplementary Tables S1 and S2.
DNA extraction
For the BIOSOPE samples, DNA was extracted with the AllPrep kit (Qiagen, Chattsworth, CA, USA) as described previously (Manes et al., 2010). For the Equatorial Pacific samples, DNA was extracted from the filters as described previously (Suzuki et al., 2001b). DNA concentrations were measured by the Quant-it Picogreen assay (Invitrogen, Carlsbad, CA, USA). For qPCR, the BIOSOPE samples were diluted to 500 pg μl−1.
Construction of 16S rRNA gene and 16S-ITS-23S rRNA gene-clone libraries
16S rRNA gene clone libraries were constructed for samples collected from the surface and deep chlorophyll maximum depths from stations MAR (15 and 40 m), HNL (5 and 80 m), GYR (5 and 180 m) and UPW (5 and 35 m). 16S-ITS-23S rRNA gene-clone libraries were constructed for samples from 80 and 140 m depths at the HNL station.
16S rRNA gene-clone libraries were prepared as described previously (West et al., 2008) except a mixture of two DNA polymerases was used for amplification of the 16S rRNA genes (1U SuperTaq Polymerase (HT Biotechnology, Cambridge, UK), and 0.05 μl Advantage 2 Polymerase Mix (Clontech, Mountain View, CA, USA) in a total reaction volume of 25 μl and 20 cycles were used for the cycling conditions. For the 16S-ITS-23S rRNA gene-clone libraries, PCR products were amplified in five replicate reacions using the primers PHPICO191F (5′-TGAAATGAATTTCGCCTGAG-3′) (Suzuki et al., 2000) and 23Sr-Y (5′-GGGTTBYCCCATTCRG-3′) (Dyda et al., 2009). PCR reactions (25 μl) contained ∼1 ng DNA, 0.1 μ forward primer, 0.4 μ reverse primer, 0.2 m dNTPs, 0.025U Platinum Taq (Invitrogen), 1 × Platinum Taq buffer and 3.0 m MgCl2. Cycling conditions were 2 min at 94 °C and then 30 s at 94 °C, 30 s at 55 °C and 3 min at 72 °C for 30 cycles followed by 2 min at 72 °C. Products were pooled, purified and reconditioned as above using the 16S-23S ITS PCR reaction and cycling conditions. PCR products were then gel purified by cutting out the bands from agarose gels prepared with modified TAE buffer (40 m Tris-Acetate, pH 8.0, 0.1 m Na2EDTA) and then purified using Ultrafree-DA columns (Millipore) with a centrifugation at 5000 × g for 10 min.
PCR products for the two libraries were cloned within 24 h of PCR cleanup with the TOPO TA cloning kit pCR2.1 (Invitrogen) according to the manufacturer's instructions.
Sequencing and phylogenetic analysis
Plasmid DNA (192 clones for each library) was sequenced using the BigDye terminator kit and a 3730 xl Automatic Sequencer (Applied Biosystems, Foster City, CA, USA) by Macrogen (Seoul, Korea) using primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) for 16S rRNA gene-clone libraries and the primers M13F and M13R for the 16S-23S rRNA ITS clone libraries. Sequences were trimmed manually or using the CodonCode Aligner software (CodonCode Corporation, Dedham, MA, USA). Sequences were aligned and manually corrected using the ARB software (Ludwig et al., 2004). 16S rRNA gene alignments were improved by using secondary structure information within the ARB editor and ITS alignments were manually corrected by using secondary structure models for different Prochlorococcus ecotypes (Rocap et al., 2002). Phylogenetic trees for 16S rRNA gene sequences were constructed with the neighbor joining tree tool in ARB using near full-length sequences, and a maximum frequency filter (50%) for Prochlorococcus that removed hypervariable regions where the alignment could be ambiguous. The same filtered alignment was also used for the construction of a tree using MrBayes 3.1 (Altekar et al., 2004). Partial non-identical sequences were then added using the ADD_BY _PARSIMONY tool in ARB without changing the initial tree topology.
Sequences were submitted to Genbank under the accession numbers HQ232982-HQ233045 for the 16S rRNA gene-clone libraries and HQ233046-HQ233089 for the 16S-ITS-23S clone libraries.
Real-time PCR primer design
qPCR primers specific for the HNLC Prochlorococcus clade were designed using the Probe Design function in the ARB software. Primer pairs were selected to target the whole HNLC clade. Primer specificity was tested in silico through blastn queries.
qPCR standards
Standards were prepared from gene clones from the station HNL 16S-ITS-23S rRNA gene-clone libraries for the HNLC clade and from a BAC clone (MB11E08) for the eMED4 ecotype (Suzuki et al., 2001a). For the eMIT9312 ecotype, 16S-ITS-23S rRNA gene fragments were PCR amplified from Prochlorococcus sp. GP2 (Urbach et al., 1998) DNA and cloned as described above. Plasmid DNA was prepared from overnight Escherichia. coli cultures with the QIAprep Spin Miniprep kit (Qiagen) according to the manufacturer's instructions and eluted in EB buffer. Plasmids (2 μl) were linearized by incubation with the NotI restriction endonuclease (10 U; Promega, Charbonnières-les-Bains, France), 1 × Buffer D (Promega) and BSA (100 μgml−1 final concentration) in a final volume of 50 μl for 3 h at 37 °C. Linearized plasmids were purified with the Qiaquick PCR purification kit (Qiagen) and eluted in 50 μl EB buffer preheated to 70 °C. Concentrations of the linearized plasmids were determined in triplicate with the Quant-iT PicoGreen double-stranded DNA assay (Invitrogen) using a Victor Spectrofluorometer (Perkin Elmer, Waltham, MA, USA). Standards were serially diluted in molecular grade water from 107 down to 101 copies per μl.
qPCR
All the qPCR assays were performed using the StepOne Plus Real-Time PCR System (Applied Biosystems). qPCR reactions with standards or unknowns (samples) were ran in triplicate and negative control reactions in duplicate. HNLC and other Prochlorococcus ecotypes were quantified using the SYBR Green PCR Core Reagents kit (Applied Biosystems) with the primers listed in Table 1. Reaction mixtures (10 μl) contained 1.0 μl template DNA, 500 n primers, 3.0 m MgCl2, 200 μ each dNTP including dUTP, 0.1 U of AmpErase UNG, 0.25 U of AmpliTaq Gold DNA polymerase and 1 × SYBR Green PCR buffer. The PCR cycling conditions were 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 63 °C for 20 s and 72 °C for 20 s. The cycling program was immediately followed by a melting curve analysis to confirm the absence of nonspecific products.
Table 1. List of primers and probes used in this study.
| Target group | Primera | Sequence (5′–3′) | Conc. NM | MgCl2 m | Ann. temp. °C | Reference |
|---|---|---|---|---|---|---|
| All HNLC Pro | HNLC F2 | ATGCAGATTTATTATTGGTGCT | 500 | 3 | 63 | This study |
| HNLC R1b | CGTCCTTCATCGCCTCTAT | |||||
| HLII Pro clade | lowBAII3f | TCGGGGAGTTGGAAGCACA | 300 | 2.5 | 58 | (Ahlgren et al., 2006) |
| lowBAII2r | CCTATATATTCAGTTGGCCGTACAT | |||||
| Synechococcus/Prochlorococcus | PHPICO191F | TGAAATGAATTTCGCCTGAG | 500 | 5 | 59 | (Suzuki et al., 2000) |
| PHPICO420R | AGAAAAGAGGTTTACAGCCCAG | |||||
| (PHPICO283F) | CAGTAGCTGGTCTGAGAGGATGATC | |||||
| Bacteria | BACT1369F | CGGTGAATACGTTCYCGG | 1500 | 3 | 59 | (Suzuki et al., 2000) |
| PROK1492R | GGWTACCTTGTTACGACTT | 1000 | ||||
| (TM1389F) | CTTGTACACACCGCCCGTC | 500 |
Abbreviations: HL, high light; HNLC, high nutrient and low chlorophyll.
TaqMan probe names are in brackets.
For HNLC sets, different combinations of forward and reverse primer concentrations and different annealing temperatures were tested to obtain maximum specificity (highest difference in Ct between the positive and negative control) and maximum amplification efficiency. Clone BIOS04_ITS_HNL_80m005 was used as the positive control and the cloned 16S-ITS-23S rRNA fragment from the eMIT9312 ecotype Prochlorococcus sp. strain GP2 was used as a negative control. Tests for specificity were performed with 107 copies of each standard DNA. Specificity of the HNLC primers was demonstrated as with the optimized conditions, the negative control amplification threshold was 21 Ct higher than the positive control meaning that less than 10 copies were detected from a total of 107 copies.
Detection of the eMED4 and eMIT9312 ecotypes was performed as described previously (Ahlgren et al., 2006) using the same primers (Table 1), reaction conditions and cycling program, except that the reaction volumes were 10 μl and the SYBR Green reagents were the same as that used for the HNLC clade.
The quantification of total bacteria or total prokaryotic picophytoplankton 16S rRNA genes for the normalization of the Prochlorococcus surface ecotype concentrations was performed as described previously (Suzuki et al., 2000) using the BACT2 and PHPICO sets of primers and TaqMan probes, respectively (Table 1), with the same reaction conditions, and cycling programs except that reaction volumes were 10 μl.
Real-time PCR amplification baselines were set automatically and threshold values were set manually. The R2 values of standard curves were always above 0.99. Gene copy numbers were considered quantifiable when the Ct values were below those of the negative controls (no DNA) and when the SYBR Green melting curves showed a single peak.
Statistics
Statistical analysis of the data was performed with SigmaPlot (Systat Software Co., San Jose, CA, USA). Tests for normality demonstrated that the data was not normal, therefore Spearman Rank Order correlations were carried out for the abundance of the ecotypes versus environmental parameters. Data from all sampled depths in the euphotic zone were analyzed (n=21).
Results
Phylogenetic diversity of Prochlorococcus in mesotrophic and oligotrophic waters
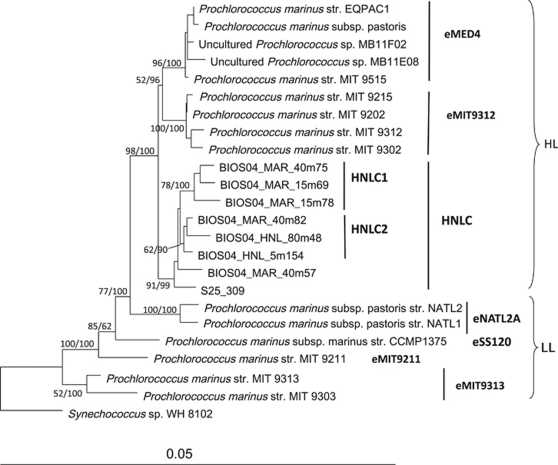
Phylogenetic analysis of bacterial 16S rRNA gene-clone libraries revealed striking differences in the diversity of the HL and LL adapted Prochlorococcus ecotypes between the mesotrophic stations MAR and HNL, and the oligotrophic station GYR. For the HL-adapted Prochlorococcus, only sequences from the well-known eMIT9312 ecotype were recovered in surface waters at the GYR station, whereas only clones belonging to a novel clade of sequences was detected at stations MAR and HNL (Supplementary Table S3). Based on complementary phylogenetic analysis of full-length Prochlorococcus sp. 16S rRNA gene sequences by neighbor joining and Bayesian analyses, this novel clade clustered most closely to HL ecotypes and could be further divided into two subclades (Figure 2). Both the position of the novel clade within the HL clade and the presence of two sub-clades were well supported by the Bootstrap values (neighbor joining) and probability values (Bayesian analysis; Figure 2). As the waters surrounding the HNL station are defined as a HNLC (Claustre et al., 2008), the novel clade was named HNLC and the two subclades HNLC1 and HNLC2. The relative abundance of HNLC1 sequences in the MAR and HNL libraries were roughly equal at the surface and deep chlorophyll maximum depths but HNLC2 had slightly lower abundances than HNLC1 at the surface (Supplementary Table S3). For the LL-adapted ecotypes, eNATL2 (clade LLI) sequences and another closely related cluster of sequences were found at 180 m at GYR, whereas more deeply-branching sequences were the most abundant at HNL at 140 m (Supplementary Table S3).
Figure 2.
Evolutionary relationship of the HNLC clade to other Prochlorococcus clades inferred from near full-length 16S rRNA gene sequences (1353 alignment positions). The tree was constructed using distance (Jukes and Cantor correction) and Neighbor joining (NJ). Trees inferred by Maximum likelihood, Parsimony and Mr Bayes gave the same topology for the HL-adapted clades. Bootstrap values for NJ and percentage confidences for Bayesian analysis are indicated.
Phylogenetic analysis of the 16S-23S rRNA ITS sequences revealed that the HNLC sequences were separated into two distinct clades as found for the 16S rRNA gene sequences (Supplementary Figure S1). Lengths of the ITS sequences ranged from 534–542 bp for HNLC1 and 539–547 bp for HNLC2, which are the ranges observed for other HL Prochlorococcus species (Rocap et al., 2002). Consistent with the well-reported niche separation of HL- and LL-adapted clades in surface and deep water, only LL sequences were recovered at 140 m (Supplementary Table S3).
Distribution of HNLC sequences in the global ocean sampling study
Blastn results of a query to the NCBI nt database using full-length HNLC clade sequences yielded 14 sequences with over 99% identity. However, after a more rigorous phylogenetic analysis only one of the sequences (Clone S25_309; EF57396) fell into the HNLC clade. Further analysis of sequences in the NCBI env_nt database revealed similar sequences belonging to the HNLC clade at several stations of the Global Ocean Sampling study (Rusch et al., 2007) in the Equatorial Pacific. Our bioinformatic analysis indicated that the greatest number of normalized sequence reads assembling into scaffolds containing 16S rRNA gene sequences of the HNLC clade occurred at station GS47, which shares the geographical coordinates with HNL (Supplementary Table S4).
Specificity and performance of qPCR primers for the HNLC clade
To examine the distribution of the HNLC clade along the BIOSOPE transect in more detail, and to determine whether it was present in other HNLC regions, a qPCR assay was developed. As PCR primers with sufficiently discriminatory mismatches were not identified in 16S rRNA gene sequences, 16S-ITS-23S sequences were targeted by these assays.
Primers for the whole HNLC clade (Table 1) were designed from a 16S-ITS-23S rRNA ARB database containing sequences from environmental clones retrieved by this study, cultured Prochlorococcus species (Rocap et al., 2002) and other environmental clones (Martiny et al., 2009c). Primers were designed to have the greatest number of mismatches to other ecotypes and to target the maximum number of HNLC sequences (Table 1). Primer HNLC-F2 had perfect matches to the majority of the HNLC sequences (33/47), whereas there were two mismatches with the eMED4 and eMIT9312 ecotype sequences and at least nine mismatches with the LL ecotypes' sequences. Reverse primer HNLC-R1b, targeted 45/47 HNLC sequences and has a single mismatch to all other Prochlorococcus clades, localized at its 3′ end.
For the HNLC-F2 and -R1b pair, a qPCR efficiency of 0.92±0.006 was achieved with a detection limit of less than 10 copies per reaction. Examination of the melting curves confirmed the presence of a single peak and hence of the specificity of the amplifications. The 13 HNLC sequences with mismatches to the primer had single mismatches at different positions and therefore we can not rule out that abundance estimates of the HNLC ecotype might be somewhat underestimated by these assays and its relative abundance of total bacteria values presented should be considered as a minimum percentage abundance.
Distribution of the HNLC clade along the BIOSOPE transect and in the Equatorial Pacific
The HNLC clade and the other HL ecotypes were quantified by qPCR using the optimized conditions described here or elsewhere (Ahlgren et al., 2006) and their abundances were expressed as a percentage of total bacteria or total picocyanobacteria 16S rRNA gene copies.
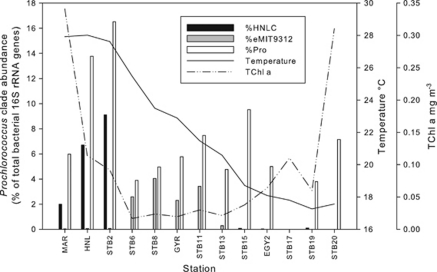
For the BIOSOPE transect, the HNLC clade increased steadily in relative abundance in surface waters from about 2 to 9% of total bacteria from station MAR to STB2, but decreased substantially in abundance, east of STB2, never exceeding 0.09% of total bacteria at these stations (Figure 3). The eMIT9312 ecotype dominated in the central gyre from stations STB6-STB11 accounting for 2.6–4% of total bacteria decreasing to less than 0.3% at STB13 and did not rise above 0.02% for the remainder of the transect. Abundances of HNLC and eMIT9312 were also calculated as gene copy per ml from the known volumes of water filtered and quantity of DNA used in the qPCR (Supplementary Figure S2) assuming complete DNA extraction and one ribosomal operon per Prochlorococcus cell. The maximum abundance of the HNLC clade was also observed at STB2 at 1.7 × 105 gene copies per ml, which corresponded well with the flow cytometry cell abundance of 1.9 × 105 cells per ml (Supplementary Figure S2).
Figure 3.
Relative abundance of HNLC and eMIT9312 clades (percent total bacterial rRNA genes) in surface samples along the BIOSOPE transect in the South Pacific Ocean. The relative abundance of Prochlorococcus cell counts (Pro) was calculated from flow cytometry measurements (Grob et al., 2007a). Meso, mesotrophic; oligo, oligotrophic.
One surprising result for the eMED4 primer set (Ahlgren et al., 2006) was that although we used the previously described reaction conditions and cycling program, we consistently obtained high amplification from a eMIT9312-negative control plasmid and thus quantification of eMED4 abundances was not further pursued. As we were most interested in understanding the factors influencing the abundance and distribution of the HNLC ecotype and eMIT9312 in the latitudinal band of 10 °N-25 °S, we did not consider the absence of data on eMED4 critical as this ecotype is most abundant outside of this band (Johnson et al., 2006; Zwirglmaier et al., 2007, 2008).
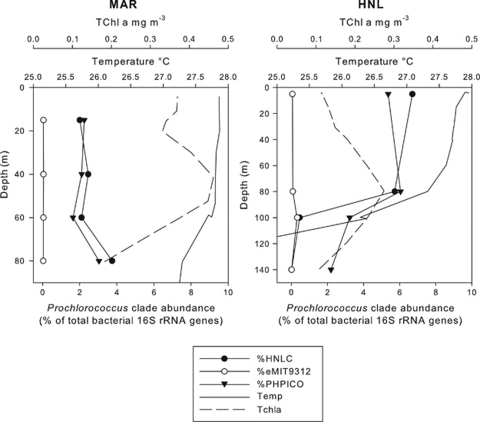
HNLC and eMIT9312 ecotypes were also quantified in vertical profiles at stations MAR and HNL relative to total picocyanobacteria measured with the PHPICO (Suzuki et al., 2000) primer set (Figures 4a and b). As expected for a HL-adapted organism, the HNLC ecotype showed constant and high relative abundances from surface water down to 80 m before decreasing significantly in the HNL depth profile between 80–100 m corresponding to the base of the euphotic zone. In addition, at station STB2 where surface HNLC abundances were the highest, HNLC only accounted for ∼0.6% of total bacterial 16S rRNA genes at 130 m (data not shown).
Figure 4.
Vertical profiles of the relative abundance (% total bacterial rRNA genes) of clades HNLC and eMIT9312 compared with relative abundance of picocyanobacterial rRNAs (percent total bacterial rRNA genes) at stations MAR and HNL.
To determine if the HNLC clade was present in significant abundance in other HNLC waters, this ecotype was quantified in samples from a cruise in the Equatorial Pacific using our qPCR assay, and compared with the abundance of eMIT9312, the dominant Prochlorococcus ecotype known to exist in low-latitude waters (Johnson et al., 2006; Zwirglmaier et al., 2007). In two North–South transects along the longitude lines of 155 °W and 170 °W, the HNLC clade was the dominant clade at the most southerly stations (8° S) accounting for 6 and 16%, respectively, of total bacterial abundance (Figure 5). In contrast, this trend was reversed towards the equator where the eMIT9312 ecotype was the most abundant reaching 3.6% of total bacteria. When considering the sum of both Prochlorococcus clades, lower relative abundances were observed at the Equator sites (Figure 5).
Figure 5.
Relative abundance of eHNLC and eMIT9312 clades (percent total bacterial rRNA genes) in surface samples along 155W and 170W transects in the Equatorial Pacific.
Relationship of the Prochlorococcus HNLC and eMIT9312 clade distributions to environmental parameters
To investigate the environmental factors influencing the differential distributions of these two clades, Spearman Rank Order correlations were performed with the wide range of environmental data available for the BIOSOPE transect. Although both clades were positively correlated with temperature, they exhibited very different patterns of correlations for the other environmental parameters (Table 2). Whereas the eMIT9312 ecotype was negatively correlated with inorganic nutrients and positively correlated with salinity, the HNLC clade only showed significant positive correlations with total chlorophyll-a, Synechococcus cell counts, and ammonium uptake and regeneration rates.
Table 2. Spearman rank order correlations for Prochlorococcus clades and environmental parameters for the BIOSOPE dataset.
| Clade | Temp. | Sal. | Tchla | NO3 | NO2 | NH4 | PO4 | ρNO3 | ρNH4 | rNH4 | Syn | PEuk |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HNLC | 0.475* | NS | 0.443* | NS | NS | NS | NS | NS | 0.539* | 0.468* | 0.505* | NS |
| eMIT9312 | 0.662* | 0.936** | −0.482* | −0.688* | −0.691* | −0.514* | −0.555* | NS | NS | NS | NS | −0.710* |
Abbreviation: NS, not significant.
*significance level <0.05; **Significance level <0.0001.
ρNH4, ammonium uptake (nmoles N l−1 d−1).
ρNO3, nitrate uptake (nmoles N l−1 d−1).
rNH4, ammonium regeneration (nmoles N l−1 d−1).
Syn, Synechocococcus concentrations (cells per ml).
PEuk, picoeukaryote concentrations (cells per ml).
A more limited number of environmental parameters were measured during the Equatorial Pacific cruise (inorganic nutrients, temperature, salinity and Chla; Supplementary Table S2). When Spearman Rank Order correlations were performed, no significant correlations were found for either the HNLC or eMIT9312 ecotype with any of these parameters.
Discussion
Here, we report on the characterization and quantification of a novel third HL clade named HNLC because of its presence in HNLC waters. A very recent publication also reported on a novel Prochlorococcus clade in HNLC regions, indentified by a metagenomic approach (Rusch et al., 2010) and analyzed in the context of modeled environmental parameters. The discovery of the HNLC ecotype means that the two HL clade paradigm must now be adjusted to accommodate this third clade into the phylogeny and ecology of Prochlorococcus.
Distribution of HNLC
Usually, the quantification of the eMED4 and eMIT9312 clades in surface waters agrees closely with flow-cytometric analyses of total Prochlorococcus cells (Zinser et al., 2006). However, notable exceptions include the lack of a HL ecotype at the west end of the BIOSOPE transect (Zwirglmaier et al., 2008). The results from our 16S rRNA and 16S-ITS-23S rRNA clone libraries explain the absence of hybridization to existing HL probes at MAR and HNL (BIOSOPE) by the almost exclusive presence of the HNLC clade.
Although the HNLC clade was the dominant Prochlorococcus clade in surface waters at the mesotrophic stations MAR and HNL, its abundance as a proportion of total bacteria was greatest at the moderately oligotrophic STB2 station. In addition, at this station, Prochlorococcus cell counts were the highest in the entire transect at 1.9 × 105 cells per ml, whereas its abundance decreased about 10-fold (0.2 × 105 cells per ml) towards the Gyre (Grob et al., 2007a; Supplementary Table S1). The dominance of eMIT9312 at the gyre stations (STB6-STB11) is in complete agreement with previous dot blot hybridization data (Zwirglmaier et al., 2008). HNLC Prochlorococcus was also present in the Equatorial Pacific North–South transects with slightly higher relative abundances compared with those at STB2. Therefore, on the basis of our qPCR results and from the distribution of HNLC sequences in the Global Ocean Sampling dataset, the HNLC clade appears to be most prevalent in a 8°N–13°S latitudinal band that corresponds to surface temperatures greater than about 27 °C.
The presence of HNLC in this region maps roughly onto the Pacific Equatorial Divergence domain (Longhurst, 1998) shown in Figure 1. Although the HNLC clade was most abundant and dominated HL Prochlorococcus at the southern boundary of this province, eMIT9312 dominated at the equator. These results contrast to the relative abundances of these two clades assessed from fragment recruitment rates that showed HNLC to be the dominant clade throughout the Equatorial Pacific and eMIT9312 to dominate in the oligotrophic gyres (Rusch et al., 2010).
Apart from the Equatorial Pacific, there is also some evidence that the HNLC ecotype may be present in the Indian Ocean (Rusch et al., 2010) together with the eMIT9312 ecotype (Bouman et al., 2006). Future studies could use our qPCR assays with different samples in the Indian Ocean in order to confirm the presence of the HNLC ecotype in this region.
The co-occurrence of both HNLC1 and HNLC2 clades at MAR and HNL at the surface and deeper depths raises the question as to the ecological significance of these two subclades (99.0% similar) and why they coexist. The other two HL clades eMIT9312 and eMED4 that show a similar degree of divergence, exhibit very different biogeographical distributions, only coexisting at similar abundances at the latitudes where one ecotype replaces the other (Johnson et al., 2006). Intriguingly, HNLC2 sequences were recovered in lower numbers at the surface depths compared with the deeper samples. Although the number of sequences analyzed is too low to be statistically significant, this distribution may hint at a preference for the HNLC2 clade for lower light and/or lower temperature. Future studies quantifying these clades separately might clarify the ecological significance of HNLC1 and HNLC2.
Influence of environmental factors
The somewhat restricted geographical distribution of HNLC Prochlorococcus is in contrast to the widespread distribution of the other HL ecotypes, which may suggest that several environmental conditions must be satisfied for HNLC to dominate the Prochlorococcus population. The features that characterize in general the Pacific Equatorial Divergence domain are warmer temperatures (approximately 27–28 °C), higher inorganic nutrients (approximately 5 μ N and Si, and 0.5 μ P) and lower iron (Fe) concentrations (<1 n). When the HNLC clade distribution is examined in the context of the environmental parameters, this clade seems quite atypical when compared with the other HL-adapted ecotypes. Whereas the eMIT9312 and eMED4 ecotypes are often negatively correlated with inorganic nutrient concentrations, as shown here and in other studies (Bouman et al., 2006; Johnson et al., 2006) HNLC did not show any significant correlations with nitrate, nitrite or phosphate. However, HNLC Prochlorococcus was correlated to ammonium uptake and regeneration rates, which were in fact measured in the same samples in the BIOSOPE cruise, whereas there were no significant relationships of eMIT9312 to these parameters. Although we did not have actual measurements for the Equatorial Pacific samples where the HNLC clade was present, other studies have shown high rates of ammonium uptake and regeneration in this region (Raimbault et al., 1999). Along with the positive correlation of HNLC Prochlorococcus to Chla and Synechococcus, these observations lead us to hypothesize that HNLC is adapted to less oligotrophic environments and may be dependent on higher ammonium availability.
Iron is thought to be the major limiting nutrient of phytoplankton in the Equatorial Pacific (Martin, 1994; Coale et al., 1996). Thus, one explanation for the presence of this new clade in a HNLC region could be adaptation to low iron bioavailability. Some evidence for this hypothesis is provided by a reduction in the number of iron-containing proteins predicted from consensus genomes of the HNLC clade assembled from metagenomic data (Rusch et al., 2010). This was suggested to give a selective advantage to the HNLC ecotype over eMIT9312 in Fe-limited and nutrient-rich environments by allowing it to better compete because of a reduced iron quota. Although we understand that such an adaptation may explain partly the success of the HNLC clade in this oceanic region, our qPCR data suggest that adaptation to low iron bioavailability is not necessarily the major factor influencing the distribution of the HNLC clade and eMIT9312. For example, eMIT9312 was the dominant Prochlorococcus ecotype at the centre of the Equatorial region at both 155°W and 170°W (Figure 5). Compared with the 8–13°S area, this area is a more typical HNLC region where excess nitrate and phosphate inputs from upwelling, and large distances from iron sources, leads to iron limitation of phytoplankton.
HNLC abundances were positively correlated with temperature and this clade only dominated the Prochlorococcus populations at temperatures above 27 °C. Temperature is thought to be the major parameter influencing the distributions of the other HL ecotypes (Johnson et al., 2006; Zwirglmaier et al., 2008) and thus may also be important in determining the relative distributions of eMIT9312 versus HNLC. Both the qPCR data from the BIOSOPE transect and the Equatorial Pacific transects support this idea. The abundance of eMIT9312 increased in the equatorial upwelling waters (∼27.2 °C) and towards the South Pacific Gyre (∼22 °C), whereas HNLC showed its maximum abundance at the stations with the highest temperatures (>28 °C, Equatorial Pacific cruise; >27.6 °C BIOSOPE). Analogous to the differential growth rates of eMIT9312 and eMED4 at different temperatures (Johnson et al., 2006), perhaps HNLC is able to grow faster than eMIT9312 at more elevated temperatures. Thus, obtaining an isolated strain in this clade remains as an important target of future efforts.
Conclusions
Here, we demonstrate the presence of a third HL clade of Prochlorococcus that dominates surface Prochlorococcus populations in the sub Equatorial Pacific. We propose that the HNLC clade is more adapted for growth at higher temperatures than eMIT9312 and less adapted for growth in oligotrophic environments, requiring higher ammonium availability provided by rapid regeneration of nutrients. The apparently more fastidious nature of the HNLC clade may explain its restriction to a relatively small geographical area compared with that of the two other HL ecotypes. The isolation of a strain from this ecotype will be crucial for comparative genomics and physiological studies to better understand the distribution and ecology of the HNLC clade. It will also be important to incorporate the HNLC clade into marine ecosystem models (Follows et al., 2007) to predict how their distributions may be affected by future climate change.
Acknowledgments
We thank I Obernosterer and P Catala for sample collection during the BIOSOPE cruise, C-L de O. Manes for technical help and C Lepère for providing the Prochlorococcus sp. GP2 DNA. The nutrient data from the Equatorial Pacific were collected under NASA and NOAA grants to F. Chavez, MBARI. We also thank A Martiny and D Rusch for fruitful exchanges and D Scanlan for the critical reading of the manuscript. This research was supported by the following programs: BIOSOPE cruise (CNRS-INSU LEFE) and PICOFUNPAC (ANR Biodiversité 06-BDIV-013). The acquisition of BIOSOPE data was funded by CNRS-INSU.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ahlgren N, Rocap G, Chisholm S. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel metropolis coupled markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Bouman H, Ulloa O, Scanlan D, Zwirglmaier K, Li W, Platt T, et al. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science. 2006;312:918–921. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- Chavez F, Buck K, Bidigare R, Karl D, Hebel D, Latasa M, et al. On the chlorophyll a retention properties of glass-fiber GF/F filters. Limnol Oceanogr. 1995;40:428–433. [Google Scholar]
- Claustre H, Sciandra A, Vaulot D. Introduction to the special section bio-optical and biogeochemical conditions in the South East Pacific in late 2004: the BIOSOPE program. Biogeosciences. 2008;5:679–691. [Google Scholar]
- Coale KH, Johnson KS, Fitzwater SE, Gordon RM, Tanner S, Chavez FP, et al. 1996A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean Nature 383495–501.doi: 10.1038/383495a0 [DOI] [PubMed] [Google Scholar]
- Dyda RY, Suzuki MT, Yoshinaga MY, Harvey H. The response of microbial communities to diverse organic matter sources in the Arctic Ocean. Deep Sea Res II: 2009;56:1249–1263. [Google Scholar]
- Ferris M, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–228. [Google Scholar]
- Follows M, Dutkiewicz S, Grant S, Chisholm S. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315:1843–1846. doi: 10.1126/science.1138544. [DOI] [PubMed] [Google Scholar]
- Grob C, Ulloa O, Claustre H, Huot Y, Alarcon G, Marie D. Contribution of picoplankton to the total particulate organic carbon concentration in the eastern South Pacific. Biogeosciences. 2007a;4:837–852. [Google Scholar]
- Grob C, Ulloa O, Li W, Alarcon G, Fukasawa M, Watanabe S. Picoplankton abundance and biomass across the eastern South Pacific Ocean along latitude 32.5 degrees S. Mar Ecol Prog. 2007b;332:53–62. [Google Scholar]
- Johnson Z, Zinser E, Coe A, McNulty N, Woodward E, Chisholm S. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- Longhurst A. Ecological Geography of the Sea. Academic: San Diego; 1998. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Kumar Y, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes C, West N, Rapenne S, Lebaron P.2010Dynamic bacterial communities on reverse-osmosis membranes in a full-scale desalination plant BiofoulingIn Press. [DOI] [PubMed]
- Martin J. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature. 1994;371:123–129. [Google Scholar]
- Martiny A, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009a;11:1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- Martiny A, Kathuria S, Berube P. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. PNAS. 2009b;106:10787–10792. doi: 10.1073/pnas.0902532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny A, Tai A, Veneziano D, Primeau F, Chisholm S. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol. 2009c;11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- Moore L, Post A, Rocap G, Chisholm S. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr. 2002;47:989–996. [Google Scholar]
- Moore L, Rocap G, Chisholm S. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Partensky F, Hess W, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimbault P, Garcia N. Evidence for efficient regenerated production and dinitrogen fixation in nitrogen-deficient waters of the South Pacific Ocean: impact on new and export production estimates. Biogeosciences. 2008;5:323–338. [Google Scholar]
- Raimbault P, Garcia N, Cerutti F. Distribution of inorganic and organic nutrients in the South Pacific Ocean—evidence for long-term accumulation of organic matter in nitrogen-depleted waters. Biogeosciences. 2008;5:281–298. [Google Scholar]
- Raimbault P, Slawyk G, Boudjellal B, Coatanoan C, Conan P, Coste B, et al. Carbon and nitrogen uptake and export in the equatorial Pacific at 150°W: evidence of an efficient regenerated production cycle. J Geophys Res Oceans. 1999;104:3341–3356. [Google Scholar]
- Ras J, Claustre H, Uitz J. Spatial variability of phytoplankton pigment distributions in the Subtropical South Pacific Ocean: comparison between in situ and predicted data. Biogeosciences. 2008;5:353–369. [Google Scholar]
- Rocap G, Distel D, Waterbury J, Chisholm S. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch D, Halpern A, Sutton G, Heidelberg K, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. Plos Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA. 2010;107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto C, Friederich G, Codispoti L. Monterey Bay Aquarium Research Institute Technical Report 90-2. Monterey Bay Aquarium Research Institute, Moss Landing, CA, USA; 1990. MBARI procedures for automated nutrient analyses using a modified Alpkem series 300 rapid flow analyzer. [Google Scholar]
- Suzuki M, Beja O, Taylor L, DeLong E. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol. 2001a;3:323–331. doi: 10.1046/j.1462-2920.2001.00198.x. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Preston C, Chavez F, DeLong E. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat Microbiol Ecol. 2001b;24:117–127. [Google Scholar]
- Suzuki M, Taylor L, DeLong E. Quantitative analysis of small subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol. 2000;66:4605–4614. doi: 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach E, Scanlan D, Distel D, Waterbury J, Chisholm S. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria) J Mol Evol. 1998;46:188–201. doi: 10.1007/pl00006294. [DOI] [PubMed] [Google Scholar]
- West NJ, Obernosterer I, Zemb O, Lebaron P. Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ Microbiol. 2008;10:738–756. doi: 10.1111/j.1462-2920.2007.01497.x. [DOI] [PubMed] [Google Scholar]
- West N, Scanlan D. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol. 1999;65:2585–2591. doi: 10.1128/aem.65.6.2585-2591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Coe A, Johnson Z, Martiny A, Fuller N, Scanlan D, et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol. 2006;72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwirglmaier K, Heywood J, Chamberlain K, Woodward E, Zubkov M, Scanlan D. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol. 2007;9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.