Abstract
The first step of nitrification, oxidation of ammonia to nitrite, is performed by both ammonia-oxidising archaea (AOA) and ammonia-oxidising bacteria (AOB) in soil, but their relative contributions to ammonia oxidation and existence in distinct ecological niches remain to be determined. To determine whether available ammonia concentration has a differential effect on AOA and AOB growth, soil microcosms were incubated for 28 days with ammonium at three concentrations: native (control), intermediate (20 μg NH4+-N per gram of soil) and high (200 μg NH4+-N per gram of soil). Quantitative PCR demonstrated growth of AOA at all concentrations, whereas AOB growth was prominent only at the highest concentration. Similarly, denaturing gradient gel electrophoresis (DGGE) analysis revealed changes in AOA communities at all ammonium concentrations, whereas AOB communities changed significantly only at the highest ammonium concentration. These results provide evidence that ammonia concentration contributes to the definition of distinct ecological niches of AOA and AOB in soil.
Keywords: ammonia-oxidising archaea, ammonia-oxidising bacteria, nitrification, ammonia concentration, amoA
Introduction, Results and Discussion
Recent studies have provided strong evidence of a major role for organisms within the domain Archaea in the global nitrogen cycle, oxidising ammonia to nitrite in the first stage of nitrification (Francis et al., 2007; Prosser and Nicol, 2008). Metagenomic studies first revealed that soil and marine thaumarchaea possess homologues of bacterial genes encoding subunits of the enzyme ammonia monooxygenase (Venter et al., 2004; Treusch et al., 2005), which oxidises ammonia to hydroxylamine, before its conversion to nitrite. Laboratory cultivation subsequently confirmed the existence of an autotrophic, ammonia-oxidising metabolism within the domain Archaea (Könneke et al., 2005). Quantification of the functional marker gene encoding ammonia monooxygenase subunit A (amoA) indicates that putative ammonia-oxidising archaea (AOA) are generally more abundant than putative ammonia-oxidising bacteria (AOB) in soil (for example, Leininger et al., 2006; He et al., 2007; Nicol et al., 2008). However, potential differences in cell size, specific cell activity and other physiological characteristics make it difficult to assess accurately the relative contributions of these two groups to soil nitrification. AOA and AOB may be active under the same conditions with both groups competing directly, there may be functional redundancy or fundamental physiological differences may result in distinct ecological niches.
There is evidence that differences in affinity for ammonia may lead to differential growth of AOA and AOB, contributing to niche separation. For example, in the open ocean, where ammonia concentration is relatively low (⩽10 μ), AOA are much more abundant than AOB and AOA abundance correlates well with inferred or measured rates of nitrification (for example Wuchter et al., 2006; Beman et al., 2008). The AOA isolate Nitrosopumilus maritimus also exhibits high specific ammonia oxidation rates at very low ammonia concentrations that are typical of the open ocean (Martens-Habbena et al., 2009). In soil, AOA and AOB have been demonstrated to exhibit some level of functional redundancy (Schauss et al., 2009). However, growth of AOB, and not of AOA, has been linked to nitrification activity following amendment with high levels of ammonium, either directly as mineral fertiliser or as urea, which is rapidly hydrolysed to ammonium (Di et al., 2009; Jia and Conrad, 2009). In contrast, growth of AOA is associated with nitrification in soils with continual supply of ammonia at low concentration through mineralisation of organic matter (Offre et al., 2009). The aim of this study was to determine whether ammonia concentration influences the relative growth of AOA and AOB in soil microcosms with low, intermediate and high ammonium supply, approximately equivalent to the conditions in unfertilised soils and those receiving moderate and high levels of inorganic fertiliser.
Soil was sampled in triplicate from the upper 10 cm of a sandy loam soil maintained at pH 7.5 (Craibstone, Scotland) before pooling and sieving. Details of the soil site and soil characteristics are described in Nicol et al. (2008). Soil microcosms were established in 30-ml glass vials, each containing 10 g soil and amended with 0.5 ml water, 2 or 20 mg ammonium sulfate per ml, giving respective final concentrations of <0.5, 20 or 200 μg NH4+-N per gram of soil and a final water content of 30% (w/w). Microcosms were then capped with loose-fitting lids, to allow air exchange, and were incubated at 30 °C in the dark. At intervals of 3.5 or 7 days, microcosms were either destructively sampled in triplicate and stored at −20 °C for subsequent analysis or supplemented (‘spiked') with water or ammonium sulfate to restore target ammonium concentrations. The volume of liquid added was equal to that lost through evaporation since the last amendment. Ammonium (the pool of ammonium+ammonia) and nitrite+nitrate concentrations were determined colorimetrically by flow injection analysis of soil-KCl extracts as described by Offre et al. (2009).
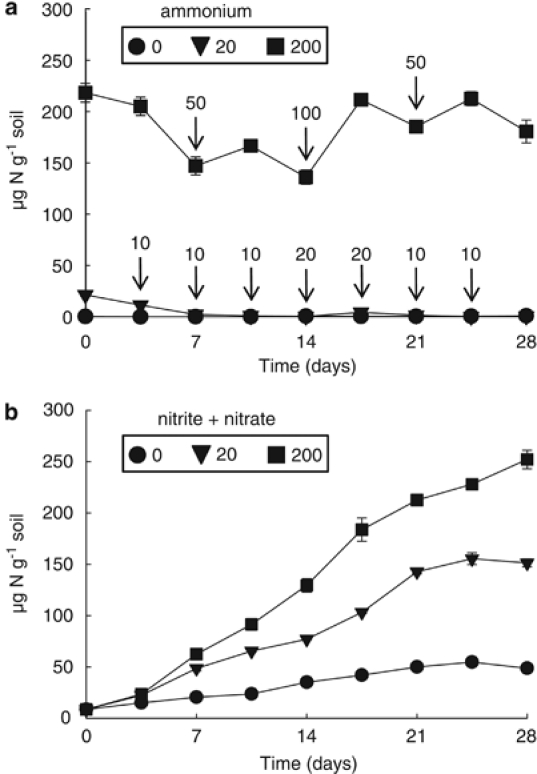
The regular microcosm amendments aimed to establish three levels of ammonium supply and influenced the nitrification rate (Figure 1). Preliminary experiments showed that ammonium added at the intermediate level (giving an ammonium concentration of 20 μg NH4+-N per gram of soil) was almost fully oxidised by 3.5 days, whereas ammonium concentration remained high after amendment to 200 μg NH4+-N per gram, despite some oxidation. The frequency of spiking was therefore greater for the intermediate than for the high level of amendment (Figure 1a). Ammonium concentration in the water-only microcosms remained below the detection level (0.1 μg NH4+-N per ml), as ammonia released through mineralisation was oxidised immediately, resulting in a relatively constant nitrification rate of 1.5–2 μg per gram of soil per day (Figure 1b). Nitrification rates were greater in microcosms with intermediate and high ammonium concentration (targeted at 20 and 200 μg NH4+-N per gram of soil), at ∼5 and ∼10 μg per gram of soil per day, respectively. This twofold difference suggests that under these conditions the maximum nitrification rate for this soil is reached with an ammonium input lower than 200 μg NH4+-N per gram. Nitrification resulted in decreases in pH from 7.5, at day 0, to 7.4, 6.8 and 6.4 at day 28 in the 0, 20 and 200 μg NH4+-N per gram of soils, respectively.
Figure 1.
Nitrification kinetics in soil microcosms incubated for 28 days at 30 °C with regular additions of ammonium to maintain target concentrations of 0, 20 and 200 μg NH4+-N per gram of soil. (a) Ammonium concentration in soil microcosms that were either destructively sampled or amended with water or ammonium sulfate solution at intervals of 3.5 days. Arrows indicate the times at which ammonium was added to soil microcosms and associated numbers describe the increase in ammonium concentration (μg N per gram of soil). (b) Nitrite+nitrate concentration in soil microcosms produced from oxidised ammonia. Each point and error bar represents the mean and standard error of triplicate microcosms, with some error bars smaller than the symbol size.
Nucleic acids were extracted as described previously (Nicol et al., 2006) and amoA gene abundance was determined by quantitative PCR (qPCR) in a DNA Engine Opticon 2 System (GRI, Braintree, UK) using the Qiagen Quantifast SYBR Green PCR Master Mix (Qiagen, Crawley, UK), according to the manufacturer's instructions. AOA and AOB amoA gene abundances were quantified using primers CrenamoA23f and CrenamoA616r (Tourna et al., 2008) and amoA-1F and amoA-2R (Rotthauwe et al., 1997), respectively. Standard curves were generated using a serial dilution of a 2105-bp amplicon (108–102 amoA copies) derived from fosmid 54d9 (containing complete amoA and amoB genes; Treusch et al., 2005) and Nitrosospira multiformis genomic DNA (107–102 amoA copies), for AOA and AOB assays, respectively. All PCR efficiencies were between 87–95% with r2>0.99.
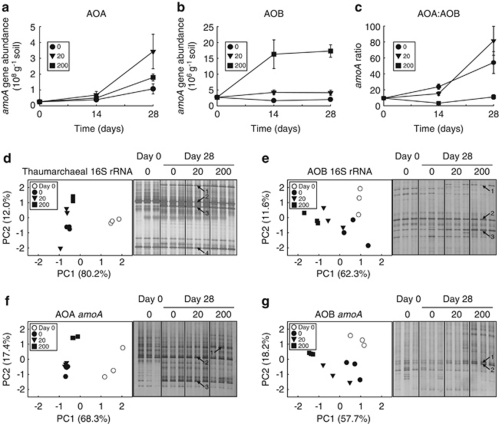
AOA amoA gene abundance increased significantly (one-way ANOVA) in microcosms during the 28-day incubation period, from 2.5 × 107 to 1.1 × 108 (P=0.06), 3.4 × 108 (P=0.04) and 1.8 × 108 (P=0.002) per gram of soil for the 0, 20 and 200 μg NH4+-N per gram of soil amendments, respectively, representing 4-, 13.5- and 7-fold increases (Figure 2a). The differential response to low, intermediate and high ammonium levels was not significant (P=0.11 at day 28), but a ‘20>200>0' trend was observed after incubation for both 14 and 28 days. Although difficult to determine reliably from three time points, an assumption of exponential growth throughout the incubation period gives specific growth rates of 0.052, 0.094 and 0.070 per day at low, intermediate and high ammonium amendments. Thus, AOA growth was not strongly correlated with different rates of ammonia oxidation. In contrast, AOB amoA abundance was not affected in the water-only (native ammonia) microcosms (P=0.22) and did not increase significantly (1.5-fold; P=0.17) at the intermediate ammonium concentration after incubation for 28 days (Figure 2b). However, it increased 6-fold, from 0.27 to 1.6 and 1.7 × 107 copies per gram of soil at days 14 (P=0.04) and 28 (P=0.002), respectively, in microcosms with high ammonium concentration and the greatest ammonia oxidation rate. AOB abundance may have reached a plateau after 14 days, but variability in abundance data may have prevented detection of further growth, and there was no evidence of exponential growth with the sample frequency employed. AOA always outnumbered AOB, but the dominant AOB response to the high ammonium level was evident by comparing AOA:AOB ratios (Figure 2c). The potential contributions of AOA and AOB to the observed nitrite+nitrate production were estimated using the approach adopted by Boyle-Yarwood et al. (2008) to estimate maximum nitrate production from each group as the product of mean amoA abundance during the incubation period and cell activities from pure cultures of bacterial ammonia oxidisers and Nitrosopumilus maritimus. This analysis indicated that either AOA or AOB were capable of the nitrite+nitrate production rates observed at all three ammonia concentrations and variability in abundance values, and uncertainties arising from the dynamic nature of the communities and the lack of cell activities for soil archaea prevented more detailed interpretation.
Figure 2.
Characterisation of AOA and AOB communities in soil microcosms receiving ammonium amendments to maintain different target ammonium concentrations of 0, 20 and 200 μg NH4+-N per gram of soil. Changes in abundance were determined by qPCR analysis of the AOA (a) and AOB (b) amoA genes and (c) changes in AOA:AOB amoA abundance ratio, in soil microcosms destructively sampled after incubation for 0, 14 and 28 days. Each point and error bar represent the mean and standard error of triplicate microcosms, with some error bars being smaller than the symbol size. Changes in AOA (d, f) and AOB (e, g) community structure were determined by DGGE analysis of 16S rRNA and amoA genes derived from microcosms sampled after incubation for 0 and 28 days, with each lane representing an individual microcosm. Analysis of thaumarchaeal 16S rRNA gene sequences assumes that those organisms targeted by this assay also possess amoA genes and are putative AOA. PCR products derived from triplicate microcosms incubated for 28 days with either 20 or 200 μg NH4+-N per gram of soil were pooled and cloned with the sequences of predominant band positions (highlighted with numbered arrows) determined by screening and sequencing multiple clones with identical migration patterns before performing phylogenetic analysis (Supplementary Figures 2 and 3). Gel images were digitised and analysed by principal component analysis as described previously (Nicol et al., 2008).
For denaturing gradient gel electrophoresis (DGGE) analysis of changes in community structure after incubation of microcosms for 28 days, AOB and thaumarchaeal 16S rRNA genes were amplified using a nested PCR approach, and amoA gene amplicons were obtained by a single PCR, as described previously (Nicol et al., 2008). The sequences of predominant band positions in the DGGE profiles at day 28 were determined by cloning, screening and sequencing multiple clones representative of each band position of interest (Supplementary Figures 2 and 3). DGGE profiles at day 0 (Figure 2) were similar to those obtained previously for the pH 7.5 soil at the same site (Nicol et al., 2008), and bands appearing after incubation were not representative of those found in soils at this site with a lower pH (Supplementary Figure 1). Phylogenetic analysis of AOA 16S rRNA and amoA gene sequences revealed that although the thaumarchaeal community was initially dominated by group 1.1b organisms, a number of group 1.1a populations became the predominant component of the communities after 28 days (Supplementary Figure 2). Analysis of AOB sequences demonstrated that Nitrosospira cluster 3 phylotypes were the most responsive in the highest amendment. Organisms within this lineage are commonly found in agricultural soils (Stephen et al., 1996), with some populations being tolerant to relatively high ammonia concentrations (Webster et al., 2005). Interestingly, in the 16S rRNA gene DGGE profiles, an additional faint band (highlighted band 1) increased in relative intensity in the highest amendment, which belonged to an organism placed within the Nitrosomonas oligotropha lineage with high sequence similarity (98%) to Nitrosomonas ureae. Visual inspection of DGGE gels and principal component analysis (PCA) of digitised images (Nicol et al., 2008) of DGGE profiles of both genes revealed contrasting responses of the AOA and AOB community structures, which correlated with abundance measurements. At all ammonium concentrations, AOA communities showed similar changes after incubation for 28 days (Figures 2d and f), compared with day 0 microcosms. AOB profiles (Figures 2e and g) showed much smaller changes. There was little detectable change in AOB community structure after no ammonium amendment, but evidence for a large change in AOB community structure following amendment to 200 μg NH4+-N per gram. Thus, ammonium concentration was a more important factor in determining AOB community structure than that of AOA.
Conclusion
AOA grew at low, intermediate and high ammonia input, although without significant difference between ammonia concentrations. However, AOB only grew significantly at high ammonia concentrations. This suggests different growth responses to ammonia concentration, and indicates that AOA and AOB occupy separate ecological niches. The results, obtained under controlled conditions, are consistent with field observations that indicate a higher AOA:AOB ratio in soil ecosystems with low-to-intermediate ammonium concentration. The data also suggest that AOA may contribute significantly to nitrification of ammonia released through mineralisation, whereas growth of AOB is favoured at high ammonium concentration, which is more typical of agricultural soils receiving high inorganic nitrogen input. Further studies are required to determine the extent to which this niche separation results from different affinities for ammonia, inhibition by high ammonium concentration or other factors.
Accession numbers
All sequences obtained in this study were deposited in the GenBank database with accession numbers HQ285255–HQ285290.
Acknowledgments
We thank Mr Lawrence Maurice and the SAC Craibstone Estate (Aberdeen) for access to the Woodlands Field pH plots. DTV is funded by a BBSRC standard research grant (BB/I009647/1) and GWN by an NERC Advanced Fellowship (NE/D010195/1).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Boyle-Yarwood SA, Bottomley PJ, Myrold DD. Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas r in Oregon. Environ Microbiol. 2008;10:2956–2965. doi: 10.1111/j.1462-2920.2008.01600.x. [DOI] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- Francis CA, Beman JM, Kuypers MMM. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007;1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- He J-Z, Shen J-P, Zhang L-M, Zhu Y-G, Zheng Y-M, Xu M-G, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Tscherko D, Chang L, Hammesfahr U, Prosser JI. Crenarchaeal community assembly and microdiversity in developing soils at two sites associated with deglaciation. Environ Microbiol. 2006;8:1382–1393. doi: 10.1111/j.1462-2920.2006.01031.x. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, et al. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol. 2009;11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- Stephen JR, McCaig AE, Smith Z, Prosser JI, Embley TM. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia oxidising archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Leininger S, Kietzin A, Schuster SC, Klenk H-P, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Webster G, Embley TM, Freitag TE, Smith Z, Prosser JI. Links between ammonia oxidiser species composition, functional diversity and nitrification kinetics in grassland soils. Environ Microbiol. 2005;7:676–684. doi: 10.1111/j.1462-2920.2005.00740.x. [DOI] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.