Abstract
Integron cassette arrays in a dozen cultivars of the most prevalent group of Vibrio isolates obtained from mucus expelled by a scleractinian coral (Pocillopora damicornis) colony living on the Great Barrier Reef were sequenced and compared. Although all cultivars showed >99% identity across recA, pyrH and rpoB genes, no two had more than 10% of their integron-associated gene cassettes in common, and some individuals shared cassettes exclusively with distantly-related members of the genus. Of cassettes shared within the population, a number appear to have been transferred between Vibrio isolates, as assessed by phylogenetic analysis. Prominent among the mucus Vibrio cassettes with potentially inferable functions are acetyltransferases, some with close similarity to known antibiotic-resistance determinants. A subset of these potential resistance cassettes were shared exclusively between the mucus Vibrio cultivars, Vibrio coral pathogens and human pathogens, thus illustrating a direct link between these microbial niches through exchange of integron-associated gene cassettes.
Keywords: integrons, coral, Vibrio, gene cassettes, microbial defense
Introduction
Corals are reef-building animals that depend on phototrophic dinoflagellate endosymbionts in the genus Symbiodinium (commonly called zooxanthellae) for much of their fixed carbon and molecular oxygen (Rosenberg et al., 2007). The importance of this symbiosis is perhaps most apparent in coral bleaching, which generally entails loss of zooxanthellae (Glynn, 1993). Severe bleaching can result in significant coral mortality (Hoegh-Guldberg, 1999). Recently, a complex role in coral health has also been claimed for the prokaryotic component of the coral microbiome, itself complex and diverse (Rohwer et al., 2001, 2002; Pantos et al., 2003; Bourne and Munn, 2005; Koren and Rosenberg, 2008). This diversity extends to location-specific differences but healthy coral includes a Vibrionaceae component (Kvennefors et al., 2010). Conversely, nitrogen-fixing bacteria (Lesser et al., 2004; Olson et al., 2009) and chitin degraders (Ducklow and Mitchel, 1979) thought to supply metabolic intermediates from these pathways to coral, pathogens that can cause bleaching (Kushmaro et al., 1996, 1997; Ben-Haim and Rosenberg, 2002; Ben-Haim et al., 2003) and commensals that might serve as ‘probiotics' (Reshef et al., 2006) defending the coral from pathogens by production of antimicrobial compounds (Koh, 1997; Castillo et al., 2001; Ritchie, 2006). Overall it is clear that coral/bacterial associations can be either detrimental or beneficial to the former.
Unlike vertebrate animals, invertebrates do not accumulate an adaptive immune response to infection, but rely on a first-line innate identification of pathogens, localized inflammation of the injured area, and the synthesis of specific antimicrobial compounds at the cell surface or within cells to target the microbe (Fearon and Locksley, 1996). In marine invertebrates such endogenous antimicrobial effectors include glycosides, brominated phenols, polyphenolics, polyketides, ribosomal and nonribosomal peptides, alkaloids, fatty acids and terpenoids (Blunt et al., 2004).
The coral probiotic hypothesis holds that the coral prokaryotic microbiome is also adaptive for the coral, providing resistance to pathogen-mediated bleaching. (Reshef et al., 2006). Studies of coral disease caused by Vibrio coralliilyticus in Pocillopora damicornis (Geffen and Rosenberg, 2005) show that resistance to the pathogen is mediated by the coral's mucus, a protective layer that coats all types of coral and is densely colonized with bacteria (Brown and Bythell, 2005; Rosenberg et al., 2007), a significant fraction of which produce antimicrobials (Koh, 1997; Castillo et al., 2001; Ritchie, 2006). Moreover, corals once susceptible to bleaching caused by a specific bacterial pathogen can become immune to it, a phenomenon called ‘experience-mediated tolerance'. For instance, Rosenberg and colleagues demonstrated that the coral Oculina patagonica, once strongly affected by bleaching events caused by Vibrio shiloi, can become resistant to reinfection by this bacterium (Reshef et al., 2006). Similarly, immunity to Aurantimonas coralicida, the putative bleaching agent responsible for the 1995 white plague outbreak in corals situated in the Florida Keys (Denner et al., 2003) has been observed. The coral probiotic hypothesis suggests that such instances of acquired immunity reflect changing patterns of antimicrobial production by commensal nonpathogenic bacteria (Brown et al., 2000; Rosenberg et al., 2007).
Pathogens may in turn develop antimicrobial resistance, recruiting resistance determinants from a ‘resistome' (D'Costa et al., 2006) through lateral gene transfer (LGT). One genetic element that facilitates this evolutionary arms race is the integron. This system comprises an integrase gene (intI) and an associated integration site (attI), at which the integrase protein (IntI) can catalyze gene cassette insertion or removal (Stokes and Hall, 1989). Gene cassettes are small mobile elements, usually composed of a single gene and a recombination site (attC). There are good reasons to suppose that integrons might have a significant role in coral bleaching and resistance. First, integrons are known to be crucial in the spread of antibiotic resistance in many human and animal pathogens (Hall et al., 1999). Second, integrons are especially prominent and well studied chromosomal elements in pathogenic and environmental Vibrio: indeed all sequenced Vibrio have at least one such integron array, often bearing more than 100 cassettes (reviewed in (Stokes and Hall, 1989; Rowe-Magnus et al., 2003; Wright, 2007)). Third, many species of Vibrio, such as V. shiloi and V. corallilyticus, are known or suspected coral pathogens and a role for Vibrio in bleaching can be inferred from an increasing proportion of Vibrio 16S rRNA genes sequenced during bleaching events (Bourne et al., 2008). But, fourth, there are also rich commensal vibrio populations associated with healthy corals (Arboleda and Reichardt, 2009; Kvennefors et al., 2010).
Here, we describe the diversity and function of encoded genes within gene cassettes associated with chromosomal integrons of Vibrio isolates cultivated from mucus expelled by a healthy Pocillopora damicornis colony living in the Great Barrier Reef off the coast of Australia. Integron sequences were obtained from twelve cultivars, which collectively share a pairwise 99% average nucleotide identity in housekeeping genes. We show that integron arrays in these coral Vibrio are extraordinarily dynamic (compared even with arrays in other members of the genus) and that this dynamic nature likely reflects LGT. Antibiotic resistance might be inferred as prominent function of these cassettes. We also show that, although the gene cassette repertoires of (a) coral mucus vibros, (b) Vibrio cholerae isolates, (c) other marine vibrio and (d) non-vibrio from other (especially polluted) environmental sites are clearly distinguishable, cassettes are shared and presumably exchanged between them.
Materials and methods
Sample collection and cultivation
Coral mucus samples were collected from a physically stressed scleractinian coral, Pocillopora damicornis, sampled from Davies Reef (Latitude 18°50.9′S/Longitude 147°41′E) within the Great Barrier Reef Marine Park off the coast of Queensland, Australia in March, 2006. Mucus was obtained by exposure of the colony to the air inducing excessive mucus production (milking), which was collected in sterile tubes. This coral mucus was plated on both Marine Agar 2216 (Difco Laboratories, Detroit, MI, USA) and thiosulfate citrate bile sucrose media (McLauglin, 1995). Plates were incubated over night at 37 °C and then colonies randomly picked and re-streaked three times for pure cultures.
Gene amplification, cloning and sequencing
PCRs were carried out in a final volume of 25 μl containing 1–5 ng of template DNA, 1.0 m of each primer and 12.5 μl of PCR Master Mix (PROMEGA, Alexandria, New South Wales, Australia). The reactions were performed with an initial denaturation step at 94 °C for 2 min, followed by 30 cycles with a denaturation at 94 °C for 30 s, primer annealing at 55 °C for 30 s and primer extension at 72 °C for 1 min. PCR products were gel purified with the MinElute kit (QIAGEN, Doncaster, Victoria, Australia) and cloned in TopoTA (INVITROGEN, Mulgrave, Victoria, Australia). Clones were sequenced from both strands.
Colony screening for IntI by PCR
Pure colonies were picked and used as template DNA in PCR reactions specific for the intI gene. This reaction was performed using class 1-specific primers HS463A and HS464 (Gillings et al., 2009). Colonies were also screened for gene cassettes with degenerate primers (HS287 and HS286) targeting attC regions (Stokes et al., 2001). PCR products by these reactions were cloned and sequenced as described above.
Fosmid library construction, screening and sequencing of Vibrio cultivars
Fosmid libraries were constructed from genomic DNA using the EPIFOS kit (EPICENTRE, Gymea, New South Wales, Australia). Purified genomic DNA was run on a low-melt 1% agarose gel (AMRESCO, Gymea, New South Wales, Australia) in a pulsed-field gel electrophoresis apparatus (Bio-Rad, Gladesville, New South Wales, Australia) and the DNA of ∼40 kb was purified from the agarose, ligated to the fosmid vector, packaged in phage capsids and used to infect E. coli as described in the EPIFOS kit manual. A total of 480 colonies from each library were picked from agar plates and used to inoculate 96-well blocks containing 1 ml of LB broth with 12.5 μg ml−1 of chloramphenicol in each well. These cultures were grown overnight and glycerol stocks were made by mixing 140 μl of culture from each well with 60 μl of 50% glycerol in a 96-well plate. PCR screening was done using primers targeting the intI gene as described above. Clones positive for the intI gene were re-streaked onto LB agar plates containing 12.5 μg ml−1 of chloramphenicol, which were used to inoculate a liquid culture for extraction of pure fosmid DNA. These clones were used for shotgun library construction, sequence and assembly performed by Macrogen Inc. (Seoul, South Korea).
Housekeeping-gene sequence acquisition from Vibrio cultivars and genomes
In the case of Vibrio cultivars, housekeeping-gene sequences were amplified using recA, pyrH and rpoB primers (Thompson et al., 2005) and sequenced as described above. The same sequences were retrieved from whole Vibrio genome sequences stored in the public database NCBI.
Taxonomic assignment of Vibrio cultivars by recA phylogeny
recA DNA sequences were amplified from 128 Vibrio coral mucus cultivars and combined with 210 reference sequences collected from the NCBI database. These were aligned together in MUSCLE (Edgar, 2004) and manually edited. The final alignment of 725 nucleotide positions was used as an input file for the Maximum Likelihood-based (ML) phylogenetic program PHYML (Guindon and Gascuel, 2003). The general time reversible nucleotide substitution model was implemented with the proportion of invariable sites and the gamma parameter of across-site rate variation (using four categories) estimated from the dataset. Bootstrap support values were calculated with the same parameters (100 replicates).
Vibrio concatenated gene phylogeny
The phylogenetic tree of coral mucus Vibrio isolates whose integrons were sequenced, in addition to reference strains of known taxonomic affiliation, was reconstructed using PHYML (Guindon and Gascuel, 2003) with the general time reversible nucleotide substitution model, the proportion of invariable sites and the gamma parameter of across-site rate variation (using four rate categories) estimated from the dataset. Bootstrap support values were calculated with the same parameters (100 replicates). The sequence dataset used for the analysis is a concatenation of the recA, pyrH and rpoB genes amplified from Vibrio coral mucus isolates and retrieved from reference strains publically available at NCBI. These were aligned using MUSCLE (Edgar, 2004) and manually edited to a final length of 2144 nucleotide positions.
Integron annotation
Assembled fosmid arrays encoding integrons were uploaded into the integron database: Annotation of Cassette and Integron Data (ACID) (Joss et al., 2009). ACID contains publicly available integron sequences and users can annotate integron-related components including and associated cassette open reading frames (ORFs).
Cassette attC sites were identified using a string-search-scoring method. The total score is composed of several individual categories that reflect attC structural components (Bouvier et al., 2009). Specifically, one point per match between inverse core/core sites of the attC site was assigned. The total possible score of this property referred to as R, is four as described by Joss et al. (2009). Briefly, one point is scored per match between the inner repeats of the attC site. These base-pairing events were weighted to make them more meaningful. Specifically, potential secondary structures of this simple site were considered as two 3 bp regions. These properties, referred to as LA and LB, when multiplied, produce a maximum score of 9. These are multiplied so that pairing is required in each of these two domains to produce a score. For example, LA=3 and LB=0 would produce a score of 0 (3 × 0). However, if each LA and LB had two base-pairing events, the score would be four. This weighting strategy ensures that the extra-helical base essential for IntI–attC interaction is considered. In addition, one point is scored for the presence of the typical extra-helical base, defined as E. Finally, one point is awarded for each mismatch in the 2 bp region located upstream from R′, referred to as site S. These mismatches produce the bulge necessary for IntI catalysis of the hairpinned attC substrate (Mazel, 2006). Final query scores are divided by the maximum possible score and the cut off was set to 75%, which results in the overall lowest rate of false identifications (Joss et al., 2009).
Cassettes identified by ACID were extracted from this database and uploaded to the Meta Genome Rapid Annotation using Subsystem Technology (MG-RAST (http://metagenomics.nmpdr.org)) server for functional assignment. MG-RAST was built as a modified version of the RAST server (Aziz et al., 2008) and is a high-throughput pipeline and provides automated functional assignments of sequences by comparing both protein and nucleotide databases. Finally, given that cassettes are mobile, the Basic Local Alignment Search Tool (BLASTx) algorithm (Altschul et al., 1990) was used to taxonomically define cassettes with the E-value threshold set to 10−5. These results were graphically illustrated using the software package MEGAN (Huson et al., 2007).
Cassette ecology
Cassette sequences were extracted from ACID in both nucleotide and amino acid FASTA formats, as were attC nucleotide sequences. These files were formatted for BLAST analysis with the formatdb freeware package (Altschul et al., 1990). Freely available blastall (Altschul et al., 1990) software was employed for BLAST ‘all versus all' analysis for each of the three datasets. This BLAST output was used as an input file for the metagenomic distance-based operational taxonomic units and Richness determination toolbox (Schloss and Handelsman, 2008). Using the furthest neighbor algorithm (Legendre and Legendre, 1998), this software package was used to arbitrarily define cassette family and attC cluster operational taxonomic units as two or more cassette or attC nucleotide sequences producing a pairwise BLAST Score Ratio (BSR) (Rasko et al., 2005) of >0.70.
When analyzing these estimates with greater scrutiny, however, it was clear that the estimate continued to grow. Specifically, the range of the Chao1 95% confidence intervals was analyzed as a function of sampling effort for these gene cassettes as suggested in (Schloss and Handelsman, 2005). There was a positive correlation with sequencing effort (R2=0.722) for these cassettes suggesting that the uncertainty of the Chao1 estimate increases with additional sampling and thus the 95% confidence intervals is artificially low for cassette sequence data and therefore more sampling of this gene pool is required for more accurate richness estimates (data not shown).
Operational taxonomic unit clustering
The relationship of shared cassettes or attC sites was visualized with Cytoscape 2.6 network visualization software (Shannon et al., 2003). The cassette/attC networks are based on the families/clusters defined above. The network was generated using a spring-embedded algorithm based on a ‘force-directed' paradigm (Kamada and Kawai, 1989). Network nodes are treated like physical objects that repel each other, and the connections between nodes attract their end points such that nodes sharing more connections will be placed more closely (relative to others sharing less) in three-dimensional space.
Statistical support for attC clustering
The UniFrac implemented G-test (Lozupone et al., 2006) was employed to assess whether or not the observed counts of attCs are distributed evenly across Vibrio lineages. The input data was in the form of an ML tree calculated using RAxML (Stamatakis, 2006). The best scoring tree of selected attC nucleotide sequences was obtained from 200 tree-building iterations using the GAMMA+P-Invar model of rate heterogeneity with an ML estimate of the alpha parameter.
Cassette phylogeny
Nucleotide alignments for each cassette family were subjected to ML-based phylogeny with PHYML (Guindon and Gascuel, 2003). The general time reversible nucleotide substitution model was implemented with the proportion of invariable sites and the gamma parameter of across-site rate variation (using four categories) estimated from the dataset. Bootstrap-support values were calculated with the same parameters (100 replicates).
Vibrio cassette dendrogram assembly
Selected Vibrio spp. were clustered according to the heuristic tree searching algorithm available in PAUP* (Swofford, 2003) with the search criterion set to distance (UPGMA) and the data (presence-absence) originating from a matrix of shared cassette families with >0.70 BSR values. Bootstrap support values were calculated with the same parameters (100 replicates). The cassette families themselves were clustered according to shared Vibrio spp. using the same algorithm and therefore the result is a two-dimentional clustering of shared cassette families.
Results
Pocillopora damicornis mucus contains a variety of Vibrio species
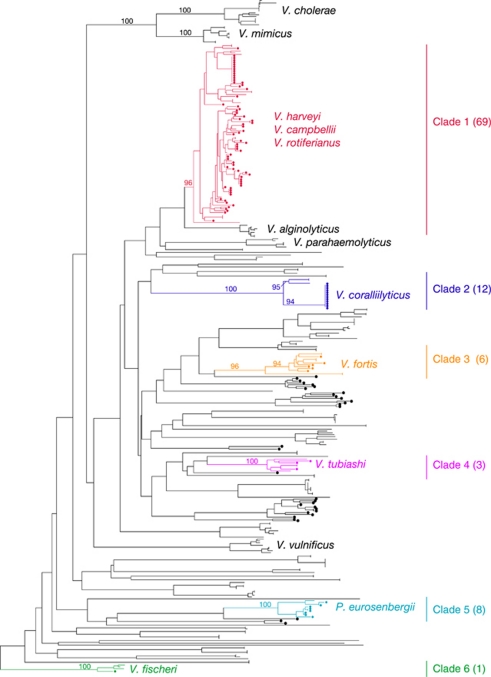
A total of 128 Vibrio cultivars were isolated on rich marine and Vibrio-selective growth media from Pocillopora damicornis mucus (see Materials and methods) and the complete cassette arrays from 12 of these isolates were sequenced and are shown diagrammatically in Figure 1. The taxonomic distribution of the 128 Vibrio isolates, according to their PCR-amplified recA nucleotide sequences, was determined by maximum-likelihood phylogeny (Guindon and Gascuel, 2003). Figure 2 shows that a diversity of bacteria from the genus Vibrio was recovered, including many whose recA nucleotide sequences grouped with reference sequences from the completed genomes of V. fischeri, P. eurosenbergii, V. tubiashi, V. fortis and V. coralliilyticus. In addition to these, the clade most represented in this dataset of cultivars (referred to as Clade 1) also contained V. alginolyticus, V. harveyi, V. campbellii and V. rotiferianus recA reference sequences.
Figure 1.
Integron gene-cassette arrays and array dynamics of Vibrio isolates cultured from coral mucus. Integron gene-cassette arrays are drawn to scale with each cassette boxed. Dark gray cassettes represent those that have no known homologs. The light gray ORFs that flank some of the integrons correspond to putative acetyltransferase (A), hypothetical unknown (H) and sodium-solute symporter (S) proteins. IntI ORFs are shown in red, whereas the white cassettes are those that have pairwise nucleotide sequence BSR values >0.70 to cassettes encoded by other more evolutionary distant Vibrio spp. not accounted for in this figure (none of these are like any other in this figure). White circles represent sequence gaps ∼10 kbp for V5H5 and an unknown value for V5D6. Cassettes that are shared between the different Vibrio isolates cultured from coral mucus are colored accordingly and are assigned a family number that is consistent with how cassette families are named in this study. Colored bars connecting cassettes common to different isolates indicate the dynamic nature of integrons between closely related bacteria.
Figure 2.
Taxonomic distribution of recA nucleotide sequences amplified from cultivars isolated from coral mucus samples. Circles at the tips of branches represent the 128 isolates cultured from mucus samples collected from a Pocillopora damicornis colony living in the Great Barrier Reef.
Most of the Vibrio recovered in this study were part of a single monophyletic group (Clade 1, Figure 2). Thus, to examine the tempo and mode of cassette sharing within a population of closely related Vibrio living in the same environment, the 12 isolates for which integron cassette arrays were sequenced all came from Clade 1.
Mucus Vibrio-cassette arrays are highly dynamic
The 12 isolates selected had a nucleotide identity of at least 99% across 2144 nucleotide positions sequenced for the recA, pyrH and rpoB genes, placing them well within even the most stringent ‘species' definitions (Doolittle and Zhaxybayeva, 2009). Despite this, the number of shared cassettes encoded in their integron gene cassette arrays ranges from only ∼1–10%. Synteny maps show that even cassettes shared between arrays have shuffled to randomness in position in the time required for 1% genomic sequence divergence to accumulate in housekeeping genes (Figure 1).
In all, 10 of the integrons from these coral mucus-associated Vibrio isolates are downstream from ORF H, encoding a conserved hypothetical protein of unknown function (Figure 1). The other two, while also associated with this same gene, had undergone genomic rearrangements. In one instance (V5B10), the intI is downstream from ORF H, but the associated array appears inverted relative to the intI gene. In the second, V5H5, the array is not only inverted but interrupted by ∼10 kbp of genomic sequence encoding ORFs without any flanking attC sequences. Interestingly, Vibrio species, V. parahaemolyticus RIMD 2210633 and V. sp. Ex25, both had their integrons inserted adjacent to ORF H, whereas the integrons associated with the two V. vulnificus strains (YJ016 and CMCP6) and all V. cholerae isolates were at a different site (downstream from the LSU ribosomal protein L20p).
Mucus Vibrio-cassette arrays evolve more rapidly than those of V. cholerae
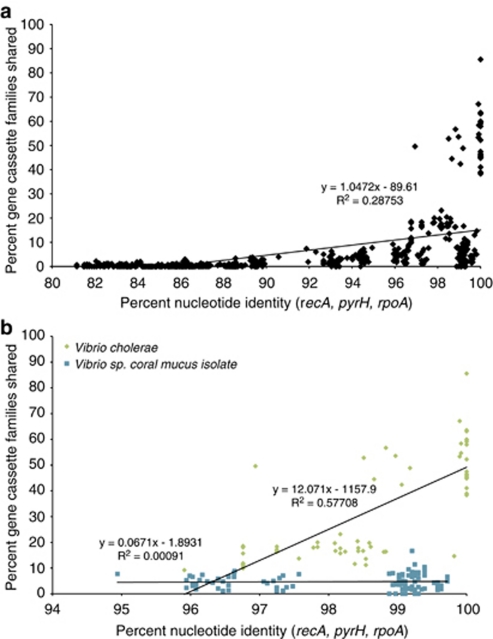
To assess whether the mucus Vibrio-cassette arrays are evolving more rapidly than the core genome in terms of structure and composition, available chromosomal integrons from any member of the Vibrio genus were compared in a pairwise manner, correlating similarity in cassette array composition to divergence in sequence of the three housekeeping genes. These analyses revealed only a weak positive correlation overall (R2=0.3) between the percent of gene cassette families shared between any two Vibrio isolates and their evolutionary relatedness as defined by percent nucleotide identity of concatenated housekeeping genes recA, pyrH and rpoA (Figure 3a), although V. cholerae isolates (upper cluster, Figure 3a) appeared different in this regard. Indeed, the same analysis focused at a finer evolutionary scale revealed a distinction between V. cholerae and coral mucus isolates specifically (Figure 3b). Six strains of the former (Thompson et al., 2004), showed a strong correlation of shared gene-cassette content and evolutionary relatedness (R2=0.6) compared with the genus level analysis (Figure 3b). Conversely, analysis of Clade 1 coral mucus Vibrio cultivars (Figure 1), together with the six most closely related strains of the V. harveyi species group, showed no correlation between shared cassettes and evolutionary relatedness (R2=0; Figure 3b).
Figure 3.
Pairwise comparison of the genetic identity versus integron gene-cassette array conservation in Vibrio isolates. (a) All Vibrio isolates for which gene cassettes have been sequenced. (b) Vibrio cholerae isolates as well as Vibrio strains collected from coral mucus and their close relatives. The trend line was obtained through a linear regression analysis of all data points, each point representing a pairwise comparison of isolates based on their combined recA, pyrH and rpoA housekeeping-genes nucleotide identity and the gene-cassette families they share. The R2 values were calculated using the square of the Pearson product moment correlation coefficient through the given data points.
Possible LGT among mucus Vibrio cassettes can be examined phylogenetically. In total, 404 cassettes were sequenced from these twelve Vibrio isolates, with sixty instances of related cassettes occurring more than once (that is, having BSR values >0.70) corresponding to 70% of amino acid sequence identity corrected for length). Phylogenetic analysis (Guindon and Gascuel, 2003) was performed on the basis of nucleotide sequences of all eleven individual cassette families that had four or more members (Supplementary Figure S1). Five of these trees presented conflicting topologies. Although it is possible that these trees reflect gene duplication and differential loss of integron gene cassettes, LGT between members of this Vibrio population seemed the more parsimonious explanation. Many Vibrio species are transformation competent, and high rates of transformation have been observed in marine populations (Meibom et al., 2005). Phylogenetic analyses of the attC sites linking cassettes in these strains (data not shown) reveal little of the strain-specific signal that might be expected if within-strain integrase catalyzed rearrangements, rather than between-strain LGT, were responsible for the shuffling shown in Figure 1.
Resistance to antimicrobials may be an important function of mucus Vibrio gene cassettes
If natural antimicrobials have a determining role in coral microbiome ecology (Shnit-Orland and Kushmaro, 2009), we would expect a significant fraction of antibiotic-resistance (or synthesis) determinants among integron cassettes. Indeed, of the approximately 26% of gene cassettes encoding a protein that can be ascribed a function, nearly 48% were implicated in biochemical processes previously identified in antibiotic resistance (Supplementary Table S2). Most prevalent among these are 39 cassette-encoded acetyltransferases, an enzyme class implicated in resistance to different aminoglycosides (Wohlleben et al., 1989). A total of 12 families were resolved from these acetyltransferase amino acid sequences when clustered at the BSR threshold >0.70. Of these, four families in particular were highly similar to four different acetyltransferase genes that have been shown to confer resistance to streptothricin and virginiamycin (Supplementary Figure S2) (Liu and Pop, 2009). In addition to these acetyltransferases, eleven cassette-encoded proteins were identified as putative homologs of glyoxalase/bleomycin-resistance proteins implicated in resistance to the glycopeptide bleomycin. This abundance of cassette-encoded acetyltransferases and glyoxalases is of interest given that glycoproteins or ‘mucins' are the primary constituents of the coral mucus matrix and may have antimicrobial activity (Brown and Bythell, 2005).
Among other cassette-encoded proteins, FtsZ and DNA topoisomerase I were also identified. FtsZ, a bacterial protein essential for cell division (Errington et al., 2003), was recently targeted with a new class of antibiotic (Haydon et al., 2008). Furthermore, DNA topoisomerase I, present in all bacteria (Forterre et al., 2007) and responsible for removal of excess transcriptionally induced negative DNA supercoiling (Viard and de la Tour, 2007) is sensitive to a few known antibiotics. Even though DNA gyrase is considered the primary target of quinolones from E. coli, some of these have been shown to inhibit the relaxation activity of E. coli topoisomerase I (Tabary et al., 1987; Moreau et al., 1990; Tse-Dinh, 2009).
The mucus cassette repertoire overlaps with that of other environmental Vibrio, and in particular with those of pathogens
To assess whether cassettes obtained from our Vibrio cultivars were also present in known pathogens, they were compared with publicly available cassettes from V. coralliilyticus and V. shiloi (coral pathogens) as well as V. cholerae and V. vulnificus (human pathogens). Indeed, a number of the cassettes listed in Supplementary Table S3 were shared (amino acid BSR values >0.70) exclusively between Vibrio mucus cultivars, known coral pathogens and known human pathogens including V. vulnificus and V. cholerae. These cassette-encoded functions include putative resistance genes, specifically glyoxalases/bleomycin resistance, three different acetyltransferases and DinB, the damage-inducible protein, which is triggered during bacterial SOS response and has the potential to increase mutation rate (McKenzie et al., 2001) and, potentially, the rate of cassette rearrangements (Guerin et al., 2009).
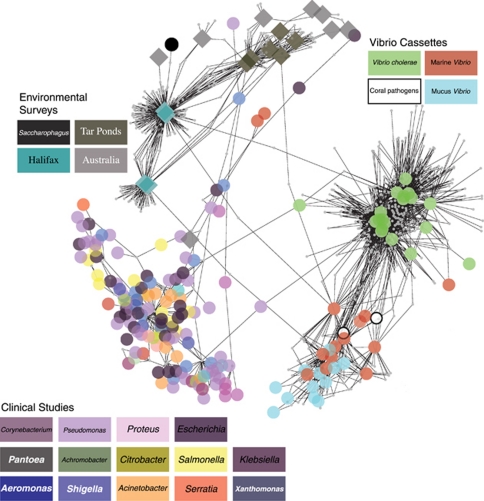
To assess the connections between mucus Vibrio cassette arrays and cassettes within a larger global cassette metagenome regardless of taxonomic assignment, 10 365 integron gene cassette sequences were extracted from the ACID database (Joss et al., 2009). These cassettes were clustered using the furthest neighbor algorithm (Legendre and Legendre, 1998) at the amino acid BSR threshold >0.70. Figure 4 displays clusters of organisms (or environments) formed on the basis of their shared cassettes. Three main clusters emerged from this analysis and were named Vibrio cassettes, clinical studies and environmental surveys. The ‘clinical studies' cluster contained the same resistance cassettes conserved in distantly related pathogens (Boucher et al., 2007). For the ‘Environmental Surveys' cluster, similar environments exhibited comparable cassette profiles (Koenig et al., 2008).
Figure 4.
Network clusters based on integron gene cassettes. Cassette sequences were collected from isolates (colored circles) or environment (colored diamonds). The network is based on cassette families (small gray-colored nodes) that have an amino acid BSR thresholds >0.70.
Some interesting subdivisions within and between clusters could be observed (Figure 4). For example, the >2000 cassettes that were sampled from the Halifax Harbour (Canada)and its vicinity form two discrete subclusters based on shared cassettes, one of these corresponding to the abundance of cassettes that were observed to be present in samples taken near sewage outfalls (Figure 4). These Halifax samples and the Australian samples share cassettes with each other but also with Pseudomonas, Saccharophagus and Shigella, specifically cassettes encoding acetyltransferases and isochorismatases-like proteins (as well as ‘conserved hypothetical' proteins).
Phylogenetic analysis of cassettes belonging to the ‘Vibrio Cassettes' cluster revealed several instances of trees, topologies of which contradict organismal phylogeny based on rpoB gene sequences and supports the notion that cassettes may be laterally transferred between these Vibrio species (Supplementary Figure S3). Furthermore, the cassettes observed to connect the ‘Vibrio cassettes' and the ‘clinical studies' clusters encode β-lactamases, chloramphenichol resistance and aminoglycoside resistance genes. Cassette-encoded amino acid sequences obtained from resistance genes were subjected to phylogenetic analysis (Guindon and Gascuel, 2003) and produced tree topologies supporting the idea that these cassettes have been exchanged recently between Vibrio cholerae and other pathogens (Supplementary Figure S4).
Discussion
Cassette arrays in vibrios are generally understood to evolve rapidly in relation to chromosomal genes (Labbate et al., 2007). This study has reinforced that observation but, remarkably, has also revealed that the integrons of coral mucus Vibrio are particularly strong evolutionary hotspots in their genomes, with a very high flux of mobile genes coming in and going out that exceeds even that seen in more free-living vibrios. This was obvious from the pairwise comparisons correlating cassette diversity with clonal divergence (Figure 3). The clear implication of this analysis is that coral mucus isolates within a single community are rapidly exchanging gene cassettes by LGT, whereas V. cholerae isolates, from different hosts and outbreaks, have more independent evolutionary trajectories. Apart from the fact that individuals in coral mucus are in close proximity, what would be the advantage of this? The diversity of cassette genes within coral mucus may represent a shared resource functioning cooperatively to help maintain the health of both the microbial and coral community. The ability to use cassette gene products as a shared resource potentially has many advantages in such a community. For example, it may allow the cooperative generation of a greater array of defense products than would be the case if all individuals had to carry the same extensive genetic burden. The fact that the coral mucus bacterial community is a dense and physically limited environment means that potentially all members could benefit from the production of adaptive diffusible gene products, even if they are only produced by a small subset of the population. This could, in theory, extend to products of complex genetic pathways where individual genes in a pathway are spread across multiple individuals.
As is the case for the majority of cassette products located within chromosomal arrays, the high degree of novelty makes identifying specific functions difficult. However, meta-analysis involving in excess of 10 000 cassettes from different organisms and environment types does suggest exchange between other vibrio types (Figure 4). In part, this is almost certainly because of relatedness of attC sites and IntI integrase preferences but more specific host–pathogen interactions may also be important. Known ecological associations of the characterized Vibrio species that have close relatives among our coral mucus cultivars are summarized in Supplementary Table S1. It can be seen that eight of the nine Vibrio species (Supplementary Table S1) that have close relatives among our coral mucus cultivars have been implicated in the pathogenesis of marine organisms or humans, and seven of these vibrios were isolated from bleached corals (Ritchie et al., 1994; Thompson et al., 2005; Bourne et al., 2008), although only one, V. coralliilyticus, has been directly proven to be an etiological agent of bleaching in P. damicornis (Ben-Haim and Rosenberg, 2002). V. coralliilyticus is also a causative agent of white syndrome disease in other coral species (Bally and Garrabou, 2007; Sussman et al., 2008, 2009; Vezzulli et al., 2010).
A function of some cassette proteins may also be for microbial defense—specifically to provide resistance to coral produced antimicrobials. This hypothesis is supported to the extent that, where putative general functions can be ascribed, a significant number relate to either resistance to antimicrobial compounds or to stress adaptation (Supplementary Table S3). These include matches to bleomycin-resistance compounds and acetyltransferases in the case of the former and damage-inducible proteins in the case of the latter.
Evidence continues to emerge that integron-encoded cassette products are likely to be crucial to adaptation of microbes to stressful environments. The outstanding example of this is the contribution mobile cassette products make to assisting human pathogens become multi-drug resistant (Mazel, 2006; Boucher et al., 2007). For bacteria, intimate association with coral mucus may be an analogous type of stress. If so, the recent observation that stress increases IntI integrase activity thereby increasing rates of cassette rearrangement may also partly explain the increase in array diversity seen here (Guerin et al., 2009). As noted above, the association between coral and vibrio may also be potentially mutually beneficial if cassette proteins contribute to coral maintenance in ways that are yet to be elucidated.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arboleda M, Reichardt W. Epizoic communities of prokaryotes on healthy and diseased scleractinian corals in Lingayen Gulf, Philippines. Microb Ecol. 2009;57:117–128. doi: 10.1007/s00248-008-9400-0. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally M, Garrabou J. Thermodependent bacterial pathogens and mass mortalities in temperate benthic communities: a new case of emerging disease linked to climate change. Global Change Biol. 2007;13:2078–2088. [Google Scholar]
- Ben-Haim Y, Rosenberg E. A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar Biol. 2002;141:47–55. [Google Scholar]
- Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol. 2003;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt JW, Copp BR, Munro MH, Northcote PT, Prinsep MR. Marine natural products. Nat Prod Rep. 2004;21:1–49. doi: 10.1039/b305250h. [DOI] [PubMed] [Google Scholar]
- Boucher Y, Labbate M, Koenig JE, Stokes HW. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 2007;15:301–309. doi: 10.1016/j.tim.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne DG, Munn CB. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol. 2005;7:1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- Bouvier M, Ducos-Galand M, Loot C, Bikard D, Mazel D. Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS Genet. 2009;5:e1000632. doi: 10.1371/journal.pgen.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BE, Bythell JC. Perspectives on mucus secretion in reef corals. Mar Ecol-Prog Ser. 2005;296:291–309. [Google Scholar]
- Brown BE, Dunne RP, Goodson MS, Douglas AE. Bleaching patterns in reef corals. Nature. 2000;404:142–143. doi: 10.1038/35004657. [DOI] [PubMed] [Google Scholar]
- Castillo I, Lodeiros C, Nunez M, Campos I. In vitro study of antibacterial substances produced by bacteria associated with various marine organisms. Rev Biol Trop. 2001;49:1213–1221. [PubMed] [Google Scholar]
- D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- Denner EB, Smith GW, Busse HJ, Schumann P, Narzt T, Polson SW, et al. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. Int J Syst Evol Microbiol. 2003;53:1115–1122. doi: 10.1099/ijs.0.02359-0. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- Ducklow HW, Mitchel R. Bacterial populations and adaptations in the mucus layers on living corals. Limnol Oceanogr. 1979;24:715–725. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–53. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Geffen Y, Rosenberg E. Stress-induced rapid release of antibacterials by scleractinian corals. Mare Biol. 2005;146:931–935. [Google Scholar]
- Gillings MR, Xuejun D, Hardwick SA, Holley MP, Stokes HW. Gene cassettes encoding resistance to quaternary ammonium compounds: a role in the origin of clinical class 1 integrons. ISME J. 2009;3:209–215. doi: 10.1038/ismej.2008.98. [DOI] [PubMed] [Google Scholar]
- Glynn PW. Coral reef bleaching: ecological perspectives. Coral Reefs. 1993;12:1–17. [Google Scholar]
- Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, et al. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall RM, Collis CM, Kim M-J, Partridge SR, Recchia GD, Stokes HW. Mobile gene cassettes and integrons in evolution. Ann N Y Acad Sci. 1999;870:68–80. doi: 10.1111/j.1749-6632.1999.tb08866.x. [DOI] [PubMed] [Google Scholar]
- Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshwater Res. 1999;50:839–866. [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joss MJ, Koenig JE, Labbate M, Polz MF, Gillings MR, Stokes HW, et al. ACID: annotation of cassette and integron data. BMC Bioinformatics. 2009;10:118. doi: 10.1186/1471-2105-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada T, Kawai S. An Algorithm for General Undirected Graphs. Inform Process Lett. 1989;31:7–15. [Google Scholar]
- Koenig JE, Boucher Y, Charlebois RL, Nesbo C, Zhaxybayeva O, Bapteste E, et al. Integron-associated gene cassettes in Halifax Harbour: assessment of a mobile gene pool in marine sediments. Environ Microbiol. 2008;10:1024–1038. doi: 10.1111/j.1462-2920.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- Koh EGL. Do scleractinian corals engage in chemical warfare against microbes. J Chem Ecol. 1997;23:379–398. [Google Scholar]
- Koren O, Rosenberg E. Bacteria associated with the bleached and cave coral Oculina patagonica. Microb Ecol. 2008;55:523–529. doi: 10.1007/s00248-007-9297-z. [DOI] [PubMed] [Google Scholar]
- Kushmaro A, Loya Y, Fine M, Rosenberg E. Bacterial infection and coral bleaching. Nature. 1996;380:396. [Google Scholar]
- Kushmaro A, Rosenberg E, Fine M, Loya Y. Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser. 1997;147:159–165. [Google Scholar]
- Kvennefors EC, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O. Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site- and species-specificity of common bacterial associates. PLoS One. 2010;5:e10401. doi: 10.1371/journal.pone.0010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbate M, Boucher Y, Joss MJ, Michael CA, Gillings MR, Stokes HW. Use of chromosomal integron arrays as a phylogenetic typing system for Vibrio cholerae pandemic strains. Microbiology. 2007;153:1488–1498. doi: 10.1099/mic.0.2006/001065-0. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre L. Numerical Ecol. Elsevier: New York; 1998. [Google Scholar]
- Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- Liu B, Pop M. ARDB--antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- McLauglin J.1995VibrioIn: Murray PR, Baron EJ and Pfaller MA (eds).Manual of Clinical Microbiology6th edn.American Society for Microbiology Press: Washington, DC [Google Scholar]
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- Moreau NJ, Robaux H, Baron L, Tabary X. Inhibitory effects of quinolones on pro- and eucaryotic DNA topoisomerases I and II. Antimicrob Agents Chemother. 1990;34:1955–1960. doi: 10.1128/aac.34.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ND, Ainsworth TD, Gates RD, Takabayashi M. Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J Exp Mar Biol Ecol. 2009;371:140–146. [Google Scholar]
- Pantos O, Cooney RP, Le Tissier MD, Barer MR, O'Donnell AG, Bythell JC. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ Microbiol. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Myers GS, Ravel J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Marine Ecology-Progress Series. 2006;322:1–14. [Google Scholar]
- Ritchie KB, Dennis JH, McGrath T, Smith GW. Bacteria associated with bleached and non-bleached areas of Montastraea annularis. Proc Symp Nat Hist Bahamas. 1994;5:75–80. [Google Scholar]
- Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs. 2001;20:85–89. [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics. 2008;9:34. doi: 10.1186/1471-2105-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnit-Orland M, Kushmaro A. Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol. 2009;67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Stokes HW, Holmes AJ, Nield BS, Holley MP, Nevalainen KM, Mabbutt BC, et al. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl Environ Microbiol. 2001;67:5240–5246. doi: 10.1128/AEM.67.11.5240-5246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M, Mieog JC, Doyle J, Victor S, Willis BL, Bourne DG. Vibrio zinc-metalloprotease causes photoinactivation of coral endosymbionts and coral tissue lesions. PLoS ONE. 2009;4:e4511. doi: 10.1371/journal.pone.0004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M, Willis BL, Victor S, Bourne DG. Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE. 2008;3:e2393. doi: 10.1371/journal.pone.0002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Sinauer Associates: Sunderland, Massachusetts; 2003. [Google Scholar]
- Tabary X, Moreau N, Dureuil C, Le Goffic F. Effect of DNA gyrase inhibitors pefloxacin, five other quinolones, novobiocin, and clorobiocin on Escherichia coli topoisomerase I. Antimicrob Agents Chemother. 1987;31:1925–1928. doi: 10.1128/aac.31.12.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, et al. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol. 2005;71:5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L, Previati M, Pruzzo C, Marchese A, Bourne DG, Cerrano C. Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ Microbiol. 2010;12:2007–2019. doi: 10.1111/j.1462-2920.2010.02209.x. [DOI] [PubMed] [Google Scholar]
- Viard T, de la Tour CB. Type IA topoisomerases: a simple puzzle. Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Wohlleben W, Arnold W, Bissonnette L, Pelletier A, Tanguay A, Roy PH, et al. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet. 1989;217:202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]
- Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.