Abstract
The sphingolipid metabolites ceramide and sphingosine-1-phosphate (S1P) have recently been implicated in autophagy. In this study, we report that depletion of sphingosine-1-phosphate phosphohydrolase-1 (SPP1), an endoplasmic reticulum (ER)-resident enzyme that specifically dephosphorylates S1P, induced autophagy. Although the mammalian target of rapamycin and class III phosphoinositide 3-kinase/Beclin-1 pathways were not involved and this autophagy was p53 independent, C/EBP homologous protein, BiP, and phospho-eucaryotic translation initiation factor-2α, and cleavage of procaspases 2 and 4, downstream targets of ER stress, were increased after SPP1 depletion. Autophagy was suppressed by depletion of protein kinase regulated by RNA-like ER kinase (PERK), inositol-requiring transmembrane kinase/endonuclease-1α, or activating transcription factor 6, three sensors of the unfolded protein response (UPR) to ER stress. Autophagy triggered by downregulation of SPP1 did not lead to apoptosis but rather stimulated, in a PERK dependent manner, the survival signal Akt, whose inhibition then sensitized cells to apoptosis. Although depletion of SPP1 increased intracellular levels of S1P and its secretion, activation of cell surface S1P receptors did not induce autophagy. Nevertheless, increases in intracellular pools of S1P, but not dihydro-S1P, induced autophagy and ER stress. Thus, SPP1, by regulating intracellular S1P homeostasis, can control the UPR and ER stress-induced autophagy.
Keywords: sphingosine-1-phosphate phosphatase-1, autophagy, ER stress, apoptosis, Akt
Macroautophagy (hereafter referred as autophagy) is a physiological response to cell stress that involves the degradation of proteins and organelles for the elimination of superfluous, damaged or aged cells or organelles. The sequestration of organelles and long-lived proteins in a double-membrane vesicle, called an autophagosome, is controlled by a set of evolutionarily conserved autophagy-related gene (Atg) proteins. The formation of autophagosomes is initiated by class III phosphoinositide 3-kinase (PI3K) and Atg6 (also known as Beclin-1) or through repression of the mammalian target of rapamycin (mTOR). Elongation of autophagosomal membranes is mediated by two ubiquitin-like conjugating systems that convert Atg8 (also known as microtubule-associated protein-1 light chain-3 (LC3)) from free form to a lipid-conjugated membrane-bound form (LC3-II).1, 2 Although autophagy is often a stress adaptation to avoid cell death (and suppress apoptosis), in other cellular settings, autophagy may be an alternative death pathway to cellular demise that is called autophagic cell death.3
The interconvertable sphingolipid metabolites, ceramide and sphingosine-1-phosphate (S1P), have long been proposed to have opposing functions in the regulation of cell fate.4, 5 Whereas ceramide has been associated with cell growth arrest and apoptosis, S1P stimulates cell growth and survival.4, 5, 6 Recent studies suggest that ceramide and S1P also have opposing effects on autophagy.7 Elevation of ceramide promotes accumulation of Beclin-1 and inhibition of Akt phosphorylation, leading to autophagic cell death.8 In addition to the induction of autophagy by exogenous short-chain ceramides or tamoxifen,8 the combination of the multikinase inhibitor sorafenib with histone deacetylase inhibitors also increases ceramide levels mainly by enhancement of de novo synthesis.9 Increased ceramide synthesis has been implicated in endoplasmic reticulum (ER) stress, autophagy, and apoptosis.9 Furthermore, exogenous dihydroceramide, an intermediate in the biosynthesis of ceramide, or its elevation induced by fenretinide, which stimulates serine palmitoyltransferase and inhibits the desaturase, also induces autophagy.10
Surprisingly, however, increasing endogenous S1P levels by overexpression of sphingosine kinase 1 (SphK1), one of the isoenzymes that produces it, leads to inhibition of the mTOR pathway and autophagic cell survival that protects cells from apoptosis during nutrient starvation.11 Thus, it has been proposed that ceramide-induced autophagy and S1P-induced autophagy may be mutually exclusive responses and that autophagy is the mechanism by which the sphingolipid rheostat controls cell fate decisions.7 Examination of the enzymes that regulate the balance between ceramide and S1P may help to resolve these issues. S1P levels are regulated by the balance between its synthesis catalyzed by SphK1 and SphK2 and its degradation by ER localized S1P phosphatases (SPP1 and SPP2) and S1P lyase. Interestingly, in Saccharomyces cerevisiae, the homolog of SPP1 has been shown to be a key regulator of stress responses.12, 13 Because previous studies have shown that SPP1 also has a key role in the regulation of the relative levels of S1P and ceramide and thus cell fate,14 it was important to examine its function in regulating autophagic responses leading to cell survival or cell death. Our results suggest that SPP1, by its influence on intracellular S1P levels, regulates the unfolded protein response (UPR) and ER stress-induced autophagy.
Results
Depletion of SPP1 induces autophagy
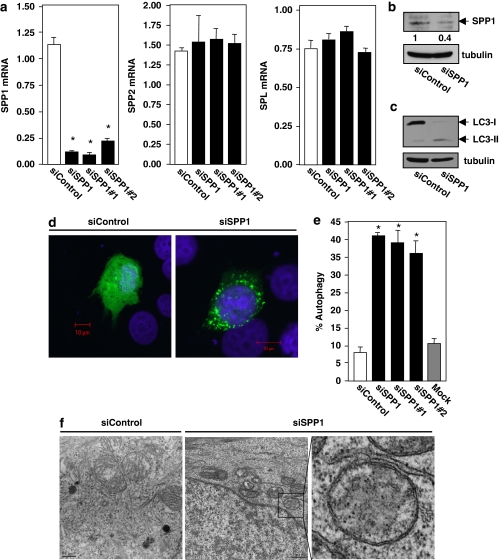
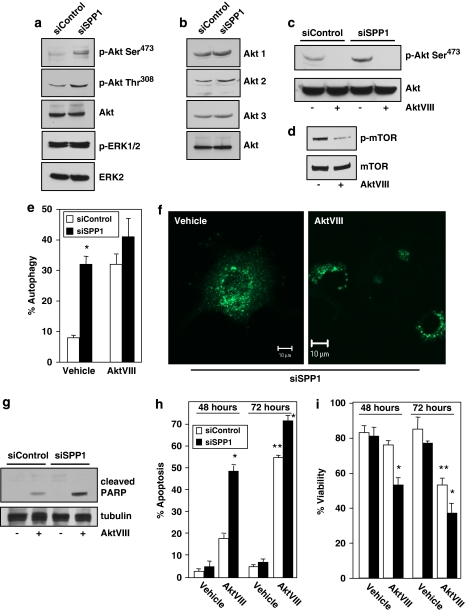
Overexpression of sphingosine kinase 1 (SphK1), which increases S1P, has been shown to induce autophagy.11 It was thus of interest to investigate the role in the autophagic process of the counteracting enzyme, SPP1, which decreases S1P levels. To this end, human breast adenocarcinoma MCF7 cells were transfected with specific siRNA targeted to SPP1, which markedly reduced its mRNA (>80%) without altering the expression of other enzymes that regulate S1P levels, including S1P phosphatase-2 (SPP2), S1P lyase (Figure 1a), and SphK1 and SphK2 (data not shown). This also effectively decreased SPP1 protein levels, determined by immunoblotting with a specific antibody (Figure 1b). Depletion of SPP1 significantly increased autophagy, as evidenced by the increased conversion of endogenous LC3/Atg815 to lipidated LC3-II, as determined by western blotting (Figure 1c), and by the redistribution of GFP-LC3 fusion protein from a diffusive cytoplasmic pattern to a punctate pattern, indicative of formation of autophagic vacuoles (Figure 1d). Almost 40% of the SPP1-depleted cells were autophagic compared with <10% of the non-targeting control siRNA transfectants or non-transfected cells (Figure 1e). Similar results were obtained with siRNAs targeted to different sequences in SPP1 (Figure 1e), suggesting that the increased autophagy was not due to off-target effects.
Figure 1.
Depletion of SPP1 induces autophagy in MCF7 cells. MCF7 cells were transfected with control scrambled siRNA or ON-TARGETplus SMARTpool siRNA targeted to SPP1 (siSPP1) or with individual siRNAs targeted to two distinct SPP1 sequences (siSPP1#1, siSPP1#2), as indicated. (a) SPP1, SPP2, and S1P lyase mRNA levels were determined by quantitative real-time PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. (b, c) Lysates from MCF7 cells transfected with siControl or siSPP1 were analyzed by immunoblotting with anti-SPP1 antibody (b), or with anti-LC3 antibody (c) and re-probed with anti-tubulin antibody to insure equal loading and transfer. Numbers indicate fold changes determined by densitometry. (d, e) Twenty-four hours after siRNA transfections as in (a), cells were transfected with EGFP-LC3. After culturing for an additional 24 h in complete medium, cells were examined by confocal fluorescence microscopy. (d) Representative images are shown. Scale bars, 10 μm. (e) Percentage of cells showing GFP-LC3 fluorescence in puncta was quantified. Mock, untransfected cells. Data are means±S.D. from three independent experiments. At least 100 cells were analyzed for each. (f) MCF7 cells transfected with siControl or siSPP1 were examined by transmission electron microscopy. Representative micrographs are shown and atypical autophagosome are boxed and shown at higher magnification. Scale bars, 0.5 μm. *P<0.01 compared with siControl
To insure that silencing SPP1 did not induce an incomplete autophagic process, we also utilized a recently developed double-tagged mCherry-GFP-LC3 construct,16 which is detected by its yellow fluorescence (green merged with red) in non-acidic autophagosomes and as red fluorescence in acidic autolysosomes due to quenching of GFP. SPP1-depleted cells displayed numerous yellow structures and some red ones (autophagolysosomes) (Supplementary Figure 1), demonstrating maturation of autophagosomes and fusion with lysosomes. Moreover, transmission electron microscopy of SPP1-depleted cells also revealed the formation of autophagosomes, identified as double-membrane vacuolar structures containing visible cytosolic content (Figure 1f).
The class III PI3K/Beclin-1 complex and mTOR are not involved in autophagy triggered by depletion of SPP1
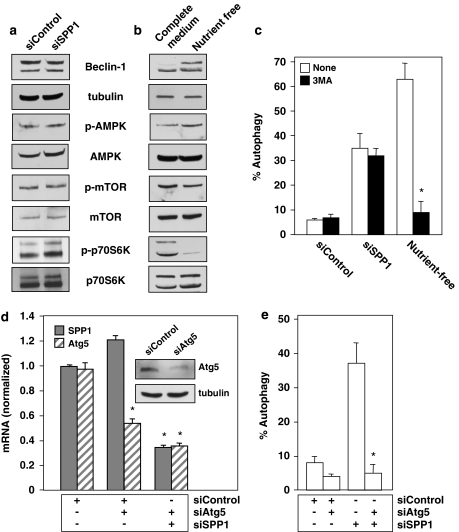
To examine the mechanism of autophagy induced by depletion of SPP1, we first investigated the canonical autophagy signaling pathways. Silencing SPP1 did not affect levels of Beclin-1 (Figure 2a), a component of the class III PI3K complex, in contrast to nutrient deprivation (Figure 2b). Moreover, downregulation of SPP1 did not affect AMP kinase (AMPK) that modulates autophagy resulting from mTOR inhibition,2 nor did it reduce the phosphorylation status of mTOR itself and that of one of its known downstream effectors, p70S6K (Figure 2a), suggesting that mTOR is not involved. However, in agreement with previous studies (reviewed by Eisenberg-Lerner et al.2), nutrient deprivation increased phosphorylation of AMPK and decreased that of mTOR and p70S6K (Figure 2b). To confirm the lack of involvement of the class III PI3K/Beclin-1 pathway, we analyzed the effects of 3-methyl adenine (3MA), a class III PI3K inhibitor, on autophagy. 3MA did not inhibit autophagy induced by silencing SPP1 (Figure 2c), yet as expected it markedly reduced autophagy induced by nutrient deprivation. Because these data suggest that depletion of SPP1 does not stimulate autophagy through the mTOR or class III PI3K/Beclin-1 signaling pathways, we next examined whether Atg5, a protein required in the early stages of autophagosome formation,3 was involved. Silencing Atg5 decreased its mRNA and protein expression without affecting SPP1 (Figure 2d), yet markedly inhibited autophagy triggered by depletion of SPP1 (Figure 2e).
Figure 2.
Downregulation of SPP1 does not activate the mTOR pathway. (a, c) MCF7 cells were transfected with siControl or siSPP1 or were cultured in nutrient-free EBSS or in complete medium, as indicated. (a, b) Equal amounts of cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Blots were re-probed for total mTOR, AMPK, p70S6K, and tubulin to insure equal loading and transfer. (c) Cells treated as indicated in (a) were transfected with EGFP-LC3. After culturing for 24 h in complete medium or in nutrient-free medium where indicated, in the absence or presence of 10 mM 3MA, autophagy was quantified by confocal microscopy. Data are means±S.D. of three independent experiments. *P<0.01 compared with untreated. (d, e) MCF7 cells were transfected with control siRNA, siSPP1, or siAtg5, as indicated. (d) SPP1 and Atg5 mRNA levels were determined by quantitative real-time PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Insert: Equal amounts of cell lysates were immunoblotted with anti-Atg5 and re-probed for tubulin to insure equal loading and transfer. (e) Autophagy was determined in duplicate cultures transfected with EGFP-LC3. *P<0.01 compared with siControl
Depletion of SPP1 triggers autophagy by ER stress
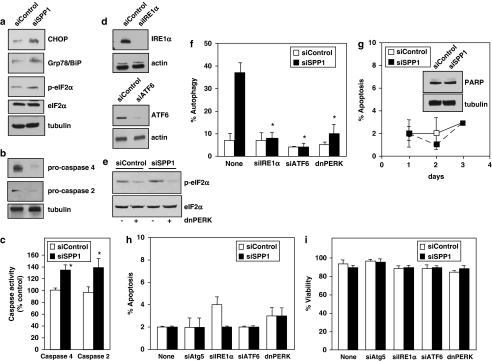
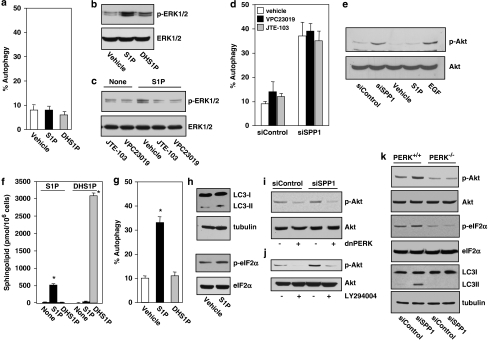
As SPP1 is located at the ER14 and abundant evidence indicates that ER stress is another potent trigger of autophagy,17, 18 the involvement of ER stress and the UPR19 in SPP1-regulated autophagy were examined. Depletion of SPP1 increased expression of transcriptional regulator C/EBP homologous protein and Grp78/BiP, a chaperone protein classically induced during UPR, as well as the phosphorylation of eukaryotic translation initiation factor-2α (eIF2α) (Figure 3a) that is usually phosphorylated by kinase-like endoplasmic reticulum kinase (PERK) during ER stress.19 Similarly, silencing SPP1 also activated caspases 2 and 4, ER-resident caspases that are activated during ER stress,20 as assessed by the disappearance of their procaspase forms (Figure 3b) and direct enzymatic assays (Figure 3c). The larger apparent loss of procaspases 2 and 4 (Figure 3b) compared with the increases in activities of caspases 2 and 4 (Figure 3c) is likely due to the different types of assays, as the first determines the disappearance of endogenous proenzymes and the latter measures in vitro cleavage of artificial fluorescent substrates.
Figure 3.
Downregulation of SPP1 induces ER stress that leads to autophagy. (a, b) Equal amounts of lysates from MCF7 cells transfected with siControl or siSPP1 were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Blots were re-probed for total eIF2α or tubulin to insure equal loading and transfer. (c) Duplicate cultures were lysed and caspase 2 and caspase 4 activities were determined by fluorescence and luminescence, respectively. Data are means±S.D. of three independent experiments each carried out in triplicate. (d) MCF7 cells were transfected with siControl or siRNAs targeted to IRE1α or ATF6, and downregulation efficiency was examined by immunoblotting. (e) MCF7 cells were also co-transfected with vector or dominant-negative PERK (dnPERK) and inhibition of PERK activity confirmed by immunoblotting with p-eIF2α antibody. (f) MCF7 cells were co-transfected with siControl, siSPP1, siIRE1α, siATF6, or dnPERK and EGFP-LC3, as indicated. Autophagy was quantified by confocal microscopy. Values are the means±S.D. of three independent experiments. (g–i) Downregulation of SPP1 does not trigger apoptosis or induce cell death. (g) MCF7 cells were transfected with siControl or siSPP1, cultured for the indicated times, and nuclei were stained with Hoechst 33342. Apoptosis was determined by scoring the percentage of cells displaying fragmented, condensed nuclei indicative of apoptosis. At least three fields were analyzed, scoring a minimum of 300 cells. Data are the means±S.D. of three independent experiments. Insert: Equal amounts of lysates from duplicate 1-day cultures were resolved by SDS-PAGE and immunoblotted with anti-PARP antibody. Blots were re-probed with anti-tubulin to insure equal loading and transfer. (h, i) MCF7 cells were transfected with siControl siRNA, siSPP1, or individual siRNAs targeted to Atg5, ATF6, or IRE1α, or transfected with dnPERK. Apoptosis was quantified by Hoechst 33342 staining (h) and viability was quantified by Trypan blue exclusion (i). Data are means±S.D. of three independent experiments. *P<0.01 compared with siControl
As these data indicate that depletion of SPP1 induces ER stress and UPR, we next examined whether this mediated the autophagic response. Downregulation of the expression of two major ER stress transducers, activating transcription factor 6 (ATF6) and inositol-requiring transmembrane kinase/endonuclease (IRE1α) (Figure 3d), completely abrogated SPP1-regulated autophagy (Figure 3f). Similarly, expression of dominant-negative PERK (dnPERK), which blocked phosphorylation of eIF2α (Figure 3e), also drastically suppressed autophagy (Figure 3f).
Autophagy induced by SPP1 depletion is not accompanied by apoptosis or cell death
While autophagy is often a stress adaptation to avoid cell death and suppress apoptosis, in other cellular contexts, it may also be an alternative cell death pathway.2, 3, 18 Thus, we examined the impact of autophagy resulting from depletion of SPP1 on apoptosis and cell survival. Silencing SPP1 did not induce apoptosis at any time point examined nor did it induce cleavage of poly(ADP-ribose) polymerase (PARP) (Figure 3g), a substrate for caspase-mediated proteolysis during apoptosis. Next, we investigated whether the lack of apoptosis was a consequence of the induction of protective autophagy. However, silencing Atg5, ATF6, or IRE1α, or expression of dnPERK, each of which completely suppressed SPP1-regulated autophagy (Figures 2e and 3f), had no effect on apoptosis (Figure 3h) or cell viability (Figure 3i).
Autophagy induced by SPP1 depletion is p53-independent
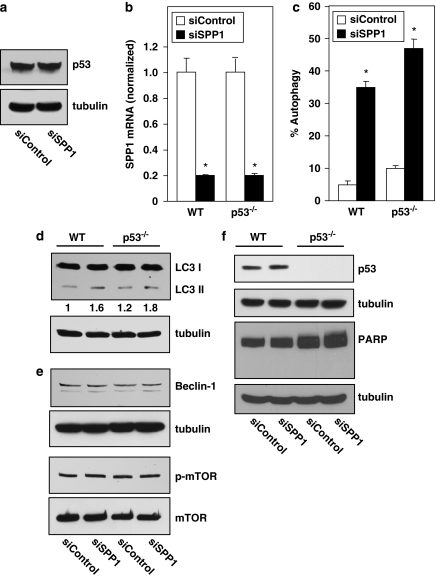
Treatment of cells with inducers of ER stress has recently been shown to stimulate proteosomal degradation of the tumor suppressor p53 resulting in autophagy.21 As we demonstrated that depletion of SPP1 triggered autophagy that was induced through ER stress, we wondered whether this was also accompanied by degradation of p53. However, in contrast to ER stress induced by tunicamycin or thapsigargin,21 silencing SPP1 in MCF7 cells did not affect cytosolic p53 levels (Figure 4a). Moreover, decreasing endogenous levels of SPP1 in human HCT116 colon carcinoma cells with siRNA (Figure 4b) also caused autophagy (Figure 4c and d). Similar autophagy (Figure 4c and d) was also induced in an isogenic cell line derived from HCT116 cells, in which p53 was inactivated by targeted homologous recombination (Figure 4c). Western blotting also showed increased conversion of endogenous LC3I to LC3II that was independent of p53 status (Figure 4d). It should be noted that only 35–45% of GFP-LC3 transfected cells were autophagic due to SPP1 depletion (Figure 4d), which explains the apparent smaller changes in endogenous LC3II formation determined by western blotting of cell lysates (Figure 4c). Because MCF7 cells are deficient in Beclin-1 expression,22 we also examined the involvement of class III PI3K/Beclin-1 complex and mTOR in Beclin-1 expressing HCT116 cells. Silencing SPP1 in wild-type or p53-null HCT116 cells did not increase levels of Beclin-1 nor did it reduce the phosphorylation status of mTOR (Figure 4e), supporting the lack of involvement of mTOR and class III PI3K/Beclin-1 pathways in autophagy triggered by depletion of SPP1. Moreover, as in MCF7 cells, autophagy was not accompanied by changes in cytosolic p53 levels (Figure 4f), nor did it lead to apoptosis as demonstrated by a lack of PARP cleavage (Figure 4f).
Figure 4.
SPP1-mediated autophagy is p53-independent. (a) MCF7 cells were transfected with control siRNA or with siSPP1. Equal amounts of lysates were resolved by SDS-PAGE and immunoblotted with anti-p53 antibody. Blots were re-probed for tubulin to insure equal loading and transfer. (b–f) Wild-type (WT) or p53−/− HCT116 cells were transfected with control siRNA or siSPP1. (b) SPP1 mRNA was determined by quantitative real-time PCR and normalized to GAPDH mRNA. (c) Twenty-four hours after siRNA transfections, as in (b), cells were transfected with EGFP-LC3 and autophagy was determined by confocal microscopy. *P<0.01 compared with siControl. (d) Autophagy was determined by western blotting with anti-LC3 antibody and with anti-tubulin as a loading control. Numbers indicate fold changes determined by densitometry. (e, f) Equal amounts of cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Blots were re-probed for tubulin or total mTOR to insure equal loading and transfer
Depletion of SPP1 induces Akt phosphorylation that protects cells from apoptosis
The function of the UPR is to mitigate ER stress by reducing the accumulation of misfolded proteins. However, cells die by apoptosis when these adaptive responses are not sufficient to relieve ER stress.23 Despite the induction of ER stress, cells depleted of SPP1 do not die. Thus, we examined whether cell survival pathways, such as Akt or ERK1/2 that counteract apoptotic signals, are activated in parallel with ER stress and UPR induced by depletion of SPP1. We utilized phospho-specific antibodies to examine phosphorylation of Akt at Thr308 in the activation loop and at Ser473 at the C terminus, which are required for full activation.24 Silencing SPP1 markedly increased phosphorylation of Akt at both Thr308 and Ser473 (Figure 5a), whereas in sharp contrast, it did not significantly affect ERK1/2 phosphorylation at Thr202/Tyr204 (Figure 5a). This was not caused by the upregulation of Akt, as expression of Akt1, Akt2, and Akt3 was unaltered (Figure 5b). We next examined the effect of AktVIII, a potent and selective Akt1 and Akt2 inhibitor that inhibited Akt phosphorylation (Figure 5c). As expected from the well-known function of Akt to activate mTOR that suppresses autophagy,25 treatment with AktVIII reduced phosphorylation of mTOR (Figure 5d) and induced autophagy (Figure 5e and f). Surprisingly, however, AktVIII treatment did not further enhance autophagy induced by depletion of SPP1 (Figure 5e). In contrast, although AktVIII alone induced little apoptosis after 48 h (<20%), as was found in another cell type,26 it significantly sensitized MCF-7 cells to SPP1 depletion-induced apoptosis, as demonstrated by cleavage of PARP (Figure 5g), increased the number of apoptotic cells (Figure 5h), and decreased viability (Figure 5i). Even after 72 h of treatment, when AktVII itself increased apoptosis and decreased viability, it still significantly enhanced the effects of SPP1 depletion (Figure 5h and i).
Figure 5.
Downregulation of SPP1 stimulates Akt, which suppresses apoptosis. (a, b) MCF7 cells were transfected with siControl or siSPP1. (c, d) Cells were cultured for 24 h in the absence or presence of AktVIII (5 μM) as indicated. (a–d) Equal amounts of cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. (e, f) MCF7 cells transfected with siControl or siSPP1 were transfected with EGFP-LC3 and cultured in the absence or presence of AktVIII (5 μM). Autophagy was determined by confocal microscopy. (f) Representative images are shown. Scale bars, 10 μm. Data are means±S.D. of three independent experiments. *P<0.05 compared with siControl. (g–i) MCF7 cells transfected with siControl or siSPP1 were cultured in the absence or presence of AktVIII (5 μM) for 48 or 72 h. (g) Equal amounts of lysates were resolved by SDS-PAGE and immunoblotted with anti-PARP antibody. Apoptosis (h) and viability (i) were quantified after 48 and 72 h in duplicate cultures by Hoechst 33342 and Trypan blue exclusion staining, respectively. *P<0.05 compared with siControl, **P<0.05 compared with vehicle
Autophagy is triggered by intracellular S1P independent of its cell surface receptors
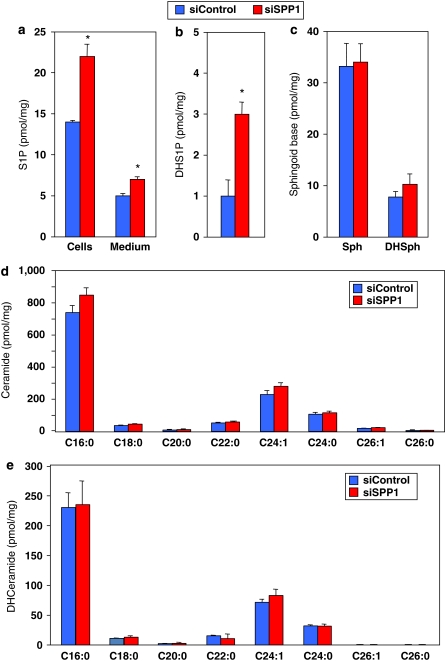
Consistent with previous studies,27, 28 silencing SPP1 not only increased intracellular mass levels of S1P (Figure 6a) and dihydro-S1P (Figure 6b), but it also slightly increased S1P released into the medium (Figure 6a), supporting the involvement of SPP1 in regulation of intra and extracellular levels of S1P. Importantly, no significant changes were observed in levels of sphingoid bases (Figure 6c), or even ceramide (Figure 6d) and dihydroceramide (Figure 6e), which have been implicated in regulation of autophagy.8, 9, 10
Figure 6.
Effect of SPP1 depletion on S1P and sphingolipids. (a–c) MCF7 cells were transfected with siControl or siSPP1. Lipids were extracted and S1P (a), dihydro-S1P (b), sphingosine and dihydrosphingosine (c), ceramide species (d), and dihydroceramide species (e) analyzed by LC-MS/MS. Numbers indicate chain length followed by the number of double bonds in the fatty acid. Data are averages of triplicate transfectants and are expressed as picomol lipid/mg of protein. *P<0.01 compared with siControl. Similar results were found in three additional experiments
Next, it was important to determine whether intra or extracellular S1P is responsible for the induction of autophagy. MCF7 cells express three of the five S1PRs, with S1P3 being the most predominant.29, 30 However, 100 nM S1P or dihydro-S1P, both of which also bind and activate all of the S1P receptors, did not trigger autophagy when added to cells (Figure 7a). Yet, this concentration of S1P and dihydro-S1P, which is much greater than the Kd values for the S1P receptors, activated ERK1/2 in these cells (Figure 7b), suggesting that the lack of effect of these sphingolipids on autophagy is not due to their inability to signal through S1P cell surface receptors. Moreover, neither VPC23019, at a concentration that antagonizes S1P3, nor the S1P2 antagonist JTE-013, blocked siSPP1-triggered autophagy in these cells (Figure 7d), although as expected, both VCP23019 and JTE-013 inhibited S1P-stimulated ERK1/2 activation (Figure 7c). Moreover, in contrast to depletion of SPP1, which increased Akt phosphorylation, exogenous S1P-stimulated ERK1/2 but not Akt in these cells (Figure 7e). To conclusively demonstrate that increased levels of intracellular S1P are responsible for induction of autophagy, we relied on previous observations that only high concentrations of exogenously added S1P and dihydro-S1P significantly increase their intracellular levels.31, 32 While S1P at 100 nM had no significant effects on autophagy (Figure 7a), treatment with 10 μM S1P, which increased the intracellular pools of S1P (Figure 7f), enhanced autophagy as shown by the increase in GFP-LC3 puncta and by the increased formation of LC3-II (Figure 7g and h), and slightly increased ER stress as shown by phosphorylation of eIF2α (Figure 7h). As expected, these effects of exogenous S1P were less pronounced than those induced by depletion of SPP1, as depletion of SPP1 increases the specific pool of S1P in the ER, whereas treatment of cells with 10 μM S1P increases S1P throughout the cell. However, a high concentration of dihydro-S1P, which as previously reported,32 increased its intracellular level to an even greater extent (Figure 7f), but did not mimic the effects of S1P on autophagy (Figure 7g). Taken together, these results indicate that intracellular S1P, rather than extracellular S1P, is responsible for induction of autophagy in response to SPP1 depletion.
Figure 7.
SPP1 depletion triggers autophagy and activates Akt by intracellular S1P independently of its receptors. (a) MCF7 cells transfected with pEGFP-LC3 were treated with 100 nM S1P or dihydroS1P (DHS1P) for 24 h, and autophagy was quantified by confocal microscopy. Data are means±S.D. of three independent experiments. (b) Cells were treated with 100 nM S1P or DHS1P for 10 min. (c) Cells were pretreated without or with 10 μM VPC23019 or 1 μM JTE-013 for 30 min, and then stimulated with 100 nM S1P for 10 min. (b, c) Equal amounts of lysates were analyzed by immunoblotting with anti-phospho ERK1/2. Blots were re-probed for ERK to insure equal loading and transfer. (d) Cells transfected with siControl or siSPP1 were transfected 24 h later with EGFP-LC3, cultured in the absence or presence of 10 μM VPC23019 or 1 μM JTE-013, and autophagy quantified by confocal microscopy. Values reported are means±S.D. of three independent experiments. (e) MCF7 cells transfected with siControl or siSPP1 or naïve MCF7 cells were treated with vehicle, S1P (100 nM), or EGF (10 ng/ml) for 10 min. Proteins were immunoblotted with anti-phospho-Akt (Ser473) and re-probed with anti-Akt antibody. (f) MCF7 cells were treated with 10 μM S1P or dihydroS1P (DHS1P) for 2 h and sphingolipids analyzed by LC-ESI-MS/MS. (g) MCF7 cells transfected with pEGFP-LC3 were treated with 10 μM S1P or DHS1P for 24 h and autophagy quantified by confocal microscopy. (h) Equal amounts of lysates from MCF7 cells treated without or with 10 μM S1P were immunoblotted with anti-LC3 or anti-p-eIF2α antibodies. Blots were re-probed for tubulin to insure equal loading and transfer. (i) MCF7 cells transfected with siControl or siSPP1 were transfected with vector or dnPERK. (j) MCF7 cells transfected with siControl or siSPP1 were treated without or with LY294004 (5 μM). Equal amounts of lysates were immunoblotted with anti-phospho-Akt (Ser473) and re-probed with anti-Akt antibody. (k) Wild-type (PERK+/+) and PERK null (PERK−/−) MEFs were transfected with siControl or siSPP1. Equal amounts of lysates were immunoblotted with the indicated antibodies. *P<0.01 compared with vehicle
Although the UPR is known to alleviate ER stress by reducing accumulation of misfolded proteins,2, 18, 19 recent studies demonstrate that the UPR also induces activation of Akt to protect cells from ER stress-induced apoptosis.33, 34, 35 Because activation of the Akt pathway by inducers of ER stress is often dependent on PERK,33, 34, 35 we examined its role in phosphorylation of Akt induced by SPP1 depletion. Overexpression of dnPERK not only suppressed phosphorylation of eIF2α (Figure 3d) and autophagy (Figure 3e), but it also reduced Akt phosphorylation (Figure 7i). LY294002, an inhibitor of PI3K, also markedly reduced Akt phosphorylation induced by SPP1 depletion (Figure 7j), suggesting that PERK induced PI3K-dependent Akt phosphorylation. Finally, although induction of Akt phosphorylation and autophagy were observed in wild-type mouse embryonic fibroblasts (MEFs), downregulation of SPP1 in PERK-null MEFs no longer induced phosphorylation of Akt or eIF2α (Figure 7k) or triggered autophagy (Figure 7k), further supporting the importance of PERK in autophagy induced by SPP1 depletion.
Discussion
In this study, we have shown that SPP1, the enzyme that specifically dephosphorylates S1P and dihydro-S1P at the ER, regulates autophagy in diverse cell types. Depletion of SPP1 in human breast cancer MCF7 cells, HCT116 colon cancer cells, or in MEFs induced autophagy, even in the presence of nutrients, as demonstrated by the formation of autophagosomes and autophagolysosomes, as well as by LC3 lipidation. In contrast to autophagy mediated by ceramide elevation,8 neither class III PI3K/Beclin-1 nor mTOR pathways were involved, as depletion of SPP1 did not increase Beclin-1 or inhibit activation of mTOR. Moreover, autophagy was not decreased by treatment with the class III PI3K inhibitor 3MA. Consistent with a ceramide-independent effect, neither ceramide nor dihydroceramide species were elevated by depletion of SPP1, yet as expected, S1P and dihydro-S1P levels were significantly increased. Our data indicate that autophagy induced by downregulation of SPP1 was mediated by intracellular S1P and not by secreted S1P, as treatment with 100 nM S1P or dihydro-S1P, concentrations that activate cell surface S1P receptors, did not cause autophagy. In contrast, increased intracellular pools of S1P, but not of dihydro-S1P, by treatment with 100-fold higher concentrations similarly induced autophagy. It should be noted that depletion of SPP1 likely increases specific pools of S1P at the ER, whereas, treatment of cells with 10 μM probably not only increases S1P at the ER but also throughout the cell. Elevation of intracellular S1P by overexpression of SphK1, like downregulation of SPP1, has previously been reported to cause autophagy.11 Although this was also characterized by a lack of robust upregulation of Beclin-1, inhibition of mTOR signaling was suggested to be involved.11 On the other hand, autophagy was not altered in S1P lyase-null MEFs, despite increased S1P and concomitant elevation of both sphingosine and ceramide.36 As the phosphatase and the lyase are important for different routes of S1P metabolism, the first for salvage of sphingolipids and the latter for generation of phosphatidylethanolamine, they likely have different functions in regulation of ER homeostasis and autophagy.
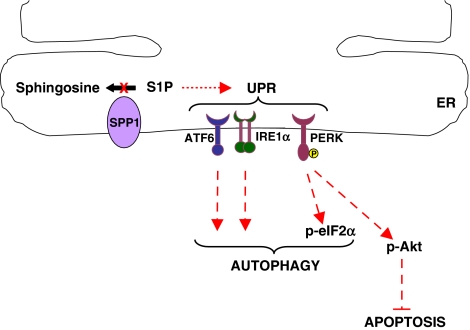
Interestingly, downregulation of SPP1 led to activation of the UPR (Figure 8), an integrated signaling pathway that senses protein-folding status in the ER to regulate protein folding capacity and is the hallmark of ER stress.19 The three essential sensors of the UPR, IRE1α, ATF6, and PERK were all involved in ER stress induced by depletion of SPP1 (Figure 8). This is unusual as, although PERK and IRE1α share functionally similar ER lumenal-sensing domains, they are selectively engaged in vivo by the physiological stress of unfolded proteins. For example, autophagy induced by the ER stressors, tunicamycin and thapsigargin, requires IRE1α expression but not PERK or ATF6.37 It is tempting to speculate that upregulation of the chaperone protein GPR78/BiP that maintains ER integrity has an important role in transducing the ER stress signals on depletion of SPP1. In unstressed cells, GRP78/BiP binds and inhibits PERK, IRE1, and ATF6. When misfolded proteins accumulate in the ER, GPR78/BiP binds to them to promote their folding, and PERK, IRE1α, and ATF6 are released from GRP78/BiP and become activated.38 Although the mechanisms that couple the UPR to autophagy in mammalian cells are not completely understood, both PERK and IRE1α have been implicated. It has been suggested that JNK activation mediated by IRE1α is involved in autophagosome formation.37 A recent report suggested that phosphorylation of the antiapoptotic protein BCL-2 at the ER membrane by JNK, possibly by ceramide elevation, may directly affect the initiation of autophagy by modulating the activity of Beclin-1.39 Activation of eIF2α by PERK causes the induction of Atg12 expression by ATF4, which likely participates in autophagy.17 In this regard, PERK has been shown to activate Akt as a mechanism to protect cells from ER stress-induced apoptosis.33, 34, 35 Indeed, depletion of SPP1-stimulated phosphorylation of Akt in a PERK-dependent manner (Figure 8), without affecting total Akt levels. Similarly, both tunicamycin and thapsigargin, inducers of ER stress, activate Akt without increasing its expression.33, 40 Not much is yet known about how PERK activates Akt, but it has been suggested that phosphorylation of eIF2α by PERK is critical to the activation of Akt, due to translation inhibition of a protein that negatively regulates PI3K activity. This leads to the activation of Akt downstream of PI3K signaling.40 In agreement, we found that inhibition of PI3K with LY294004 decreased activation of Akt induced by SPP1 depletion.
Figure 8.
Model of involvement of SPP1 in ER stress and UPR and connection to autophagy. Depletion of SPP1 and increased intracellular S1P triggers ER stress and the UPR by activation of ATF6, IRE1α, and PERK, resulting in autophagy. PERK signaling also activates Akt, which protect cells from apoptosis. Other known essential components of ATF6, IRE1α, and PERK that downstream signaling19 are not shown
Autophagy induced by depletion of SPP1 has unique characteristics compared with other inducers of ER stress. First, in contrast to tunicamycin or thapsigargin, which enhance proteosomal degradation of the tumor suppressor p53 resulting in autophagy,21 SPP1 downregulation-induced autophagy did not alter p53 levels and was independent of p53 status. Second, unlike these chemical ER stressors that also induce apoptosis,33 SPP1 depletion did not trigger apoptosis even after prolonged periods of autophagy, despite activation of caspases 2 and 4. Although a requirement for caspase 4 in ER-stress-induced apoptosis has been suggested,41 it is uncertain whether caspase 4 is required for ER-stress-induced apoptosis in general.18 Moreover, inhibition of autophagy by dowregulation of three arms of the UPR did not lead to apoptosis. Our data indicate that regardless of the cell type, depletion of SPP1 triggers p53-independent autophagy that does not lead to autophagic cell death. This is probably due to activation of Akt in a PERK-dependent manner, which has a critical role in counteracting ER-stress-induced apoptotic signaling, thereby controlling cell survival. Our results support the notion that sphingolipid homeostasis is important for proper function of the ER.
Materials and Methods
Reagents
Cell culture medium and fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA, USA). 3MA was purchased from Sigma (St. Louis, MO, USA). Monoclonal anti-Beclin-1 was from Bioscience (San Jose, CA, USA); polyclonal anti-LC3 from Novus Biologicals (Littleton, CO USA); anti-actin, anti-p53, polyclonal anti-ERK2, anti-GRP78 and anti-p70S6K were from Santa Cruz (Santa Cruz, CA USA); polyclonal anti-SPP1 was from Pierce (Rockford, IL, USA); monoclonal anti-IRE1α, anti-Akt1, anti-Akt2, anti-Akt3, anti-AMPK, anti-phospho-eIFα, anti-phospho-ERK1/2, polyclonal anti-phospho-Akt (Ser473), anti-Akt, anti-tubulin, anti-mTOR, anti-phospho-mTOR, anti-phospho-AMPK, anti-phospho-p70S6K (Thr308), and anti-eIF2 were from Cell Signaling (Danvers, MA, USA); monoclonal anti-ATF6 was from IMGENEX (San Diego, CA USA); monoclonal anti-caspase 4 from Stressgen (Farmingdale, NY, USA); and anti-Atg5 from Abgent (San Diego, CA USA).
Plasmids
Plasmid expression vectors encoding GFP-LC3 (pEGFP-LC3) were provided by T Yoshimori (National Institute of Genetics, Mishima, Japan) and K Kirkegaard (Stanford University School of Medicine, Stanford, CA). Wild-type and dnPERK (Myc-tagged PERKΔC) and pBABE tandem mCherry GFP-LC3 were a generous gift from Dr. JA Diehl (University of Pennsylvania) and Dr. Andrew Thorburn (University of Colorado), respectively.
Cell culture and transfection
Human breast cancer MCF7 cells were maintained in phenol red-free IMEM, supplemented with 10% heat-inactivated FBS and 0.25% glucose. Wild-type and p53−/− human colon cancer HCT116 cells, immortalized wild-type and PERK−/− MEFs were kindly provided by Dr. David Ron (Skirball Institute, NYU), were maintained in DMEM, supplemented with 10% FBS. For culture in nutrient-free medium, cells were cultured in Earle's Balanced Salt Solution (Sigma), which does not contain serum or amino acids.
MCF7 and HCT116 cells in six-well plates were transfected with siRNAs (50 nM) using Oligofectamine (Invitrogen). MEFs were transfected with Lipofectamine 2000 (Invitrogen). Expression of SPP1, IRE1α, and Atg5 was downregulated with ON TARGETplus SMARTpool siRNAs (Dharmacon, Lafayette, CO, USA), unless indicated otherwise, and ATF6 with siRNA from Qiagen (Valencia, CA, USA). siRNA sequences are available in Supplementary Table 1. For plasmid transfections, MCF7 and HCT116 cells were transfected using Fugene 6 (Roche, Basel, Switzerland) and Lipofectamine 2000 (Invitrogen), respectively. When co-transfected with siRNA and plasmids, cells were first transfected with siRNA and then with plasmids the following day.
Quantitative RT-PCR
Cells were disrupted in Trizol reagent (Invitrogen) and RNA was reverse transcribed with SuperScript II (Invitrogen). For quantitative PCR, premixed primer-probe sets were purchased from Applied Biosystems (Carlsbad, CA ,USA) and cDNA was amplified with ABI 7900HT (Carlsbad, CA, USA).
Autophagy assay
MCF7 cells were grown on poly-lysine (Sigma) coated glass coverslips. Briefly, after transfection with GFP-LC3 and treatments as described in figure legends, cells were washed with PBS and fixed for 20 min at room temperature with 4% paraformaldehyde. After washing, coverslips were mounted on glass slides and examined with a Zeiss (Thornword, NY, USA) LSM 510 laser confocal microscope. Autophagy was quantified in a minimum of 100 cells per preparation in three independent experiments. Cells with five or more intense GFP-LC3 puncta were considered autophagic, whereas those with diffuse cytoplasmic GFP-LC3 staining were considered non-autophagic. Percent autophagy was determined in a double-blind manner.
Electron microscopy
MCF7 cells were grown on poly-lysine-coated glass coverslips, washed with PBS and initially fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer followed by secondary fixation with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer. The samples were dehydrated through an ethanol series and embedded in EMbed-812 epoxy resin (Electron Microscopy Sciences, Hatfield, PA, USA). Samples were thin sectioned at 900 Angströms with a diamond knife using the LKB 2128 Ultrotome IV (Vienna, Austria) and then stained with 5% uranyl acetate in 50% methanol and Reynold's lead citrate. Cells were observed with a JEOL JEM-1230 transmission electron microscope (JEOL, Tokyo, Japan), and photographs were taken with a Gating UltraScan 4000 digital camera (Gatan, Pleasanton, CA, USA).
Apoptosis assay
Apoptotic and necrotic cells were quantified by fluorescence microscopy after labeling with Hoescht 33342 (10 μg/ml) and propidium iodide (10 μg/ml). Cells exhibiting blue condensed or fragmented nuclei were considered apoptotic. Red nuclei without signs of condensation or fragmentation were considered necrotic. Cells were observed with a Nikon (Tokyo, Japan) eclipse TE300 fluorescence microscope. Cell viability was also determined by Trypan blue exclusion.
Caspase activity assays
Caspase 2 and caspase 4 activities, were determined with a luminescence assay kit (Promega, Madison, WI, USA) and a fluorescence assay kit (Abcam, Cambridge, MA, USA), respectively, using a Victor X4 plate reader (PerkinElmer, Waltham, MA, USA).
Western blot analysis
Cells were lysed in buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM EDTA, 1 mM sodium orthovanadate, and 1 : 500 protease inhibitor mixture (Sigma). Equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose, incubated with the indicated primary antibodies overnight, washed, and incubated with appropriate HRP-conjugated secondary antibodies. Immune complexes were visualized by enhanced chemiluminescence (Pierce, Rockford, IL, USA).
Mass spectrometric analysis of lipids
Sphingolipids were quantified by liquid chromatography, electrospray ionization-tandem mass spectrometry with a Shimadzu (Columbia, MD, USA) LC-10 binary pump system coupled to a PerkinElmer Series 200 autoinjector and an Applied Biosystems 4000 QTRAP operating in a triple quadrupole mode, as described previously.42
Statistical analysis
Experiments were repeated at least three times. Results were analyzed for statistical significance with the Student's t-test for unpaired samples and P<0.05 was considered significant.
Acknowledgments
We are grateful to Drs. T Yoshimori (National Institute of Genetics, Mishima, Japan), K Kirkegaard (Stanford University School of Medicine, Stanford, USA), JA Diehl (University of Pennsylvania), David Ron (Skirball Institute, NYU), and Andrew Thorburn (University of Colorado) for generous gifts of reagents. This work was supported by NIGMS Grant 5R37GM043880 (SS) and in part by 2R01DK052825 (PD). Confocal and electron microscopy were supported in part by NIH Grant P30 CA16059 to the Massey Cancer Center and NINDS Center core Grant 5P30NS047463, respectively.
Glossary
- AMPK
AMP kinase
- ATF6
activating transcription factor 6
- Atg
autophagy-related gene
- dn
dominant-negative
- eIF2α
eucaryotic translation initiation factor 2α
- ER
endoplasmic reticulum
- IRE1α
inositol-requiring transmembrane kinase/endonuclease-1α
- LC3
microtubule-associated protein-1 light chain 3
- MEF
mouse embryonic fibroblasts
- 3MA
3-methyladenine
- mTOR
mammalian target of rapamycin
- PARP
poly(ADP-ribose) polymerase
- PERK
protein kinase regulated by RNA-like ER kinase
- S1P
sphingosine-1-phosphate
- SphK
sphingosine kinase
- SPP1
S1P phosphohydrolase 1
- UPR
unfolded protein response
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Piacentini
Supplementary Material
References
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, et al. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- Lavieu G, Scarlatti F, Sala G, Levade T, Ghidoni R, Botti J, et al. Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision. Autophagy. 2007;3:45–47. doi: 10.4161/auto.3416. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–1662. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, et al. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, et al. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci USA. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Saba JD, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- Le Stunff H, Galve-Roperh I, Peterson C, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. J Cell Biol. 2002;158:1039–1049. doi: 10.1083/jcb.200203123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Johnson KY, Becker KP, Mao C, Obeid LM. Role of human sphingosine-1-phosphate phosphatase 1 in the regulation of intra- and extracellular sphingosine-1-phosphate levels and cell viability. J Biol Chem. 2003;278:34541–34547. doi: 10.1074/jbc.M301741200. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Kalari SK, Usatyuk PV, Gorshkova I, He D, Watkins T, et al. Intracellular generation of sphingosine 1-phosphate in human lung endothelial cells: role of lipid phosphate phosphatase-1 and sphingosine kinase 1. J Biol Chem. 2007;282:14165–14177. doi: 10.1074/jbc.M701279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Van Brocklyn JR, Edsall L, Nava VE, Spiegel S. Sphingosine-1-phosphate inhibits motility of human breast cancer cells independently of cell surface receptors. Cancer Res. 1999;59:6185–6191. [PubMed] [Google Scholar]
- Hadizadeh S, King DN, Shah S, Sewer MB. Sphingosine-1-phosphate regulates the expression of the liver receptor homologue-1. Mol Cell Endocrinol. 2008;283:104–113. doi: 10.1016/j.mce.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Van Brocklyn JR, Lee MJ, Menzeleev R, Olivera A, Edsall L, Cuvillier O, et al. Dual actions of sphingosine-1-phosphate: extracellular through the Gi-coupled orphan receptor edg-1 and intracellular to regulate proliferation and survival. J Cell Biol. 1998;142:229–240. doi: 10.1083/jcb.142.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giussani P, Maceyka M, Le Stunff H, Mikami A, Lepine S, Wang E, et al. Sphingosine-1-phosphate phosphohydrolase regulates endoplasmic reticulum-to-Golgi trafficking of ceramide. Mol Cell Biol. 2006;26:5055–5069. doi: 10.1128/MCB.02107-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Han Z, Couvillon AD, Exton JH. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem. 2004;279:49420–49429. doi: 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- Yacoub A, Park MA, Hanna DJ, Hong Y, Mitchell C, Pandya AP, et al. OSU-03012 promotes caspase-independent, PERK-, cathepsin B-, BID- and AIF-dependent killing of transformed cells. Mol Pharmacol. 2006;70:589–603. doi: 10.1124/mol.106.025007. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, Diehl JA. PERK-dependent regulation of IAP translation during ER stress. Oncogene. 2009;28:910–920. doi: 10.1038/onc.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colie S, Van Veldhoven PP, Kedjouar B, Bedia C, Albinet V, Sorli SC, et al. Disruption of sphingosine 1-phosphate lyase confers resistance to chemotherapy and promotes oncogenesis through Bcl-2/Bcl-xL upregulation. Cancer Res. 2009;69:9346–9353. doi: 10.1158/0008-5472.CAN-09-2198. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi S, Mounir Z, Baltzis D, Raven JF, Wang S, Krishnamoorthy JL, et al. A novel function of eIF2alpha kinases as inducers of the phosphoinositide-3 kinase signaling pathway. Mol Biol Cell. 2007;18:3635–3644. doi: 10.1091/mbc.E07-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger L, Schneider J, Rheme C, Tapernoux M, Hacki J, Borner C. Serine proteases mediate apoptosis-like cell death and phagocytosis under caspase-inhibiting conditions. Cell Death Differ. 2003;10:1188–1203. doi: 10.1038/sj.cdd.4401288. [DOI] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.