Abstract
Krüppel-like factors (KLFs) as a family of zinc-finger transcription factors involve in the regulation of many physiological processes. In these studies, KLF9 was characterized for its role in adipogenesis. The expression of KLF9 was markedly upregulated during the middle stage of 3T3-L1 adipocyte differentiation, and inhibition of KLF9 by RNAi impaired adipogenesis. Using promoter deletion and mutation analysis, we identified two KLF9-binding sites within the 0.6-kb region of the PPARγ2 proximal promoter, indicating that KLF9 interacts with the PPARγ2 promoter. Furthermore, we found that KLF9 could synergistically activate PPARγ2 promoter by directly interacting with C/EBPα. In addition, overexpression of PPARγ2 rescued the impairment of adipocyte differentiation induced by KLF9 knockdown, which supports that PPARγ2 is a downstream target of KLF9. Collectively, our results indicate KLF9 as a key pro-adipogenic transcription factor through regulation of PPARγ2 expression with C/EBPα at the middle stage of adipogenesis.
Keywords: KLF9, adipogenesis, PPARγ2, C/EBPα, 3T3-L1 cell
Obesity is now a prevalent health hazard worldwide, which tends to develop a number of pathological disorders such as type-2 diabetes and cardiovascular disease.1 This excess of adipose tissue can be the consequence of both increased fat-cell number (hyperplasia) and increased fat-cell size (hypertrophy).2 The number of adipocytes present in an organism is determined to a large degree by the adipocyte differentiation process. Consequently, understanding the mechanisms regulating adipocyte differentiation may provide valuable information in limiting obesity and its pathological consequences. The 3T3-L1 cell line has been widely used as adipogenesis model to elucidate the mechanism of adipocyte differentiation, in which confluent 3T3-L1 pre-adipocytes differentiate into adipocytes upon exposure to a hormone cocktail including isobutylmethylxanthine, dexamethasone and insulin (MDI).3, 4
A number of transcription factors are involved in the regulation of adipogenesis, including the peroxisome proliferator-activated receptor (PPAR) family5, 6, 7 and CCAAT/enhancer-binding proteins.8, 9 PPARγ is a member of the nuclear receptor superfamily, and is both necessary and sufficient for adipogenesis.10 PPARγ1 and PPARγ2 are generated by alternative splicing and promoter usage and induced during adipogenesis. PPARγ1 is found in other cell types besides adipocytes, whereas PPARγ2 is expressed specifically in adipocytes.11 In 3T3-L1 pre-adipocytes, a hormonal adipogenic stimulus induces a transient increase in the expression of C/EBPβ and C/EBPδ,12 which then stimulates cell proliferation and expression of PPARγ2 and C/EBPα. Once activated, PPARγ2 and C/EBPα can cross-regulate one another to maintain their expression and other adipogenic factors in mature adipocyte.13
Krüppel-like factors (KLFs) are members of an emerging family of DNA-binding transcriptional regulators with crucial roles in development, differentiation and a number of other physiological cellular processes.14 The members of this protein family contain three C2H2 zinc fingers near their C-terminus, which recognize CACCC and related GC-rich elements in promoters and enhancers, and their N-terminal domains are highly variable and show different molecular functions.14 Previous reports showed that KLF proteins had different expression patterns during adipogenesis and several KLFs had been proved to either promote or inhibit this process.15, 16, 17, 18, 19, 20, 21
Krüppel-like factor 9 (KLF9), previously designated as basic transcription element-binding protein-1 (BTEB1), was initially isolated from a rat liver cDNA library.22 KLF9 was further identified to be important for development and other physiological functions. For example, female mice null for the Klf9 gene show some defects such as uterine growth retardation, fewer numbers of embryo implantation sites and decreased litter size.23 KLF9 could control intestinal crypt cell proliferation and villus cell migration.24 In addition, KLF9 is required for proper maturation of the central nervous system.25 Furthermore, KLF9 may function at the node of progesterone receptor and estrogen receptor genomic pathways to influence cell proliferation.26 In our previous study with mouse oligonucleotide microarray, it was identified that the expression of Klf9 was induced during 3T3-L1 adipocyte differentiation. In the present study, we found that KLF9 functioned in adipocyte differentiation through transactivating PPARγ2.
Results
The expression of KLF9 increases at the middle stage of adipogenesis
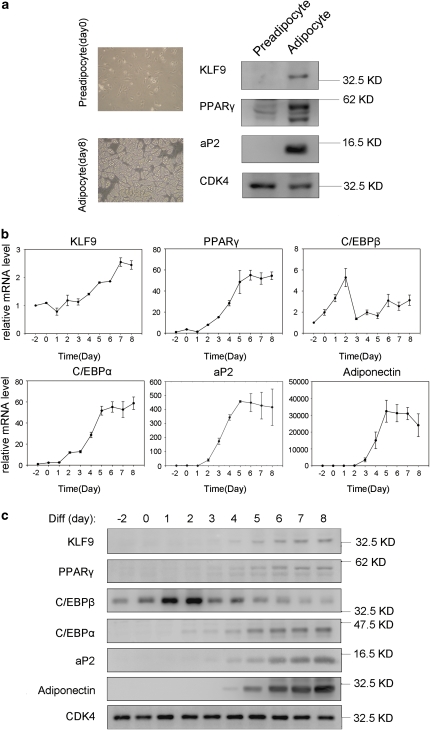
To detect whether KLF9 was related to adipogenesis, we isolated rat primary pre-adipocytes and induced these cells for differentiation. Western blot analysis showed that the protein level of KLF9 was elevated in adipocytes, but undetectable in pre-adipocytes (Figure 1a). As expected, the expression levels of other adipocyte-specific proteins such as PPARγ and aP2 were also greatly increased in adipocytes4 (Figure 1a).
Figure 1.
Expression of KLF9 during adipogenesis. (a) Rat primary pre-adipocytes (day 0) were isolated from rat adipose tissue and differentiated into adipocytes (day 8). The protein level of KLF9, PPARγ and aP2 were detected by western blotting. CDK4 is shown as loading control. (b) The mRNA expression of KLF9, PPARγ, C/EBPβ, C/EBPα, aP2 and adiponectin in 3T3-L1 cells during adipocyte differentiation. The mRNA level was determined by real-time PCR and normalized to PPIA mRNA. The numbers indicate the time points of differentiation induction. Results are expressed as means±S.D. (n=3). (c) The protein expression of KLF9, PPARγ, C/EBPβ, C/EBPα, aP2 and adiponectin in 3T3-L1 cells during adipocyte differentiation. The targeted proteins were detected by western blotting. CDK4 is the loading control. Representative blots from three independent experiments are shown
As 3T3-L1 pre-adipocytes have been widely used as an in vitro model for adipogenesis, we analyzed the expression of KLF9 during 3T3-L1 cell differentiation induced by the standard hormone cocktail MDI.3, 4 Real-time PCR analysis showed that the KLF9 mRNA significantly increased from day 4 of the MDI induction, reaching a maximum at day 7 (Figure 1b). As expected, the mRNA levels of other adipogenesis-specific marker genes such as PPARγ, C/EBPα, Adiponectin and aP2 also increased during the process of 3T3-L1 cell differentiation (Figure 1b).
Furthermore, western blot analysis showed that the KLF9 protein was first clearly detected at day 4 after the MDI induction and continuously increased during the process of adipogenesis, whereas this protein was undetectable in 3T3-L1 pre-adipocytes (Figure 1c). The expression pattern of KLF9 was similar to that of previously reported later transcription factors for adipogenesis such as C/EBPα and PPARγ,27 but different from that of early transcription factors such as C/EBPβ12 (Figure 1c). In addition, the results determined by immunofluorescence staining and sub-cellular fractionation assays showed that KLF9 proteins mainly lied in the nucleus at day 4 of adipogenesis, whereas a portion of KLF9 proteins was detected in the cytosol fraction at day 8 (Supplementary Figure 1).
To find out if overexpression of KLF9 at the early stage had any effect on adipocyte differentiation, we forced exogenous KLF9 expression in 3T3-L1 pre-adipocytes, and then induced the cells for adipogenesis with MDI. The results showed that KLF9 overexpression at the early stage of adipogenesis did not upregulate the expression level of C/EBPα or PPARγ (Supplementary Figure 2). It was not sufficient for the initiation of adipogenesis.
KLF9 functions in adipogenesis at the middle stage of 3T3-L1 adipocyte differentiation
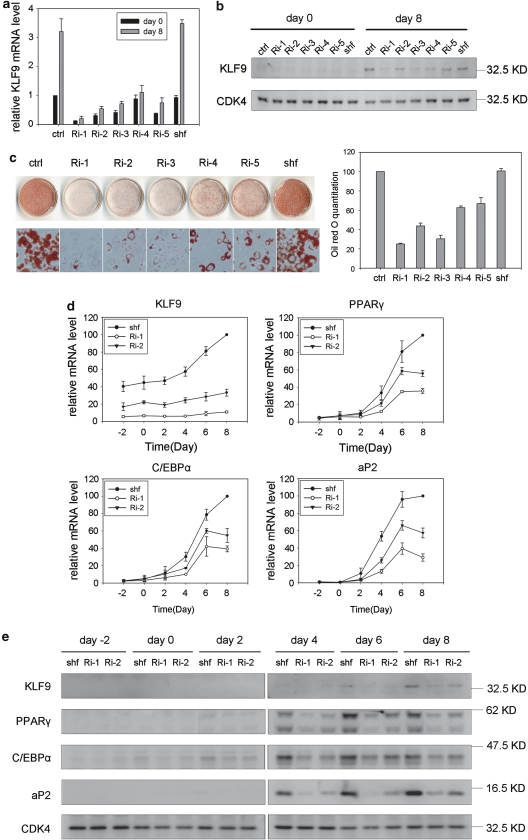
To address the function of KLF9 in adipogenesis, we established five stable cell lines containing different small interference RNA (siRNA) sequences against KLF9 (see Materials and Methods). The results showed that KLF9 was knocked down at both the mRNA and the protein level in these stable cell lines with or without the MDI induction (Figure 2a and b). Importantly, the results determined by Oil-red-O staining assay indicated that KLF9 knockdown by RNA interference (RNAi) suppressed adipocyte differentiation (Figure 2c), suggesting that KLF9 is essential for adipogenesis.
Figure 2.
Blocked adipocyte differentiation by KLF9 knockdown. (a) The KLF9 mRNA level in stable KLF9-knockdown 3T3-L1 cells. Ri-1, 2, 3, 4 and 5 were five stable cell lines generated by five siRNA sequences (see Materials and Methods for details). Ctrl indicates wild-type 3T3-L1 cells and Shuffle(shf) indicates control siRNA sequence. Endogenous KLF9 mRNA was determined by real-time PCR in pre-induction (day 0) and post-induction cells (day 8). Results are expressed as means±S.D. (n=3). (b) The KLF9 protein level in stable KLF9-knockdown 3T3-L1 cells. The KLF9 protein in pre-induction (day 0) and post-induction cells (day 8) was detected by western blotting. CDK4 is the loading control. Representative blots from three independent experiments are shown. (c) The inhibited adipocyte differentiation in KLF9-knockdown 3T3-L1 cells. The cells were induced with standard differentiation hormones. On day 8, the cell monolayer was stained with Oil-red-O. The cell monolayer and micrograph are shown on the left. The Oil-red-O staining was quantified by measuring the OD510 nm. The results were normalized to that of wild-type 3T3-L1 cells and expressed as means±S.D. (right) (n=3). (d) The mRNA expression of KLF9, C/EBPα, PPARγ and aP2 in Ri-1 and Ri-2 stable KLF9-knockdown cells during differentiation induction. The level of mRNA was determined by real-time PCR and normalized to PPIA. The numbers indicate the time points of differentiation induction. The results are expressed as means±S.D. (n=3). (e) The protein expression of KLF9, C/EBPα, PPARγ and aP2 in Ri-1 and Ri-2 stable KLF9-knockdown cells during differentiation induction. The targeted proteins were detected by western blotting. CDK4 is blotted as loading control. Representative blots from three independent experiments are shown
We further analyzed the expressions of adipocyte-specific markers in two KLF9-knockdown cell lines. Real-time PCR analysis showed that the mRNA expression levels of PPARγ, C/EBPα and aP2 were inhibited in these cells (Figure 2d). In agreement with the mRNA results, the western blot results indicated downregulation of these adipogenic protein markers (Figure 2e). Conversely, the mRNA levels of early transcription factors of adipogenesis, including C/EBPβ, C/EBPδ, KLF5 and Krox20, did not show significant changes in these KLF9-knockdown cells, compared with that of the control cells (Supplementary Figure 3), suggesting that KLF9 does not function at the early stage of adipogenesis. Taken together, these results indicate that KLF9 is required for adipocyte differentiation and the time frame of its function is at the middle stage of adipogenesis, which is in accordance with the expression pattern of KLF9 during the process of adipocyte differentiation (Figure 1).
To find out if differences in C/EBPα and PPARγ expression between the normal and KLF9-knockdown cell lines resulted from variations in mRNA stability, we analyzed the effects of KLF9 knockdown on the mRNA stability for PPARγ and C/EBPα. As shown in Supplementary Figure 4, there was no significant difference in the mRNA stability for PPARγ and C/EBPα among the control and KLF9-knockdown cells, suggesting that downregulation of C/EBPα or PPARγ mRNAs in the KLF9-knockdown cells results from transcriptional suppression rather than reduction of mRNA stability.
KLF9 could transactivate PPARγ2
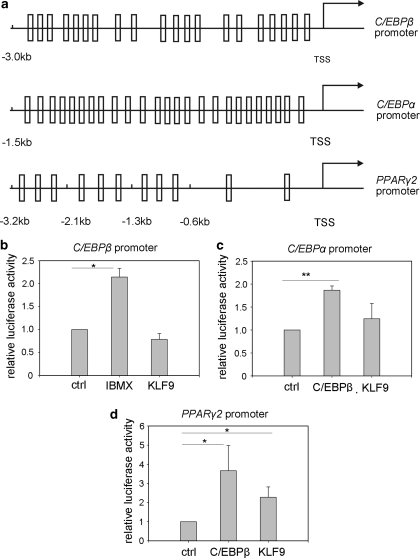
As a transcription factor, what kind of target gene(s) KLF9 might regulate? As shown in Supplementary Figure 3, downregulation of KLF9 did not affect the expression of early adipogenic factors. In addition, downregulation of KLF9 also did not influence the expression of some inhibitory factors such as DLK1, CHOP10, KLF2 or GATA211 (data not shown). In consideration of the time frame of its expression, we proposed three adipogenic transcription factors, C/EBPβ, C/EBPα and PPARγ2, as potential KLF9 targets during the process of adipogenesis. Bioinformatics analysis showed that each of the three promoters, including 3.0 kb of the C/EBPβ promoter, 1.5 kb of the C/EBPα promoter and 3.2 kb of the PPARγ2 promoter, had many potential binding sites for KLF9 (Figure 3a).
Figure 3.
Activation of PPARγ2 transcription by KLF9. (a) A schematic illustration of the murine C/EBPβ, C/EBPα and PPARγ2 promoter. Square stands for the possible KLF9 binding sites predicted using the TESS software. TSS stands for transcription start site. (b) The activation of the C/EBPβ promoter (−3 kb) by IBMX or KLF9. IBMX: HEK293 cells transfected with a luciferase reporter construct of C/EBPβ promoter (−3 kb) and treated with IBMX; KLF9: HEK293 cells co-transfected with a luciferase reporter construct of the C/EBPβ promoter (−3 Kb) and KLF9. Results are expressed as firefly luciferase activity normalized to renilla luciferase activity. Error bars indicate S.D. (n=3). Statistical significance is indicated: *P<0.05. (c) Activation of the C/EBPα promoter (−1.5 kb) by C/EBPβ or KLF9. C/EBPβ: HEK293 cells co-transfected with a luciferase reporter construct of the C/EBPα promoter (−1.5 kb) and C/EBPβ; KLF9: HEK293 cells co-transfected with a luciferase reporter construct of the C/EBPα promoter (−1.5 kb) and KLF9. Results are expressed as firefly luciferase activity normalized to renilla luciferase activity. Error bars indicate S.D. (n=3). Statistical significance is indicated: **P<0.01. (d) Activation of the PPARγ2 promoter (−3.2 Kb) by KLF9 or C/EBPβ. C/EBPβ: HEK293 cells co-transfected with a luciferase reporter construct of the PPARγ2 promoter (−3.2 Kb) and C/EBPβ; KLF9: HEK293 cells co-transfected with a luciferase reporter construct of the PPARγ2 promoter (−3.2 kb) and KLF9. Results are expressed as firefly luciferase activity normalized to renilla luciferase activity. Error bars indicate S.D. (n=6). Statistical significance is indicated: *P<0.05
To identify the KLF9-targeted promoter(s) from these three candidates, we co-transfected HEK293 cells with a plasmid expressing myc-tagged KLF9 protein and a luciferase reporter plasmid containing C/EBPα, or C/EBPβ or PPARγ2 promoters, respectively. The results showed that KLF9 could activate the PPARγ2 promoter (Figure 3d), whereas no such effect was detected on the promoters of C/EBPα or C/EBPβ (Figure 3b and c), suggesting that PPARγ2 was the downstream target of KLF9.
The small proximal promoter region of PPARγ2 is sufficient for KLF9 binding and transactivating activity
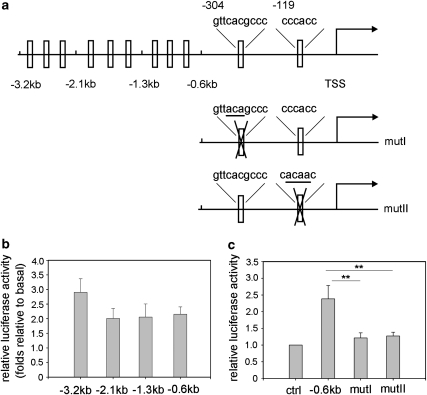
To identify the PPARγ2 promoter region responsible for KLF9 transactivation, we analyzed the activity of the PPARγ2 promoter in detail by generating a series of 5′ deletions (0.6, 1.3, 2.1 kb) within the 3.2-kb fragment (Figure 4a, upper panel). The Figure 4b showed that 0.6-kb promoter of PPARγ2 had similar activity as the 1.3-kb, or the 2.1-kb, and even the total 3.2-kb promoter in response to KLF9 activation in HEK293 cells, which was also confirmed in the NIH3T3 cell line (Supplementary Figure 5a). These results suggest that the 0.6-kb proximal promoter region of PPARγ2 is sufficient to drive the gene expression by KLF9.
Figure 4.
Two KLF9 binding sites in the PPARγ2 promoter (−0.6 kb) are responsible for the activation of the PPARγ2 promoter by KLF9. (a) A schematic representation of a deletion series derived from a luciferase reporter construct of the PPARγ2 promoter (3.2 kb) (the upper). A schematic representation of two mutant forms of the 0.6-kb upstream PPARγ2 promoter with one mutation on each site (the middle and bottom, designated as mutI and mutII). TSS stands for transcription start site. (b) PPARγ2 promoter activity. A deletion series derived from a luciferase reporter construct of the PPARγ2 promoter (3.2 Kb) was co-transfected with KLF9 into an HEK293 cell. Results were expressed as firefly luciferase activity normalized to renilla luciferase activity, then further normalized to the quantity of the basal state (promoter activation by empty vector), respectively. Error bars indicate S.D. (n=5). (c) Mutation of KLF9 binding sites in the PPARγ2 promoter inactivates the promoter. HEK293 cells were co-transfected with a luciferase reporter construct of the PPARγ2 promoter (−0.6 kb, mutI or mutII) and KLF9. Results are expressed as firefly luciferase activity normalized to renilla luciferase activity. Error bars indicate S.D. (n=5). Statistical significance is indicated: **P<0.01
Sequence analysis of the 0.6-Kb PPARγ2 promoter predicted two possible KLF9-binding sites at −304 and −119 bp, respectively (Figure 4a, upper panel). To verify if these two DNA fragments of PPARγ2 promoter are responsible for KLF9 activation, we separately mutated the two sites (Figure 4a, mutI and mutII) and then analyzed the activity of these two mutated PPARγ2 promoters by luciferase report assay. The results showed that either mutant significantly reduced the transcription activity induced by KLF9 in HEK293 cells (Figure 4c), which was also confirmed in the NIH3T3 cell line (Supplementary Figure 5b), suggesting that these two KLF9 binding sites are both functionally important.
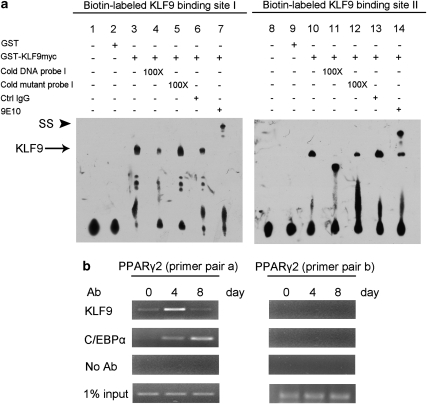
To test whether KLF9 could directly bind to the PPARγ2 promoter, electrophoretic mobility-shift assays (EMSAs) were performed to analyze these two binding sites of the PPARγ2 promoter (see Materials and Methods). Results showed that the complexes formed in vitro by GST-KLF9 with labeled binding sites I and II, respectively, could be greatly decreased or diminished by excess cold DNA probes (Figure 5a, compare lane 3 and 4, and lane 10–l1), whereas cold mutant probes did not have the same effect on these complexes (Figure 5a, compare lane 3–5, and lane 10–12). In addition, these complexes could be super-shifted by an anti-myc monoclonal antibody directed to the fused protein (Figure 5a, compare lane 3–7, and lane 10–l4), confirming the specificity of the DNA sequences binding to KLF9. Collectively, these results indicate there is specific interaction between KLF9 and the PPARγ2 promoter.
Figure 5.
Binding of KLF9 to the PPARγ2 promoter in the middle stage of adipocyte differentiation. (a) EMSA of the KLF9 binding site (−310 to −286 and −128 to −104, respectively) within the PPARγ2 promoter. Biotin-labeled DNA probes were incubated with 3′ myc-tagged KLF9 GST fusion protein. Cold competitors of the wild-type or mutated sequence were added to the reactions in lanes 4 and 5, and lanes 11 and 12. Addition of anti-myc antibody (9E10) to the reaction resulted in the formation of the supershift (SS) complex in lane 7 or lane14. (b) Binding of KLF9 to the PPARγ2 promoter in 3T3-L1 cells. 3T3-L1 cells at the indicated differentiation stages (day 0, 4 and 8) were subjected to ChIP with an anti-KLF9 antibody or an anti-C/EBPα antibody. KLF9 and C/EBPα binds to the region containing the KLF- and C/EBP-binding elements within the PPARγ2 promoter (−413 to −247 bp, left panel), but not the region of −2000 bp (right panel)
To assess whether KLF9 could bind specifically to the PPARγ2 promoter during adipogenesis, we analyzed the binding activity of KLF9 to the endogenous PPARγ2 promoter in 3T3-L1 cells by chromatin immunoprecipitation (ChIP) assays (see Materials and Methods). ChIP results showed that significant KLF9 binding to the −413 to −247 region of the PPARγ2 promoter was detected at day 4 after MDI induction (Figure 5b, left panel; Supplementary Table S1). As positive control, C/EBPα binding to the same promoter region was detectable at day 4 and peaked at day 8 (Figure 5b, left panel). As negative control, neither C/EBPα nor KLF9 could bind to the −2000-bp region of the PPARγ2 promoter (Figure 5b, right panel; Supplementary Table S1), which is in agreement with previous report that C/EBPα does not bind to the distal region of the PPARγ2 promoter,28 and our observation that KLF9 binds to the 0.6-kb proximal promoter region of PPARγ2 (Figure 4). Taken together, the results indicate that KLF9 could directly bind to the promoter region of PPARγ2 at the middle stage of 3T3-L1 adipocyte differentiation.
KLF9 acts in concert with C/EBPα to transactivate the PPARγ2 promoter
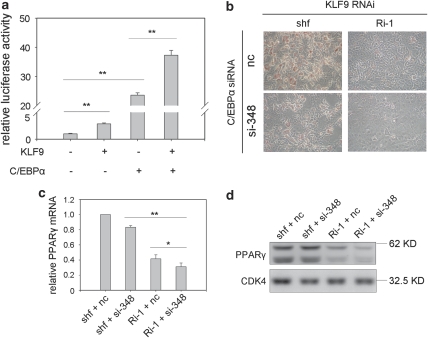
We realized that KLF9 itself stimulated modest PPARγ2 promoter activity in reporter assays (Figure 3d), and KLF9 knockdown partially inhibited the expression of PPARγ (Figure 2d). These results imply that KLF9 might interact with other transcription factor(s) to synergistically activate PPARγ2 like its family members, KLF518 and KLF15.21 The result detected by the luciferase report assay showed that when KLF9 was co-transfected with C/EBPα, the two factors acted in concert to more strongly transactivate the PPARγ2 promoter (Figure 6a).
Figure 6.
KLF9 acts in concert with C/EBPα to transactivate the PPARγ2 promoter. (a) The synergistic effect of KLF9 and C/EBPα on the transactivation of the PPARγ2 promoter. HEK293 cells were co-transfected with the pGL3-PPARγ2 promoter (0.6 kb), and KLF9 or C/EBPα, or both. Results are expressed as firefly luciferase activity normalized to renilla luciferase activity. Data shown are mean±S.D. of triplicates from a representative experiment. (b–d) KLF9-knockdown cells (Ri-1) or control shuffle cells (shf) were transfected with 100 nM siRNA targeting mouse C/EBPα (si-348) or control siRNA (nc). The cells were induced for differentiation. Oil-red-O staining was performed on day 4. A representative result from three independent experiments is shown (b). Total RNA was extracted at day 4 of differentiation and the mRNA level of PPARγ2 was determined by real-time PCR and normalized to PPIA mRNA level. Results are expressed as means±S.D. (n=3, *P<0.05, **P<0.01) (c). The protein level of PPARγ2 was detected by western blotting. CDK4 is blotted as loading control. Representative blots from three independent experiments are shown (d)
Furthermore, the result of Oil-red-O staining at day 4 of MDI-induced adipocyte differentiation showed that inhibition of C/EBPα expression with a chemically synthetic siRNA oligo in the KLF9-knockdown cells impaired adipogenesis more significantly than that of the KLF9 knockdown (Ri-1) alone or C/EBPα-siRNA (si-348) alone (Figure 6b and Supplementary Figure 6). In addition, the results of real-time PCR and western blot showed that PPARγ expression was further inhibited when expression of C/EBPα was repressed in the KLF9-knockdown cells (Figure 6c and d). Taken together, these results indicate that KLF9 should act in concert with C/EBPα to efficiently transactivate the PPARγ2 promoter.
KLF9 physically interacts with C/EBPα
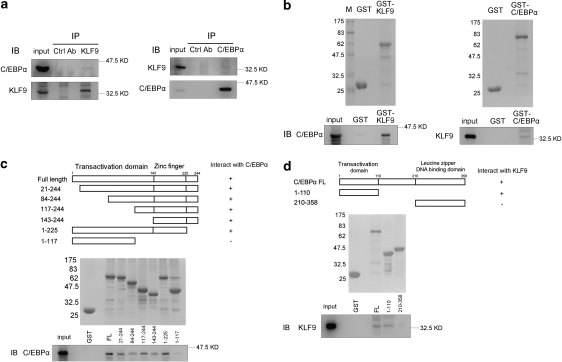
We further analyzed whether the synergistic activating effect of KLF9 and C/EBPα on the PPARγ2 promoter was due to physical interaction between these two proteins. We ectopically expressed KLF9 and C/EBPα in HEK293 cells and then detected the proteins by co-immunoprecipitation assays. The results showed there might be a physical interaction between KLF9 and C/EBPα (Figure 7a). To further confirm this observation, we purified the fusion proteins GST-KLF9 and GST-C/EBPα, and then incubated these two proteins with lysates of HEK293 cells overexpressing C/EBPα or KLF9, respectively (see Materials and Methods). The results detected by GST pull-down assay showed that the C/EBPα proteins in the cell lysate could be pulled down in the presence of the GST-KLF9 proteins (Figure 7b, left panel), whereas KLF9 in the cell lysate could be pulled down in the presence of GST-C/EBPα (Figure 7b, right panel). Taken together, these results indicate there is physical interaction between KLF9 and C/EBPα.
Figure 7.
The interaction of KLF9 with C/EBPα. (a) Immunoprecipitation of KLF9 and C/EBPα. Lysates from HEK293 cells overexpressing C/EBPα and KLF9 were immunoprecipitated with an anti-KLF9 antibody (left panel) or an anti-C/EBPα antibody (right panel), after which the immunoprecipitates were subjected to immunoblot analyses for the presence of C/EBPα (left panel) or KLF9 (right panel). (b) GST pull down of KLF9 and C/EBPα. GST-KLF9 or GST-C/EBPα proteins were incubated with lysates of HEK293 cells overexpressing C/EBPα (left panel) or KLF9 (right panel). The protein bound to the GST fusion protein was analyzed by western blotting (bottom panel). The top panel shows Coomassie blue staining of the GST fusion proteins. (c) Interaction of C/EBPα with the zinc-finger region in KLF9. A schematic illustration of KLF9 deletion mutants is shown in the top panel. Coomassie blue staining of GST fusion mutated KLF9 proteins is shown in the middle panel. C/EBPα expressed in HEK293 cells was pulled down by GST fusion mutated KLF9 proteins and detected by western blotting with an anti-C/EBPα antibody (bottom panel). (d) Interaction of KLF9 with the N-terminal domain of C/EBPα. A schematic illustration of C/EBPα deletion mutants is shown in the top panel. Coomassie blue staining of GST fusion mutated C/EBPα proteins is shown in the middle panel. KLF9 expressed in HEK293 cells was pulled down by GST fusion mutated C/EBPα proteins and detected by western blotting with an anti-KLF9 antibody (bottom panel)
To further identify the local region(s) of KLF9 responsible for its interaction with C/EBPα, we constructed six GST-fused truncated fragments of KLF9, which were subjected to GST pull-down analysis. The results showed that the C/EBPα-interaction domain of KLF9 lied in the region from amino acid 143 to 225 (Figure 7c). Conversely, to identify the C/EBPα domain interacting with KLF9, we constructed two GST-fused truncated fragments of C/EBPα and then applied the same GST pull-down assay. The results showed that the C/EBPα domain responsible for interaction with KLF9 localized between amino acid 1 and 110 of C/EBPα (Figure 7d).
Ectopic expression of PPARγ2 rescues the impairment of adipogenesis in KLF9-knockdown 3T3-L1 cells
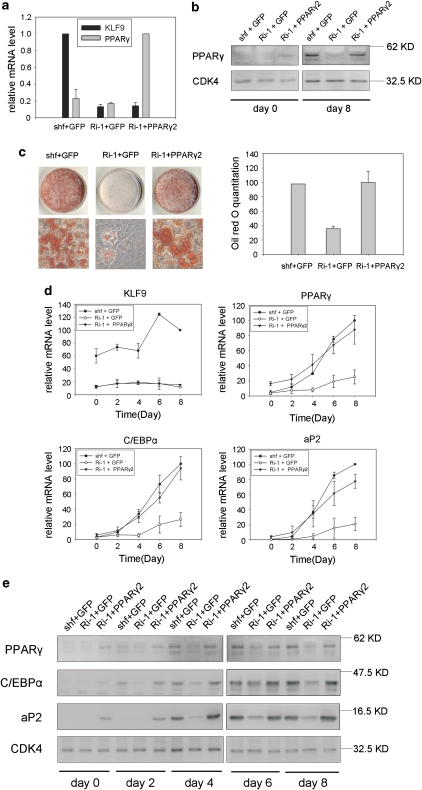
As these results indicate that KLF9 transactivates PPARγ2, we expected that overexpression of PPARγ2 could reverse the defect in adipogenesis due to KLF9 deficiency. To test this hypothesis, we infected the stable cell line of KLF9 knockdown with a retrovirus encoding PPARγ2. Real-time PCR assay confirmed both siRNA-mediated knockdown of endogenous KLF9 expression and ectopic expression of PPARγ2 in this cell line (Figure 8a), and western blot analysis showed overexpression of PPARγ2 before (day 0) and after (day 8) MDI induction (Figure 8b). These cells were subjected to adipocyte differentiation and detected by Oil-red-O staining assay. The results showed that ectopic PPARγ2 expression rescued the impairment of adipogenesis caused by KLF9 knockdown (Figure 8c). Furthermore, real-time PCR analysis showed that the mRNA levels of PPARγ, C/EBPα and aP2 in these KLF9-knockdown cells expressing ectopic PPARγ2 were recovered (Figure 8d). In agreement with the mRNA results, the western blot results indicated recovery of protein levels of these adipogenic markers (Figure 8e). Collectively, these results support the conclusion that KLF9 involves in adipogenesis regulation by transactivating PPARγ2.
Figure 8.
Rescue of inhibited adipocyte differentiation in KLF9-knockdown cells by PPARγ2 overexpression. (a) PPARγ mRNA level in the indicated 3T3-L1 cells. KLF9-knockdown (Ri-1) cells or shuffle cells (shf) were infected with retrovirus expressing GFP or PPARγ2. KLF9 and PPARγ mRNA were determined by real-time PCR. Error bars indicate S.D. (n=3). (b) PPARγ protein level in the indicated 3T3-L1 cells. Western blot was used to quantify the protein level of PPARγ2 of the indicated cell lines at day 0 and day 8 after induction. CDK4 is blotted as loading control. Representative blots from three independent experiments are shown. (c) Triglyceride accumulation in PPARγ expressed KLF9-knockdown cells. The cells as described in panel (a) were induced to differentiation. On day 8, the cells were stained with Oil-red-O (left panel) and quantified by measuring the OD510 nm (right panel). The results were normalized to that of shf+GFP cells and then expressed as means±S.D. (right) (n=3). (d) Real-time PCR analysis of the expression level of KLF9, PPARγ, C/EBPα and aP2 in shf+GFP, Ri-1+GFP and Ri-1+PPARγ2 cells at different time points after induction. The expression level was normalized to PPIA mRNA expression. Results are expressed as means±S.D. (n=3). (e) Western blot analysis of the protein expression level of adipocyte marker proteins in shf+GFP, Ri-1+GFP and Ri-1+PPARγ2 3T3-L1 cells at different time points after induction. CDK4 is blotted as loading control. Representative blots from two independent experiments are shown
Discussion
The dynamic changes of the sub-cellular localization and activities of KLF9 during the process of 3T3-L1 adipocyte differentiation
In this study, we showed that KLF9 expression at both the mRNA and protein level continuously increased during adipocyte differentiation (Figure 1b and c), whereas expression of PPARγ increased from day 2 to day 6 (Figure 1b), indicating that the expression pattern of KLF9 and PPARγ during the differentiation is not completely parallel. This phenomenon could be explained as follows: First, we showed that KLF9 acted in concert with C/EBPα to more strongly transactivate the PPARγ2 promoter (Figure 6a). In fact, the expression profile of C/EBPα is quite similar to that of PPARγ (Figure 1b). Secondly, although KLF9 protein level was much higher at day 8 than that at day 4 during the MDI-induced differentiation, KLF9 no longer bound to the PPARγ2 promoter, as shown by ChIP (Figure 5b), and a great part of KLF9 existed in the cytoplasm, as shown by immunofluorescence and sub-cellular fractionation analysis (Supplementary Figure 1).
We speculate that post-translational modifications might be involved in KLF9's sub-cellular localization and function. Post-translational modifications are widely used by transcription factors to regulate their sub-cellular localizations and activities.29 Several KLFs were reported to be phosphorylated and their transcriptional activities were changed by the modifications.30, 31, 32 We found that when several serines on the KLF9 protein were mutated to alanines, its transcriptional activities detected by luciferase report assay for the PPARγ2 promoter were greatly enhanced (unpublished data). However, whether post-translational modifications really happened to endogenous KLF9 in adipogenesis needs to be further investigated.
The relationship between KLF9 and other transcription factors for binding to the PPARγ2 promoter
Downregulation of KLF9 by retrovirus-mediated RNAi impaired adipocyte differentiation, indicating that KLF9 is required for this process. However, ectopic KLF9 overexpression in 3T3-L1 pre-adipocytes had no obvious promotions on PPARγ or C/EBPα expression at the early stages of differentiation (Supplementary Figure 2). Our results are in agreement with a previous report that KLF9 overexpression does not lead to lipid accumulation in NIH3T3 cells.21 The results from KLF9 overexpression and knockdown assays suggest that KLF9 is required but not sufficient to perform a full program of adipocyte differentiation.
The present works showed that KLF9 transactivated PPARγ2 at the middle stage of adipocyte differentiation by directly binding to the PPARγ2 promoter (Figure 5). However, overexpression of KLF9 in 3T3-L1 pre-adipocytes did not lead to upregulation of PPARγ at the early stage of differentiation (Supplementary Figure 2), which suggests that the time of KLF9 upregulation is important for its function. We speculate that the cellular environment such as the expression levels of other transcription factors might be important for KLF9's function. Because of the high similarity in their DNA-binding zinc fingers, many KLFs bind to very similar consensus sites, such as CACCC boxes.14 In fact, it is widely reported that different members of the KLF family may compete for the consensus sites of the promoter regions, perhaps due to similarity in their recognition sequences.33, 34, 35, 36, 37 For example, KLF5 could abrogate the activating effect of KLF4 on the Klf4 promoter, whereas KLF4 could abrogate the inhibiting effect of KLF5 on the same promoter, and it was speculated that this competing effect was due to physical competition of the two proteins for binding to a cognate DNA sequence.36 In another report, KLF9 competed with Sp1 for the BTE sequence in the cytochrome P4501A1 promoter.34
Importantly, a previous work identified that KLF2 bound to a GC box at the proximal region of the PPARγ2 promoter in pre-adipocytes and at the early stage of differentiation,15 which is just the KLF9-binding site-II identified in the present experiment (Figures 4 and 5). Thus, it is reasonable to speculate that the endogenous KLF2 binds to this specific region on the PPARγ2 promoter in pre-adipocytes, which made the PPARγ2 promoter unavailable for the ectopic KLF9 proteins. To test this possibility, we performed an in vitro competition transactivation assay for KLF9 and KLF2. As shown in Figure 3 and Supplementary Figure 7, KLF9 itself modestly upregulated transcription from the PPARγ2 promoter. This effect could be counteracted by expression of KLF2, and the inhibitory efficiency depends on the extent of KLF2 expression, which indicates that KLF2 and KLF9 compete for the same binding site on the PPARγ2 promoter. We noticed that a small quantity of KLF2 was enough to abolish KLF9's transactivating activity to basal level, and KLF2's repression effects on PPARγ2 promoter could hardly be reversed by increasing amounts of KLF9 (Supplementary Figure 7), which indicates that the binding affinity of KLF2 to the PPARγ2 promoter is higher than that of KLF9. So, it is reasonable to speculate that the exogenous KLF9 in pre-adipocytes may not be able to replace the endogenous KLF2, which already exists on the PPARγ2 promoter. In addition, other inhibitory factors existing on the PPARγ2 promoter before differentiation could also impede KLF9 to bind to the promoter.38 For example, GATA2 and GATA3 are two transcription factors, which are abundant in pre-adipocytes and their expressions decline after differentiation.38 Both proteins were reported to bind to the PPARγ2 promoter and their binding site was just 13 bp upstream from the KLF9's binding site-II, which may also impede KLF9's binding.38
The roles and relationships among the members of the KLF family during adipogenesis
Our present results and previous works have shown that four members of the KLF family have roles in adipocyte differentiation by regulating PPARγ2 transcription.15, 18, 21 These KLFs are expressed sequentially during adipogenesis. KLF2 is first expressed in 3T3-L1 pre-adipocytes and its expression is shut down after differentiation.15 Then, KLF5 is induced 4 h after MDI induction and its expression peaks at the early stage of adipocyte differentiation.18 The expression of KLF9 appears at the middle stage of differentiation and gradually increases throughout the middle and late stages (Figure 1). In addition, KLF15's expression pattern mimics KLF9 and is highly expressed in mature adipocytes.21
These KLFs carry out their respective functions by binding to the PPARγ2 promoter either to open up or to shut down its transcription. While the time frame for each KLF binding is different, the DNA elements they bind to are the same.15, 18 These KLFs bind to one or both of the DNA elements within the 0.6-kb PPARγ2 upstream promoter region,15, 18 except for KLF15, whose binding target on PPARγ2 promoter has not been identified.21 We also found that KLF9 and C/EBPα have physical interactions and they can synergistically activate the PPARγ2 promoter (Figure 6 and 7). This observation is coincident with previous reports that KLF5 and C/EBPβ/δ interact directly and they colocalize on the PPARγ2 promoter on day 2.5 of differentiation,18 and KLF15 cooperates with C/EBPα to synergistically activate the PPARγ2 promoter at the late stages.21 Synergistic activation of the PPARγ2 promoter by these factors and their physical interactions suggest that these molecules may form an enhanceosome,18, 39, 40 and different enhanceosomes are formed at different stages.
Obesity has been closely related to a series of human health problems such as diabetes. Adipocyte differentiation is one of the direct causes of adipose formation in vivo.41 Our finding adds a new member of the KLF family related to adipocyte differentiation and places KLF9 into a group of key pro-adipogenic transcription factors. The dissection of the mechanisms controlling KLF9 expression could lead to further identification of the mechanism that control adipogenesis, which would provide new clues against the growing incidence of obesity in the modern world.
Materials and Methods
Materials
Anti-KLF9 (C-17), anti-C/EBPα (14AA), anti-PPARγ2 (E-8), anti-CDK4 (H-22), anti-C/EBPβ (C-19) antibody and secondary antibody (horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG and donkey anti-goat IgG) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-adiponectin (19F1) was from Abcam (Cambridge, MA, USA). Anti-aP2 (AF1443) was from R&D Systems (Minneapolis, MN, USA). The Immobilon Western Chemiluminescence HRP Substrate was from Millipore Corporation (Billerica, MA, USA). Insulin, dexamethasone, 1-methyl-3-isobutylxanthine were from Sigma (St. Louis, MO, USA).
Primary pre-adipocyte isolation, culture and differentiation
Rat (normal male Sprague–Dawley rat) white adipose tissues from the epididymal, inguinal, omental and scapular fat pads were isolated and washed with PBS. The adipose tissues were then minced and digested with collagenase solution (1 mg/ml collagenase type-I, 2% BSA in PBS) for 1 h. The digested tissues were centrifuged at 200 g for 10 min. The floating fractions consisting of adipocytes were discarded and the pellets were then resuspended in erythrocyte lysis buffer (154 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 10 min to remove red blood cells. The suspension was filtered through a size 200 filter and the cells were pelleted by 200 g, 10 min centrifugation. The cells were resuspended in Dulbecco's modified Eagle's medium (DMEM)/F12 and the medium was changed 3 h after inoculation. Two days after inoculation, cells were induced to differentiate with 1 μg/ml insulin, 1 μM dexamethasone and 0.5 mM 1-methyl-3-isobutylxanthine. The medium was replaced with a medium containing 1 μg/ml insulin after 72 h, and then changed every other day.
3T3-L1 cell culture, differentiation induction and Oil-red-O staining
3T3-L1 pre-adipocytes were cultured in DMEM supplemented with 10% calf serum (Invitrogen, Carlsbad, CA, USA). The 2-day post-confluent cells were induced for adipocyte differentiation following the protocol described previously.42 Cytoplasmic triglyceride droplets were visible by day 4. The differentiated 3T3-L1 adipocyte monolayer was stained with Oil-red-O.42 The Oil-red-O in triglyceride droplets was extracted with 100% isopropanol and OD510 nm was determined.
KLF9 knockdown and PPARγ2 expression
For stable KLF9-knockdown cells, a retrovirus-based siRNA expression system was used. The targeted sequences of KLF9 were Ri-1, 5′-GTGGGAAAGTCTATGGAAA-3′ Ri-2, 5′-GGAGAGTCCCGATGAGGAT-3′ Ri-3, 5′-AGCCCATTACAGAGTGCAT-3′ Ri-4, 5′-GATTATTGCACGCTGGTCA-3′ and Ri-5, 5′-GAGAAGAGATTCATGAGGA-3′, and cloned into a pSIREN-Retro-Q Vector and shuttled into Knockout RNAi Systems (Clontech Laboratories Inc., Otsu, Shiga, Japan). A sequence of Shuffle, 5′-AACTAGAGCCACAACTACC-3′, was used as control. The retrovirus was produced in HEK293 cell following the manufacturer's protocol and infected the proliferating 3T3-L1 cells. The stable cell lines were selected in medium supplemented with 5 μg/ml puromycin.
For PPARγ2 expression in KLF9-knockdown cells, mouse PPARγ2 cDNA was cloned into a Clontech pMSCV/Hygro retrovirus expression system. The stable KLF9-knockdown cells were infected with a PPARγ2 expression retrovirus and cultured in medium supplemented with 300 μg/ml hygromycin.
Real-time quantitative PCR
Total RNA of 3T3-L1 cells was isolated using the TRIzol Reagent (Invitrogen) and reverse transcribed according to the manufacturer's protocol (Takara, Dalian, China). The SYBR Green quantitative PCR analysis reactions (TOYOBO, Osaka, Japan) were then performed using the ABI Prism 7500 sequence detection system (Life Technologies Corporation, Carlsbad, CA, USA).
Luciferase assay for promoter activities
HEK293 cells or NIH3T3 cells were plated in a 24-well plate and transfected with the promoter-luciferase reporter plasmids using lipofectamine 2000 (Invitrogen). The cells were harvested 48 h after transfection and luciferase activity was measured using the Promega (Madison, WI, USA) Dual-Luciferase Reporter Assay System. Promoter activity was expressed as arbitrary luciferase units (firefly/renilla).
Immunoprecipitation and western blotting
For immunoprecipitation, HEK293 cells transfected with plasmids expressing the targeted proteins for 48 h, and then were lysed in buffer containing 1% Triton X-100, 50 mM Tris-Cl (pH 7.4), 120 mM NaCl and protease inhibitors (cocktail; Sigma). The cell debris was removed by centrifugation at 14 000 g for 15 min at 4°C. The supernatant was collected and immunoprecipitated with appropriate primary antibodies.43 Control goat or rabbit IgG was used as the negative control (Santa Cruz Biotechnology). The immunoprecipitated samples were subjected to western blot analysis.43
GST fusion protein expression, purification and GST pull down
GST and GST fusion proteins were expressed in Escherichia coli BL21 (DE3) cells (Stratagene, Agilent Technologies, Inc., Santa Clara, CA, USA) and purified using a glutathione Sepharose 4B column (Amersham Biosciences Biotech Inc., Piscataway, NJ, USA). Purified GST or GST-fusion proteins were incubated with glutathione–Sepharose 4B beads in PBS at 4°C for 2 h. After that, the beads were washed three times with PBS and then incubated with the HEK293 cell lysate at 4°C overnight. Then the beads were collected by centrifugation and washed four times with the lysis buffer. The samples were then subjected to SDS-PAGE followed by western blot analysis with the indicated antibody.
Electrophoretic mobility-shift assay
The myc-tagged KLF9 GST fusion protein was incubated with two oligonucleotides corresponding respectively to two potential binding sites of KLF9 in the 0.6-kb promoter of the PPARγ2 gene (site-I from −310 to −286: 5′-TGTACAGTTCACGCCCCTCACAGAA-3′ and site-II from −128 to −104: 5′-GTGTTTATTCCCACCTCTCCCAAAT-3′). Two mutated oligonucleotides were also used (mutI: 5′-TGTACAGTTACAGCCCCTCACAGAA-3′ and mutII: 5′-GTGTTTATTCACAACTCTCCCAAAT-3′) (potential KLF9-binding core sequences are indicated in bold and mutated nucleotides are underlined). Biotin labeling of probes was performed using the biotin 3′-end DNA-labeling kit (Thermo Fisher Scientific Inc., Rockford, IL, USA) according to the manufacturer's instructions. Single-stranded oligos were labeled individually and annealed for 1 h at 25°C to produce double-stranded probes. A 2-μg weight of the myc-tagged KLF9 GST fusion protein was incubated at 25°C with the binding buffer (Thermo) for 20 min. Then biotin 3′-end-labeled DNA probes were added. For specific or non-specific competition or antibody supershift, the respective unlabeled DNA probes (100-fold excess) or antibody (1 μg control mouse IgG (Santa Cruz) or anti-myc 9E10 (Sigma)) were added to the reaction mixture before addition of labeled probes. Free probe and protein–DNA complexes were separated on 6% neutral polyacrylamide gel and transferred to a nylon membrane for detection using the Lightshift electrophoretic mobility shift assay kit (Thermo).
ChIP assay
ChIP assays were performed using a commercial kit according to the manufacturer's instructions (Upstate Biotechnologies Inc., Temecula, CA, USA). Chromatin samples were prepared from 3T3-L1 cells at the indicated time points after induction of adipocyte differentiation, and then immunoprecipitated with antibodies. Briefly, 2 × 106 3T3-L1 cells at the indicated day of differentiation were cross-linked with 1% formaldehyde for 10 min at 37°C, followed by cell lysis and sonication. Then proteins cross-linked with DNA were immunoprecipitated with 1 μg of anti-KLF9 antibody (C-17X; Santa Cruz) or anti-C/EBPα antibodies (14AA; Santa Cruz) and 40 μl of salmon sperm DNA/protein-A–agarose beads. The protein-A–agarose antibody–protein complexes were washed extensively and eluted, according to the manufacturer's recommendations. The cross-link was reversed and proteins were digested by proteinase-K for 1 h at 45°C. DNA was recovered by phenol/chloroform extraction and ethanol precipitation, and used as template for PCR.
Acknowledgments
We gratefully acknowledge Wei Wang and Yubo Ding for the pMSCV-hygromycin and pSIREN-RetroQ retroviral vectors, and the C/EBPα, C/EBPβ promoter vectors and C/EBPβ expression vector, respectively. This work was supported in part by the ‘973 Program' (No. 2006CB503900 to JW; 2006CB910703 to KL and JW; 2010CB912102 to KL); a grant from the National Natural Science Foundation of China (No. 30821065 to KL and JW; 30870559 to KL) grants from the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX1-YW-02, KJCX2-YW-M15 to JW) and a grant from Science and Technology Commission of Shanghai Municipality 07dz05907 to KL and JW.
Glossary
- aP2
fatty-acid binding protein 2
- CDK4
cyclin-dependent kinase 4
- C/EBP
CCAAT/enhancer-binding protein
- DEX
dexamethasone
- DMEM
Dulbecco's modified Eagle's medium
- IBMX
isobutylmethylxanthine
- KLF
Krüppel-like factor
- MDI
isobutylmethylxanthine dexamethasone and insulin
- PBS
phosphate-buffered saline
- PPARγ
peroxisome proliferator-activated receptor gamma
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- Couillard C, Mauriege P, Imbeault P, Prud'homme D, Nadeau A, Tremblay A, et al. Hyperleptinemia is more closely associated with adipose cell hypertrophy than with adipose tissue hyperplasia. Int J Obes Relat Metab Disord. 2000;24:782–788. doi: 10.1038/sj.ijo.0801227. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Freytag SO, Paielli DL, Gilbert JD. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Rosen ED.The molecular control of adipogenesis, with special reference to lymphatic pathology Ann N Y Acad Sci 2002979143–158.discussion 188–196. [DOI] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, et al. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;278:2581–2584. doi: 10.1074/jbc.M210859200. [DOI] [PubMed] [Google Scholar]
- Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, et al. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol Cell Biol. 2008;28:3967–3978. doi: 10.1128/MCB.01942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Li D, Yea S, Li S, Chen Z, Narla G, Banck M, et al. Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941–26952. doi: 10.1074/jbc.M500463200. [DOI] [PubMed] [Google Scholar]
- Kanazawa A, Kawamura Y, Sekine A, Iida A, Tsunoda T, Kashiwagi A, et al. Single nucleotide polymorphisms in the gene encoding Kruppel-like factor 7 are associated with type 2 diabetes. Diabetologia. 2005;48:1315–1322. doi: 10.1007/s00125-005-1797-0. [DOI] [PubMed] [Google Scholar]
- Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, et al. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen RC, Eason RR, McQuown JR, Linz AL, Kang TJ, Chatman L, Jr, et al. Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Kruppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J Biol Chem. 2004;279:29286–29294. doi: 10.1074/jbc.M403139200. [DOI] [PubMed] [Google Scholar]
- Simmen FA, Xiao R, Velarde MC, Nicholson RD, Bowman MT, Fujii-Kuriyama Y, et al. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1757–G1769. doi: 10.1152/ajpgi.00013.2007. [DOI] [PubMed] [Google Scholar]
- Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J Biol Chem. 1999;274:23128–23134. doi: 10.1074/jbc.274.33.23128. [DOI] [PubMed] [Google Scholar]
- Velarde MC, Zeng Z, McQuown JR, Simmen FA, Simmen RC. Kruppel-like factor 9 is a negative regulator of ligand-dependent estrogen receptor alpha signaling in Ishikawa endometrial adenocarcinoma cells. Mol Endocrinol. 2007;21:2988–3001. doi: 10.1210/me.2007-0242. [DOI] [PubMed] [Google Scholar]
- Xue JC, Schwarz EJ, Chawla A, Lazar MA. Distinct stages in adipogenesis revealed by retinoid inhibition of differentiation after induction of PPARgamma. Mol Cell Biol. 1996;16:1567–1575. doi: 10.1128/mcb.16.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SL, Robinson CE, Gimble JM. CAAT/enhancer binding proteins directly modulate transcription from the peroxisome proliferator-activated receptor gamma 2 promoter. Biochem Biophys Res Commun. 1997;240:99–103. doi: 10.1006/bbrc.1997.7627. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Veitia RA. A post-translational modification code for transcription factors: sorting through a sea of signals. Trends Cell Biol. 2009;19:189–197. doi: 10.1016/j.tcb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Slavin DA, Koritschoner NP, Prieto CC, Lopez-Diaz FJ, Chatton B, Bocco JL. A new role for the Kruppel-like transcription factor KLF6 as an inhibitor of c-Jun proto-oncoprotein function. Oncogene. 2004;23:8196–8205. doi: 10.1038/sj.onc.1208020. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Teng CT. Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function. Nucleic Acids Res. 2003;31:2196–2208. doi: 10.1093/nar/gkg310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenrieder V, Zhang JS, Kaczynski J, Urrutia R. Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J. 2002;21:2451–2460. doi: 10.1093/emboj/21.10.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa K, Kikuchi Y, Imataka H, Fujii-Kuriyama Y. Comparison of DNA-binding properties between BTEB and Sp1. J Biochem. 1993;114:605–609. doi: 10.1093/oxfordjournals.jbchem.a124224. [DOI] [PubMed] [Google Scholar]
- Kaczynski JA, Conley AA, Fernandez Zapico M, Delgado SM, Zhang JS, Urrutia R. Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem J. 2002;366:873–882. doi: 10.1042/BJ20020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Shields JM, Sogawa K, Fujii-Kuriyama Y, Yang VW. The gut-enriched Kruppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J Biol Chem. 1998;273:17917–17925. doi: 10.1074/jbc.273.28.17917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte–adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhai B, Qiu Z, Wu J, Lane MD, Liao K. c-Crk, a substrate of the insulin-like growth factor-1 receptor tyrosine kinase, functions as an early signal mediator in the adipocyte differentiation process. J Biol Chem. 2000;275:34344–34352. doi: 10.1074/jbc.M004927200. [DOI] [PubMed] [Google Scholar]
- Hong S, Huo H, Xu J, Liao K. Insulin-like growth factor-1 receptor signaling in 3T3-L1 adipocyte differentiation requires lipid rafts but not caveolae. Cell Death Differ. 2004;11:714–723. doi: 10.1038/sj.cdd.4401405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.