Abstract
Ret finger protein-like 1 (RFPL1) is a primate-specific target gene of Pax6, a key transcription factor for pancreas, eye and neocortex development. However, its cellular activity remains elusive. In this article, we report that Pax6-elicited expression of the human (h)RFPL1 gene in HeLa cells can be enhanced by in vivo p53 binding to its promoter and therefore investigated the hypothesis that hRFPL1 regulates cell-cycle progression. Upon expression in these cells, hRFPL1 decreased cell number through a kinase-dependent mechanism as PKC activates and Cdc2 inhibits hRFPL1 activity. hRFPL1 antiproliferative activity led to an increased cell population in G2/M phase and specific cyclin B1 and Cdc2 downregulations, which were precluded by a proteasome inhibitor. Specifically, cytoplasm-localized hRFPL1 prevented cyclin B1 and Cdc2 accumulation during interphase. Consequently, cells showed a delayed entry into mitosis and cell-cycle lengthening resulting from a threefold increase in G2 phase duration. Given previous reports that RFPL1 is expressed during cell differentiation, its impact on cell-cycle lengthening therefore provides novel insights into primate-specific development.
Keywords: Ret finger protein-like, cell-cycle lengthening, E3 ubiquitin ligase
The human Ret finger protein-like (hRFPL)1,2,3 genes (OMIM 605968, 605969, 605970) are recently identified targets of Pax6, which is notably a key transcription factor for pancreas, eye and neocortex development.1, 2, 3 In this line, high hRFPL1,2,3 expression is found at the onset of neurogenesis in differentiating embryonic stem cells and in the developing neocortex.4 In addition, the study of RFPL1,2,3 evolutionary history revealed that they are only found in Old World monkeys and great apes and show features believed to be important for human brain evolution.4
Yet, the cellular activity of RFPL1,2,3 is still unknown. A murine RFPL (mRFPL) protein, encoded by an ancestral gene not belonging to the RFPL1,2,3 gene subfamily,4 has also been cloned.5 Previously reported as being expressed only in testis, ovaries and oocytes,5, 6 mRFPL has been shown to interact with the destruction box motif of cyclin B1 – the Cdc2 activating partner for driving germ cells through metaphases I and II7– and with proteins of the proteasome, speculating on mRFPL ability to elicit cyclin B1 degradation to control meiosis progression.6 However, the fact that cyclin B1 and Cdc2 also form a key complex for controlling cell entry into mitosis8 and our recent observations that the RFPL genes are also expressed in tissues in which cells divide mitotically4 suggest that the RFPL proteins could regulate other aspects of cell division.
We therefore examined RFPL-mediated control of mitotic cell-cycle progression by focusing on hRFPL1. Because no endogenously hRFPL1-expressing cell type suitable for this kind of study has been reported to date, we examined the influence of hRFPL1 gain of function on HeLa cells, a reference cell system for examining cell-cycle regulation. We report that hRFPL1 is an antiproliferative gene that controls G2–M phase transition, thereby lengthens G2 phase by reducing cyclin B1 and Cdc2 accumulation. Accordingly, in Pax6-expressing cells to elicit endogenous hRFPL1 expression, we observed decreased cyclin B1 and Cdc2 levels that were prevented by hRFPL1 RNA interference, confirming that the control of cyclin B1 and Cdc2 levels is a physiologically relevant function of the endogenous hRFPL1 protein.
Results
hRFPL1 expression level can be enhanced by p53
We previously reported that Pax6 binds to the hRFPL1 promoter and elicits its transcription,4 but also induces p53 activation and nuclear translocation.9 p53 is an important transcription factor for the control of cell-cycle progression and apoptosis. Given the possible role of hRFPL1 on cell-cycle and in silico prediction of putative p53 binding sites on its promoter, we examined the influence of p53 on hRFPL1 transcriptional regulation. Upon Pax6-elicited hRFPL1 expression and p53 activation in HeLa cells,4, 9 we observed using chromatin immunoprecipitation (ChIP) that p53 interacted in vivo with hRFPL1 promoter (Figure 1a). After blocking p53 activity using either cyclic-Pifithrin-α, an inhibitor of p53 nuclear translocation10 or a p53 shRNA,11, 12 we observed that Pax6-induced hRFPL1 expression was significantly reduced (Figure 1b). We also investigated the influence of p53 RNA interference on previously identified Pax6-regulated genes.13 Among the tested genes, only FOXM1 and MCM5 showed significant expression changes on Pax6 gain of function in HeLa cells. However, p53 RNA interference did not alter their Pax6-mediated regulation (Supplementary Figure 1A), suggesting that the regulatory cross talk between Pax6 and p53 signaling pathways is restrained to specific genes.
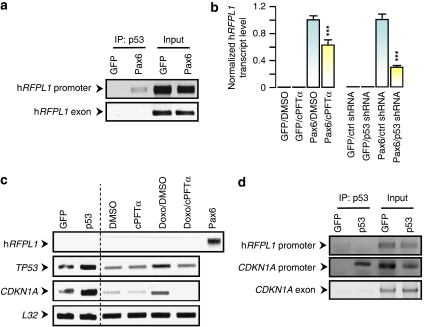
Figure 1.
p53 functions as an enhancer of Pax6-elicited hRFPL1 expression. (a) In vivo binding of p53 to hRFPL1 promoter was assessed by chromatin immunoprecipitation assay following induction of hRFPL1 expression by Pax6. After p53 immunoprecipitation, end-point PCRs were performed with specific primers for hRFPL1 promoter or exon 2 to control nonspecific immunoprecipitation. Input: dilution of input chromatin used as PCR controls. (b) Inhibition of p53 using Cyclic Pifithrin-α (cPFTα) or p53 shRNA decreases the Pax6-elicited hRFPL1 expression in HeLa cells as measured by quantitative PCR. ***P<0.001 compared with Pax6 control groups using two-way ANOVA followed by Tukey's post hoc test. (c) Assessment of hRFPL1 and CDKN1A, a p53 target encoding p21WAF1/CIP1, transcript levels by semiquantitative end-point PCR following p53 overexpression using a CMV-TP53 sequence containing pcDNA vector or doxorubicin. Use of the p53 inhibitor, Cyclic Pifithrin-α (cPFTα), serves to control that these observations result from the impact of doxorubicin on p53 regulation. (d) Assessment of p53 in vivo binding to hRFPL1 and CDKN1A promoters independently of Pax6 signaling was assessed by chromatin immunoprecipitation assay following p53 overexpression. End-point PCRs were performed similarly to (a)
We next examined whether p53 could elicit hRFPL1 expression independently of Pax6 signaling. However, p53 overexpression or the use of the p53 inducer, doxorubicin, did not elicit hRFPL1 expression (Figure 1c). By contrast, these two approaches increased TP53 expression and that of one of its downstream effectors, the CDKN1A target gene encoding p21WAF1/CIP1 (Figure 1c). We therefore assessed p53 ability to bind to hRFPL1 promoter independently of Pax6 signaling using ChIP. p53 gain of function did not lead to its binding to hRFPL1 promoter, whereas it bound to that of CDKN1A (Figure 1d). Given the reduced stability of p53 in HeLa cells due to human papilloma virus E6 protein,14 we investigated whether ectopic p53 expression could elicit hRFPL1 expression in other human cell lines presenting different p53 status. However, p53 overexpression did not induce hRFPL1 expression in HEK 293, p53-deficient PC3 or SH-SY5Y cells whereas CDKN1A expression was augmented in all these cells (Supplementary Figure 1B). Interestingly, Pax6 could not induce hRFPL1 expression in any of these other cell lines either (Supplementary Figure 1C).
Altogether, these data indicate that Pax6-mediated regulation of hRFPL1 expression is cell dependent. Further, in HeLa cells, p53 activation downstream of Pax6 signaling allows its binding to the hRFPL1 promoter to enhance its transcriptional expression.
hRFPL1 decreases cell number through its RDM and SPRY domains
To address the potential hRFPL1 role on cell cycle, we next examined the impact of hRFPL1 gain of function on cell number in HeLa cells. hRFPL1 significantly decreased cell number from day 2 after cell transduction (Figure 2a). Such an effect was also observed following hRFPL1 gain of function in other human cell lines (Supplementary Figure 1D), indicating that its effect was not restricted to specific cell types.
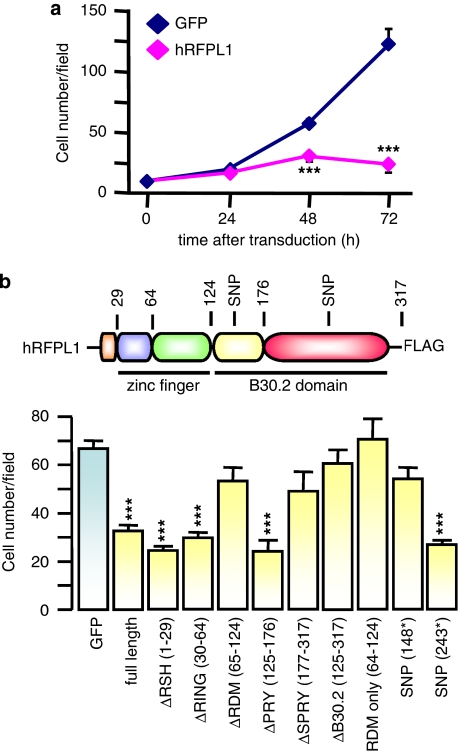
Figure 2.
hRFPL1 alters cell number increase over time. (a) Time course of HeLa cell number transduced with eGFP or hRFPL1. ***P<0.001 compared with eGFP-expressing cells using two-way ANOVA followed by Tukey's post hoc test. (b) Identification of protein domains involved in hRFPL1 effect on cell number 3 days after transduction. Color legend: RFPL-specifying helix (RSH), orange; RING finger, blue; RFPL-defining motif (RDM), green; PRY, yellow; SPRY, red. The RING finger associated to the RDM form a predicted larger zinc finger of the RING/U-Box superfamily, and the presence of both PRY and SPRY form a B30.2 domain. ***P<0.001 compared with eGFP-expressing cells using one-way ANOVA followed by Tukey's post hoc test
Then, we performed a structure–function study by truncating the different hRFPL1 protein domains to determine those involved in this effect. In addition to its zf-C3HC4 RING-type zinc-finger domain and C-terminal PRY/SPRY-forming B30.2 domain that are present in other proteins, hRFPL1 is also formed of an RFPL1,2,3-specifying helix (RSH), which is an N-terminal α-helix loop restricted to the RFPL1,2,3 proteins, and of an RFPL-defining motif (RDM), between the RING and the B30.2 domains, that is found exclusively in the RFPL proteins and predicted to form together with the RING domain a more complex zinc finger of the RING/U-Box superfamily. We controlled by immunofluorescence that the deletion of either domain was not altering the localization of truncated, FLAG-tagged hRFPL1 forms (Supplementary Figure 1E). Neither RSH, nor RING, nor PRY were involved in hRFPL1 effect on cell number (Figure 2b). By contrast, we observed the implication of RDM and SPRY N-terminal part (Figure 2b).
Interestingly, hRFPL1 presents the rare feature of having two nonsense single nucleotide polymorphisms (SNPs) leading to premature stop codons (R148* SNP: rs16987628; Q243* SNP: rs12484086). Although the Q243* SNP did not alter hRFPL1 effect on cell number, the protein encoded by the R148* SNP sequence lost this effect (Figure 2b).
hRFPL1 reduces the number of mitotic events
To understand how hRFPL1 limits the increase in cell number over time, we determined the behavior of hRFPL1-expressing HeLa cells by performing quantitative live-cell imaging studies. When considering only the cycling cells between 24 and 72 h after transduction, we observed that hRFPL1-expressing HeLa cells showed a decreased cell division rate. On the other hand, some cells showed defect in performing mitosis (either they did not perform any during the recorded period or this event was not completed) and some cells showed typical morphological changes of entry into apoptotic cell death (cell shrinkage, blebbing; Table 1). Apoptosis was confirmed by quantitative immunocytochemistry (0.36±0.36% of GFP-positive cells were immunoreactive for cleaved caspase-3 versus 32.8±4.0% of hRFPL1-transduced cells, P<0.001), similar to the time-lapse video-imaging data. Thus, hRFPL1 decreases cell proliferation that may lead to apoptotic cell death.
Table 1. hRFPL1 reduces the number of mitotic events.
| Events and cell-cycle parameters | GFP-expressing cells (%) | hRFPL1-expressing cells (%) |
|---|---|---|
| 3 mitotic divisions | 23.3±3.5 | 1.7±0.3 |
| 2 mitotic divisions | 65.5±6.7 | 18.0±2.0 |
| 1 mitotic division | 9.5±2.5 | 24.6±4.0 |
| Incomplete mitosis | 0 | 13.1±1.2 |
| No mitotic division | 0 | 13.1±2.7 |
| Apoptosis | 1.7±0.5 | 29.5±3.5 |
The different parameters were quantified during 48 h using live-cell imaging experiments in GFP- or hRFPL1-expressing HeLa cells. Apoptosis was defined by typical morphological changes (cell shrinkage, blebbing).
PKC and Cdc2 regulate hRFPL1 activity
To assess the specificity of hRFPL1 effect on cell proliferation and apoptosis, we assessed the influence of kinase-dependent post-translational mechanisms on hRFPL1 activity following in silico identification of putative PKC (S200) and Cdc2 (S287) phosphorylation sites.
In accordance with these predictions, we observed that FLAG-tagged hRFPL1 was present in the purified fraction of phospho-proteins from HeLa cells (Figure 3a), and that the levels of phospho-hRFPL1 were reduced when the cells were treated with Gö6983, a PKCα/β/γ inhibitor, or the Cdc2 inhibitor (Figure 3a; Supplementary Figure 1F). Interestingly, Gö6983 also prevented Pax6-elicited hRFPL1 expression but not the Cdc2 inhibitor (Figure 3b), suggesting that the influence of the PKC pathway was not limited to controlling hRFPL1 phosphorylation status, but was also involved in Pax6 signaling mediating hRFPL1 transcription.
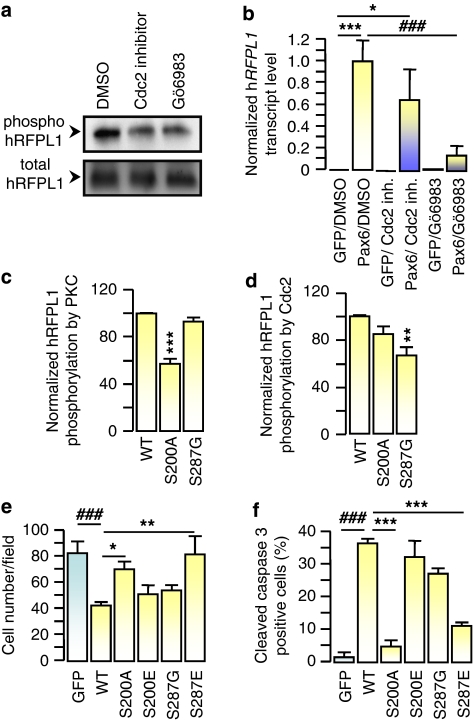
Figure 3.
hRFPL1 activity is regulated by classical PKCs and Cdc2. (a) Phosphorylation status of FLAG-tagged hRFPL1 was determined by purification of phosphoproteins versus extraction of whole cell lysates in HeLa cells treated with DMSO (control), Cdc2 inhibitor or the PKCα/β/γ inhibitor, Gö6983, and immunoblotting using a FLAG antibody. (b) Influence of PKC and Cdc2 kinases on Pax6-elicited hRFPL1 expression was examined by quantitative PCR. *P<0.05, ***P<0.001 versus eGFP/DMSO- or ###P<0.001 versus Pax6/DMSO-treated cells using one-way ANOVA followed by Tukey's post hoc test. (c, d) Phosphorylation of hRFPL1 Ser200 and Ser287 residues was determined by in vitro PKC and cyclin B1/Cdc2 kinase assays. **P<0.01, ***P<0.001 versus [32P]ATP incorporation in wild-type (WT) hRFPL1 using one-way ANOVA followed by a Tukey's post hoc test. (e) Comparison of the impact of wild-type (WT) or mutant hRFPL1 on HeLa cell number 3 days after transduction. We controlled by immunoblotting using a FLAG antibody that all hRFPL1 proteins were expressed at the same level (data not shown). ###P<0.001 versus eGFP- or *P<0.05, **P<0.01 versus WT hRFPL1-expressing cells using one-way ANOVA followed by Tukey's post hoc test. (f) Comparison of the impact of wild-type (WT) or mutant hRFPL1 on apoptosis induction in HeLa cells determined by quantitative cleaved caspase-3 immunocytochemistry 3 days after transduction. ###P<0.001 versus eGFP- or ***P<0.001 versus hRFPL1-expressing cells using one-way ANOVA followed by Tukey's post hoc test
Concerning PKC implication on hRFPL1 post-translational regulation, we observed that radiolabeled ATP incorporation in hRFPL1 using an in vitro PKCα/β/γ kinase assay was significantly decreased in the S200A mutant in comparison to wild-type hRFPL1 (Figure 3c). Further, the loss of S200 residue (S200A mutant) reversed hRFPL1-mediated reduction in cell number (Figure 3e) and caspase 3 activation (Figure 3f). At the opposite, mimicking permanent phosphorylation on this residue (S200E mutant) led to effects similar to wild-type hRFPL1 (Figure 3e and f). As for Cdc2 involvement, we observed that Cdc2 phosphorylated hRFPL1 S287 residue in an in vitro cyclin B1/Cdc2 kinase assay (Figure 3d). The loss of this phosphorylation site (S287G mutant) did not alter hRFPL1-mediated decrease in cell number and caspase 3 activation, whereas the expression of the S287E mutant reversed these effects (Figure 3e and f).
Altogether these data suggested that classical PKCs and Cdc2 exert opposite regulatory effects on hRFPL1 activity by promoting or preventing its effect on cell number, respectively. Further, these observations indicated that (1) hRFPL effect on cell proliferation in our gain-of-function approach is specific as it was reversed when regulatory post-translational sites were mutated; and (2) apoptosis, which was also reversed with these mutations, may therefore be a consequence of hRFPL1 action on cell-cycle machinery, rather than a toxic, nonspecific effect due to protein overexpression (even if we cannot rule out that apoptosis could be less prominent if hRFPL1 expression level were closer to an endogenous one).
hRFPL1 alters G2–M phase transition
From there, we investigated the influence of hRFPL1 on cell-cycle regulation. Using FACS analysis to examine the cell-cycle phase altered during hRFPL1 antiproliferative activity, we observed that hRFPL1 significantly increased the population of cycling cells in G2–M phase (Figure 4a). We then examined the impact of hRFPL1 on the levels of proteins regulating cell-cycle progression: cyclin D1/CDK6 (regulating G1 phase progression), cyclin A/CDK2 (G1–S transition) and cyclin B1/Cdc2 (G2–M transition). In line with the data on cell-cycle analysis, hRFPL1 influence on cyclin/CDK complexes was restricted to cyclin B1 and Cdc2 as it elicited a decrease in their protein levels (Figure 4b; Supplementary Figure 2A), whereas their transcriptional expression remained steady (Figure 4c). We also examined the impact of hRFPL1 on other proteins associated with cell-cycle check point (p53 and p21WAF1/CIP1) or known to regulate G2–M transition, namely cdc25C (a Cdc2-activating phosphatase), PLK1 (a cdc25C-activating kinase) and Emi1 (a F-Box protein required to induce M-phase entry by stimulating cyclin B1 accumulation). hRFPL1 did not alter the expression of any of these proteins, suggesting that it does not alter the p53 pathway, nor the complete machinery involved in G2–M phase transition, but preferentially regulates cyclin B1 and Cdc2 levels (Figure 4b; Supplementary Figure 2A). Further, both hRFPL1-mediated cyclin B1 and Cdc2 downregulations were precluded by MG-132, a proteasome inhibitor (Figure 4d; Supplementary Figure 2B), suggesting that hRFPL1 reduces cyclin B1 and Cdc2 levels by triggering their ubiquitin-mediated proteasomal degradation. In addition, neither S200A nor S287E hRFPL1 mutants led to cyclin B1 and Cdc2 downregulations (Supplementary Figure 2C), in line with our observations that these mutations alter hRFPL1 ability to reduce cell number and induce apoptosis.
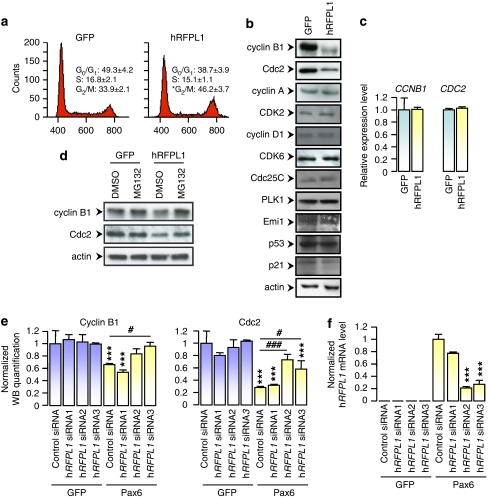
Figure 4.
hRFPL1 increases cell population in G2/M phase by inducing cyclin B1 and Cdc2 proteasomal downregulations. (a) Cell-cycle analysis of eGFP- or hRFPL1-expressing HeLa cells was assessed by FACS. *P<0.05 versus eGFP cells using Student's t-test. (b) Levels of cell-cycle-associated proteins in eGFP- or hRFPL1-transduced HeLa cells were determined by immunoblotting with the antibodies indicated. (c) CCNB1 and CDC2 transcript levels were determined by quantitative RT-PCR in eGFP- and hRFPL1-expressing HeLa cells. (d) Cyclin B1 and Cdc2 levels in eGFP- and hRFPL1-expressing HeLa cells were assessed by immunoblotting following cell treatment with DMSO (control) or MG-132, a proteasome inhibitor. (e, f) Quantification of cyclin B1 and Cdc2 protein levels from immunoblotting experiments in eGFP- or Pax6-expressing HeLa cells transfected using one control and three hRFPL1 siRNA sequences. Normalization was performed using the amount of the actin reference protein in each condition and band intensities were measured using the ImageJ software. *P<0.05 versus control cells using one-way ANOVA followed by Tukey's post hoc test. (f) Assessment of the eGFP- or Pax6-induced endogenous expression of hRFPL1 and of the efficiency of the siRNA sequences. hRFPL1 transcript levels were determined by RT–PCR in eGFP- or Pax6-expressing HeLa cells transfected using one control and three hRFPL1 siRNA sequences. ***P<0.001 versus GFP-expressing cells transfected with the same siRNA sequence; #P<0.05, ###P<0.001 versus Pax6-expressing cells transfected with the control siRNA sequence using one-way ANOVA followed by Tukey's post hoc test. Note that the hRFPL1 siRNA sequence 1, which does not reverse the Pax6-provoked cyclin B1 and Cdc2 downregulations, does not alter the Pax6-elicited hRFPL1 expression either and serves as a second control sequence
We next examined the ability of endogenously expressed hRFPL1 to control levels of cyclin B1 and Cdc2. Expression of Pax6 in HeLa cells elicited hRFPL1 expression and induced cyclin B1 and Cdc2 downregulation (Figure 4e and f). Using RNA interference, we observed that Pax6-mediated hRFPL1 expression was significantly diminished with two of the three tested hRFPL1 siRNA sequences, which also significantly reversed effect of Pax6 on cyclin B1 and Cdc2 in a partial or total manner (Figure 4e and f). Thus, it confirmed that the hRFPL1-mediated control of cyclin B1 and Cdc2 levels is physiologically relevant.
We also observed that effect of hRFPL1 on cell number and on cyclin B1 and Cdc2 levels was shared by hRFPL2, hRFPL3 and mRFPL (Supplementary Figures 2D and 2E), indicating that this activity was conserved during the evolution of the RFPL gene family.
hRFPL1 reduces the accumulation of cyclin B1 and Cdc2 during interphase
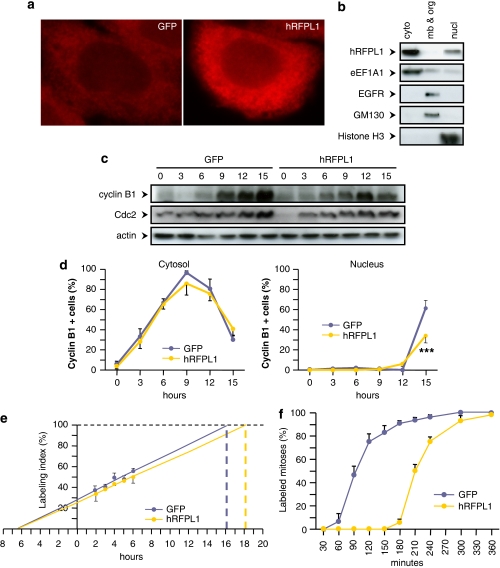
Using immunofluorescence and subcellular compartment fractionation followed by immunoblotting, we detected that hRFPL1 was mostly localized in the cell cytoplasm (Figure 5a and b), similar to cyclin B1 and Cdc2 subcellular localization during S and G2 phases.15 Thus, we hypothesized that hRFPL1 could alter cyclin B1 and Cdc2 accumulations during this period. To address this question, we synchronized HeLa cells in G1 phase (cell population going from 47-18-35 to 71-18-11% after mimosine treatment, in G1, S and G2–M phase, respectively) and examined by immunoblotting cyclin B1 and Cdc2 levels over time following release from cell-cycle block. Cyclin B1 accumulation was markedly decreased in hRFPL1-expressing cells as compared to GFP-transduced cells (Figure 5c; Supplementary Figure 2F). Changes in Cdc2 expression were less prominent during cell-cycle progression, but its protein accumulation was also reduced in hRFPL1-expressing cells (Figure 5c; Supplementary Figure 2F). Hence, hRFPL1 alters cyclin B1 and Cdc2 protein levels by reducing their accumulation.
Figure 5.
Cytoplasm-localized hRFPL1 reduces cyclin B1 and Cdc2 accumulation to delay mitosis entry. (a, b) In FLAG-tagged hRFPL1-transduced HeLa cells, subcellular localization of hRFPL1 was determined by (a) immunocytochemistry using a pan-hRFPL1,2,3 antibody and (b) immunoblotting after subcellular compartment fractionation using a FLAG antibody. In (a), a similar subcellular localization was obtained by immunocytochemistry after endogenous induction of hRFPL1 expression by Pax6 overexpression (data not shown). In (b), the following abbreviations were used: cyto, cytosolic proteins; mb & org, proteins in membranes and organelles; nucl, nuclear proteins. Immunoblotting using EGFR, Golgi marker GM-130, eEF1A1 and Histone H3 antibodies was performed to control the fractionation process. (c, d) Cyclin B1 and Cdc2 accumulations (c) and number of cyclin B1-positive cells expressed as a percentage of the total number of cells (d) over the time were examined by immunoblotting and quantitative immunofluorescence, respectively, in eGFP- and hRFPL1-expressing HeLa cells following release from mimosine-induced G1 phase block. In (d), ***P<0.001 versus eGFP-expressing cells using two-way ANOVA followed by Tukey's post hoc test. (e, f) Cell-cycle parameters were assessed in eGFP- and hRFPL1-expressing HeLa cells using (e) EdU cumulative labeling and (f) determination of the percentage of labeled mitosis
Using quantitative immunofluorescence, we then examined whether this reduction was caused by a delayed onset of accumulation or a slower rate of accumulation. In normal conditions, cyclin B1 highly accumulates in the cytosol during interphase until a threshold concentration is reached to allow cyclin B1/Cdc2 activation and nuclear translocation, thereby cell entry into mitosis.16, 17 We therefore focused on this component to compare its cytosolic and nuclear accumulation profile in hRFPL1- versus eGFP-expressing cells, following cell-cycle block release. The number of cells positive for cytosolic cyclin B1 increased following cell-cycle block release until it reached a plateau between 9 and 12 h. Both eGFP- and hRFPL1-expressing cells showed a similar kinetics (Figure 5d), suggesting that the onset of cyclin B1 accumulation was not altered by hRFPL1. Cyrosolic cyclin B1 sharply decreased at 15 h whereas we detected an augmentation in the number of cells positive for nuclear cyclin B1. However, the number of cells positive for nuclear cyclin B1 was significantly diminished in hRFPL1-expressing cells in comparison to control cells (Figure 5d), suggesting that hRFPL1 decreased cell entry into mitosis. Altogether, our kinetics experiments suggest that hRFPL1 decreases the rate of accumulation of cyclin B1 rather than its onset, and this possibly results in a delay in mitosis entry.
hRFPL1 induces an increase in G2 phase length
We therefore investigated the impact of hRFPL1 on cell-cycle kinetics by assessing the duration of the different phases of the cell cycle. Using the cumulative 5-ethynyl-2′-deoxyuridine (EdU) labeling approach to determine the duration of S phase and of total cell cycle, we observed that hRFPL1 gain of function lengthened the cell cycle by 2 h without modifying S-phase duration (Figure 5e; Table 2). We then examined the percentage of labeled mitosis to measure the duration of G2 and M phases and thereby deduce G1 phase duration. Cells expressing hRFPL1 had a striking threefold increased G2 phase duration with no modification of M or G1 phase (Figure 5f; Table 2). Thus, hRFPL1 alters cell-cycle kinetics by increasing G2 phase duration to elicit its antiproliferative activity.
Table 2. hRFPL1 alters cell-cycle duration by increasing G2 phase length.
| Cell-cycle parameters | GFP-expressing cells (h) | hRFPL1-expressing cells (h) |
|---|---|---|
| TG1+TG2+TM | 16.1 | 18.3 |
| TS | 6.5 | 6.5 |
| TC | 22.6 | 24.8 |
| TG2 | 1 | 3 |
| TM | 2 | 2 |
| deduced TG1 | 13.1 | 13.3 |
Length of the different cell-cycle phases were measured by EdU cumulative labeling (TG1+TG2+TM, TS and TC) and percentage of labeled mitosis (TG2 and TM) experiments in GFP- or hRFPL1-expressing HeLa cells.
Discussion
Cell-cycle regulation is a crucial and tightly regulated cellular mechanism involved in a wide range of biological processes. Here we show that hRFPL1 transcriptional level can be regulated downstream of Pax6 signaling by the prototypical cell-cycle-regulating p53 transcription factor in a cell-specific manner and hRFPL1 exerts a phosphorylation-dependent antiproliferative effect that is positively regulated by PKC and abrogated by Cdc2. We further report that hRFPL1 controls G2–M phase transition by preventing the accumulation of cyclin B1 and Cdc2 proteins during interphase through a proteasome-dependent mechanism. Thus, such an activity lengthens G2 phase duration, which accumulates cell population in G2 phase and delays cell entry into mitosis.
So far, little was known about the cellular activity of the RFPL proteins. The reported expression of the murine (m)RFPL in germ cells, its putative role as an E3 ubiquitin ligase based on its predicted secondary structure and its biochemical interaction with cyclin B1 as well as with HR6A, PMSB1 and ubiquitin B proteins of the proteasomal degradation pathway led to the conclusion that it could regulate meiosis progression.5, 6 However, the expression of RFPL genes in other tissues4 suggested that they could regulate other aspects of cell division. Upon gain of function in HeLa cells, we report here that hRFPL1 elicited cyclin B1 and Cdc2 downregulations that were precluded by a proteasome inhibitor, in line with the previous biochemical identification of mRFPL-interacting proteins.6 However, in contrast to the classical view that cyclin B1 degradation occurs during anaphase to allow mitosis exit and cell-cycle progression,18 our study data indicate that hRFPL1 prevented the accumulation of cyclin B1 and its partner Cdc2 during interphase, leading to an antiproliferative activity by an increase in G2 phase duration, a cell accumulation in G2 phase and thus a delay of cell entry into mitosis.
Despite our observations were primarily found in an hRFPL1-overexpressing cell system, we also observed, in a model where Pax6 directly induces hRFPL1 endogenous expression and elicits a decrease in cyclin B1 and Cdc2 levels, that the latter was prevented by hRFPL1 RNA interference, supporting the physiological relevance of this hRFPL1 activity. Given that these data were obtained following Pax6 gain of function, it is noteworthy that, in progenitor cells endogenously expressing Pax6, the physiological levels of this latter are highly dynamic during cell-cycle progression, and an increase in Pax6 during G2 phase can notably occur to specify their differentiation.19 Increase in Pax6 levels, either physiological or ectopic, has been shown to direct cell-cycle exit and differentiation.19, 20, 21 Accordingly, we previously observed that ectopic Pax6 expression provoked a postmitotic G0/G1 cell-cycle arrest in HeLa cells.9 This was however not altered by hRFPL1 RNA interference (data not shown), suggesting that hRFPL1 influence on cell-cycle progression is not sufficient to produce cell-cycle exit and differentiation in Pax6-expressing cells.
Indirectly supporting the specificity of hRFPL1 cellular function, we also observed that hRFPL1 did not alter the protein levels of cyclins or CDKs controlling the progression of other cell-cycle phases, nor those of other regulators of G2–M phase transition.
In respect with our data, it has been previously reported that cells failing to accumulate cyclin B1 have an increased cell population in G2/M phase due to an altered G2–M phase transition.22, 23 Such a failure in cyclin B1 accumulation could rely on p53 activity22 or on interphasic activation of E3 ubiquitin protein ligases.23 We however did not find any change in the protein levels of p53 or of its canonical effector, p21WAF1/CIP1.
We also observed that the decreased cell proliferation elicited by hRFPL1 was accompanied by apoptotic cell death. Although apoptosis may be considered as a nonspecific, toxic effect of hRFPL1 overexpression, it is known that cyclin B1 inhibition, by arresting cells in G2 phase, leads to apoptosis.24, 25 Hence, hRFPL1-mediated cell death could be the consequence of its influence on cyclin B1 levels. The fact that Cdc2 levels were concomitantly regulated by hRFPL1 is also of importance. Indeed, Cdc2 is the most important CDK for cell-cycle regulation: it is the only one controlling cell-cycle progression in simple eukaryotes26 and it has been shown in multiple knockout mice that Cdc2 is the only mandatory CDK to promote the mammalian cycle whereas the lack of other CDKs can be overcome by compensating mechanisms.27 This indicates that hRFPL1-mediated control of cell cycle through the regulation of Cdc2 protein level could not be overruled by other cyclin/CDK-dependent processes, therefore making it a very tight and precise mechanism.
This also emphasizes the importance of Cdc2 regulation of hRFPL1 activity. Such a post-translational regulatory mechanism is traditionally described to pinpoint the oscillatory activity of proteins controlling cell-cycle progression.28 Accordingly, we propose that hRFPL1 activity would oscillate during cell-cycle progression to exert a finely tuned effect, first through a PKC-dependent phosphorylation allowing its activation during S and G2 phases to prevent cyclin B1 and Cdc2 accumulation. Then, a Cdc2-mediated phosphorylation would terminate hRFPL1 action, probably on M phase triggering cyclin B1/Cdc2 activation. Moreover, hRFPL1 regulation could reach another degree of complexity as we also observed that PKCs could be involved in Pax6 signaling mediating hRFPL1 transcription.
Our observations also showed that hRFPL1 expression level could be regulated by a Pax6-dependent p53 activation. Pax6-mediated hRFPL1 expression appeared to be cell specific and could rely, among other hypotheses, on chromatin state of the hRFPL1 locus or on the presence of specific interacting partners. Although p53 could not transactivate hRFPL1 promoter in any tested cell line, it was able to enhance Pax6-elicited hRFPL1 levels in HeLa cells. Such an enhancing activity offers a first mechanistic hypothesis to support the previously reported importance of RFPL1,2,3 gene-dosage effect.4 It also suggests the involvement of other transcription factors or protein partners for recruiting p53 activity. Fitting with this hypothesis, it has been previously reported a transcriptional cooperation between p53 and a ligand-bound estrogen receptor to result in a regulatory cross talk synergistically transactivating VEGFR1/FLT1, whereas p53 alone has relatively no effect.29, 30 In addition, Pax6-downregulated FOXM1 and MCM5 genes were not affected by p53 RNA interference despite it has been reported in other cell systems that p53 could also repress their expression independently of Pax6 signaling.31, 32 Thus, cross talk between Pax6 signaling and p53 activity appears to regulate specific genes in a given environment as the identification of either Pax6 or p53 target genes greatly varies depending on the cell system used.4, 13, 33, 34, 35
Although we observed that hRFPL1 impact on cell number and on cyclin B1 and Cdc2 levels was shared by hRFPL2, hRFPL3 and mRFPL, indicating that this activity was conserved during the evolution of this gene family, it is noteworthy that such an activity involved the RDM domain. This RDM, present in all RFPL proteins but restricted to them, is the result of a neofunctionalization event that occurred after the emergence of the RFPL gene ancestor.4 Thus, RDM acquisition likely retained the functionality of the ancestral gene after its emergence by conferring it its antiproliferative activity. Further, RDM forms together with the RING domain a large zinc-finger structure of the RING/U-Box superfamily. RING-containing proteins are known to exert an E3 ubiquitin protein ligase activity with the zinc-finger structure being mandatory for binding to the E2 ubiquitin-conjugating enzyme.36 Hence, our study data suggest that the C-terminal part of the RING-RDM-formed zinc finger is crucial for hRFPL1 antiproliferative activity, fitting with the concept that the RFPL proteins function as E3 ubiquitin protein ligases. Although this domain was crucial, it was not sufficient to exert any intrinsic effect as hRFPL1-mediated antiproliferation also relied on SPRY N-terminal part that has been reported to be the region in which cyclin B1 binds to mRFPL.6 Thus, SPRY would function as the recognition domain for proteasome-targeted substrates.
In line with a possible influence of hRFPL1 on development and cell differentiation, it has been reported that hRFPL1 protein expression is induced during chemically mediated differentiation of human gastric carcinoma cells.36, 37 We also detected a striking increase in hRFPL1 transcript level in differentiating human embryonic stem cells at the onset of neurogenesis,4 during which regulation of cell-cycle lengthening is a key event to trigger transition from proliferation to differentiation.38, 39 The potential ability of hRFPL1 to trigger cell death is also of interest, as it has been reported that some cell-cycle regulators can elicit a key pro-apoptotic activity during diverse processes such as developmental morphogenesis40 or tumor suppression.41 Thus, this report that the primate-restricted RFPL1 modulates G2–M phase transition could provide novel insights into understanding developmental changes over the course of evolution.
Material and Methods
Cell culture
Human HeLa-TAT cell lines were obtained from the National Institutes of Health AIDS Research and Reference Reagent, HEK 293T cells were kindly provided by Dr. P Salmon (Department of Fundamental Neurosciences, University of Geneva, Switzerland). Cells were cultured at 37°C with 5% CO2 in DMEM medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin (all from Invitrogen-Gibco, Basel, Switzerland).
Cloning and plasmid constructions
Genes of interest were cloned by PCR using Pfx DNA polymerase (Qiagen, Hombrechtikon, Switzerland) and primers for Gateway B1-B2 elements (Invitrogen, Basel, Switzerland) containing a minimal Kosak (gCCACC) sequence and a C-terminal FLAG tag sequence (5′-gACTACAAggACgACgACgACAAg-3′). The CMV promoter was cloned using primers for Gateway B4-B1 elements. PCR products were then purified using Qiagen PCR purification kit and recombined into Gateway pDONR 221 plasmid (for the genes of interest), or pDONR P4-P1 vector (for the CMV promoter) according to the manufacturer's instructions (Invitrogen). Clones were sequenced (Macrogen, Seoul, Korea) and the correct entry vectors were used for recombination reactions using Gateway LR Plus Recombinase (Invitrogen) into a modified Gateway pLenti6 lentiviral destination vector.42 hRFPL1 was cloned from cDNA of HeLa cells transduced with Pax6. The hRFPL1-truncated proteins were also cloned by PCR. Domains located at the terminal parts of the proteins were simply suppressed and internal domains were replaced by the gCCggC sequence coding Ala and Gly residues. Briefly, amplicons before and after these domains were obtained with specific primers containing the gCCggC sequence on each appropriate end, then digested using KasI and ligated together using T4 DNA ligase according to the manufacturer's instructions (Promega, Dübendorf, Switzerland). All the constructs encoding the truncated proteins had a C-terminal FLAG tag to check their stability by FACSCalibur four-color flow cytometer (Becton Dickinson, Allschwil, Switzerland) from transduced HeLa cells using a FLAG antibody (data not shown).
Lentiviral vector production and transduction
The HIV-1-based lentiviral particles were produced following calcium chloride-based transient transfection of 2 × 106 HEK 293T cells in a 10-cm dish with 3 μg pMD2G (encoding the VSV G envelope protein), 8 μg psPax2 (encoding Gag, Pol, Tat and Rev proteins) and 10 μg of transfer vector. The media were changed the following day. The lentivector-containing supernatant was then collected 48 h later, filtered through 0.45-μm pore-sized polyethersulfone membrane to eliminate cell debris. Target HeLa cells were plated in six-well plates and infected with 1 ml viral supernatant or were plated in a 10-cm dish and infected with 10 ml viral supernatant.
Chromatin immunoprecipitation assay
At 2 days following eGFP, Pax6 or p53 overexpression, HeLa cells were treated for 8 min at room temperature by adding to the culture medium 5 mM HEPES (pH 7.9), containing 10 mM NaCl and 1.1% formaldehyde. Fixation was stopped by adding 180 mM glycine. After one wash with PBS, cells were lyzed by pipetting up and down in ice-cold 10 mM Tris-HCl (pH 8.0), 0.5% NP-40 supplemented with complete protease inhibitor (Roche, Rotkreuz, Switzerland). Nuclei were pelleted and lyzed in 10 mM Tris-HCl (pH 8.0), 500 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate supplemented with complete protease inhibitor. Cross-linked chromatin was sheared into 200–600 bp fragments by sonication, cleared by centrifugation at 15 000 × g for 15 min and stored at −80°C. Chromatin supernatants (20 μg) were diluted (1 : 10) in ChIP buffer (200 mM HEPES (pH 7.9), 2 M NaCl, 20 mM EDTA) supplemented with 200 μg/ml salmon-sperm DNA and complete protease inhibitor. After preclearing the supernatants by rotating incubation for 30 min at 4°C with protein A–sepharose beads (Pierce, Lausanne, Switzerland), half the chromatin was stored at −20°C as control chromatin input. The rest of the supernatant was then incubated overnight at 4°C with 20 μl p53 antibody (Calbiochem, Rotkreuz, Switzerland), cleared by centrifugation at room temperature for 10 min at 8000 × g before a 2 h rotating incubation at room temperature in 20 μl protein A–sepharose beads. The beads were washed as follows: twice in ChIP buffer; twice in ChIP buffer supplemented with 300 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate; twice in Tris-HCl (pH 8.0), 250 mM LiCl, 2 mM EDTA, 0.5% sodium deoxycholate; once in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.1% NP-40. Immune complexes were eluted by incubating 10 min at 65°C in 111 mM Tris-HCl (pH 8.0), 1.11% SDS. Cross-links of the immunoprecipitated DNA and the chromatin input were reversed by incubating for 2 h at 42°C after addition of 20 mg/ml Proteinase K (Qiagen) and 100 mM NaCl, and further incubation overnight at 67°C. After extraction with phenol–chloroform–isoamyl alcohol (25 : 24 : 1) and chloroform–isoamyl alcohol (24 : 1), DNA was precipitated with 100% ethanol in the presence of 20 μg glycogen (Fermentas, Le Mont-sur-Lausanne, Switzerland) and 3 M sodium acetate. Chromatin pellets were resuspended in 100 μl TE buffer. The immunoprecipitated DNA and the input chromatin were analyzed by semiquantitative end-point PCR (40 cycles) using Taq DNA Polymerase in a Q-solution-supplemented buffer according to the manufacturer's instructions (Qiagen) and promoter- and exon2-specific primers (Supplementary Table 1). Design of the primers was performed according to the p53 putative binding sites identified in silico in the proximal region of the hRFPL1 promoter with the MatInspector server (http://www.genomatix.de).
Impact of p53 on hRFPL1 expression
Pharmacological inhibition of p53: HeLa cells were treated with DMSO (0.1% final) or 10 μM Cyclic Pifithrin-α (Sigma, Buchs, Switzerland) and transduced the next day with eGFP- or Pax6-expressing lentivectors. DMSO and Cyclic Pifithrin-α were added to the viral supernatant and the media were replaced every day with fresh medium containing DMSO or Cyclic Pifithrin-α. At 2 days after transduction, the cells were collected to assess the expression of hRFPL1 by real-time PCR as described below.
p53 knock down: HeLa cells were transduced with a control shRNA or p53 shRNA.11, 12 Transduced cells were selected for a week using 5 μg/ml blasticidin before being transduced with eGFP or Pax6. The cells were collected 2 days later, before examining the expression of hRFPL1 or other Pax6 target genes by real-time PCR.
p53 overexpression: The different cell lines were transfected with a pcDNA3 vector containing either a CMV-p53 sequence or a CMV-eGFP sequence as control. After 2 days, the cells were collected and hRFPL1 expression was assessed by semiquantitative end-point PCR as previously described43 or by real-time PCR as described below.
Pharmacological induction of p53 expression: We elicited p53 expression using 1 μg/ml doxorubicin for 4 h. Again, the cells were collected to examine the level of hRFPL1 transcript. To control that the changes observed were resulting from p53-induced expression, we co-treated the cells with DMSO (0.1% final) or 10 μM Cyclic Pifithrin-α.
Real-time quantitative RT-PCR
Total RNA from cell lines was isolated using RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Residual genomic DNA was removed by incubating the RNA solution with 30 u RNase-free DNase for 15 min at room temperature using RNase-Free DNase Set (Qiagen). Total RNA (1 μg) was reverse transcribed using the Superscript II kit according to the manufacturer's instructions (Invitrogen). Real-time PCR reactions were performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Rotkreuz, Switzerland) and a Chromo 4TM Real-Time system (Bio-Rad, Reinach, Switzerland). The amplification efficiency of each pair of primers was determined by comparison with a standard curve generated with serially diluted cDNA of fetal brain. Quantification was performed at a threshold detection line (Ct value). The Ct of each target genes was normalized against that of eEF1A1 used as a housekeeping gene. The 2−ΔΔCt method was used to determine the relative level of expression of each gene.44 The list of the primers used is given in Supplementary Table 1. Data were expressed as mean±S.E.M.
Cell proliferation
HeLa cells were transduced with eGFP and the cells were selected using 5 μg/ml blasticidin for 5 days to establish a pure cell population stably expressing eGFP. To assess the impact of hRFPL1 on cell number, we plated the cells on coverslips in six-well culture dishes and transduced with either eGFP- or hRFPL1-expressing lentivectors. Cells were fixed from day 0 to day 3 using PBS containing 2% formaldehyde, mounted on slides using FluorSave (Calbiochem) and counted under a Zeiss Axioplan microscope equipped for epifluorescence in five randomly selected fields per experiment (Zeiss, Munich, Germany). For the structure–function study, the cells were transduced with eGFP-, wild-type or truncated hRFPL1-expressing lentivectors. At 72 h after transduction, the cells were fixed and counted in five randomly selected fields per experiment.
Live-cell imaging by confocal microscopy
eGFP-positive HeLa cells plated in 35-mm dishes were transduced as described above and placed the following day in a chamber mounted into a home-made stage microincubator. Cells were kept in complete DMEM medium at 37°C with 5% CO2 supply. The fluorescence images of the eGFP-expressing cells were acquired every 5 min for 4 h using the UltraviewRS spinning disk confocal microscope equipped with a 12-bit cooled charge-coupled device interlined camera (ORCA ER; Hamamatsu, Solothurn, Switzerland) mounted on a Nikon Eclipse T2000-U epifluorescence microscope (Nikon, Bern, Switzerland). Excitation by the 488 nm laser line (Argon/Krypton laser; Melles Griot, Bensheim, Germany) was used and fluorescence images were acquired with the × 10 PlanFluor objective were collected through a 535±20 nm bandpass filter. Stacks of images were then generated using the MetaMorph software (Molecular Devices, Downingtown, PA, USA) to quantify the number of rounds of mitosis the cells performed during the experiments.
Phosphorylation state
The phosphorylation state of hRFPL1 and the impact of PKC and Cdc2 inhibitors were assessed using the PhosphoProtein Purification Kit (Qiagen). Briefly, HeLa cells were transduced in a 10-cm dish with FLAG-tagged hRFPL1 and treated the next day with DMSO (0.1% final), PKC inhibitor (12 nM Gö6983; Calbiochem) or 12 μM Cdc2 inhibitor (Calbiochem). At day 2, cells were collected and the purification of phosphoproteins was performed according to the manufacturer's instructions. At the end, the phosphoproteins were concentrated by centrifugation at 4000 × g for 30 min at 4°C using 10 kDa cutoff Vivaspin-2 columns (Vivascience, Buchs, Switzerland). A sample of total proteins was kept before the purification to control that the cells were expressing the same amount of hRFPL1 in each condition. A western blot of the phosphoproteins and total proteins was then performed using a FLAG antibody (1 : 200; Sigma).
Kinase assays
Identification of putative phosphorylation sites and of the kinases involved was carried out using NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/), NetPhosK 1.0 (http://www.cbs.dtu.dk/services/NetPhosK/) and PhosphoELM (http://phospho.elm.eu.org/) servers. Kinase assays were then performed as follow: HeLa cells were transduced with wild-type FLAG-tagged hRFPL1, FLAG-tagged Ser200Ala hRFPL1, FLAG-tagged Ser287Gly hRFPL1 (see next paragraph for the construction of the hRFPL1 mutants). The cells were collected 24 h later and the proteins were extracted overnight in a nondenaturing buffer containing 25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.5% NP-40 and supplemented with a complete protease inhibitor cocktail (Roche). The protein extracts were then centrifuged at 14 000 × g for 15 min at 4°C and proteins from the supernatant were quantified using Bradford protein assay (Bio-Rad). Proteins (200 μg) were immunoprecipitated using a FLAG antibody (Sigma) overnight at 4°C using a rotating wheel. In parallel, proteins from nontransduced HeLa cells were prepared and 200 μg was immunoprecipitated using either a PKCα,β,γ (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or a cyclin B1 antibody (Sigma) to precipitate activated Cdc2 proteins. The immune complexes were then incubated with equilibrated protein A-Sepharose beads (Pierce) for 4 h at 4°C using a rotating wheel. Each hRFPL1 sample was mixed with one PKC-immunoprecipitated sample or one cyclin B1/Cdc2-immunoprecipitated sample and washed four times at 4°C with 25 mM Tris-HCl (pH 7.6), 150 mM NaCl. For the PKC kinase assay, the beads were then incubated for 30 min at 30°C under agitation in assay dilution buffer II, PKC Lipid Activator and Magnesium/ATP cocktail (Upstate, Zug, Switzerland) complemented with [γ-32P]ATP (Amersham, Dübendorf, Germany). For the Cdc2 kinase assay, we incubated the beads for 30 min at 30°C under agitation in assay dilution buffer and Magnesium/ATP cocktail (Upstate) complemented with [γ-32P]ATP (Amersham). The reactions were stopped by deposing the beads on P81 Whatmann chromatography paper (Upstate), which was then immersed in 0.75% orthophosphoric acid (Prolabo, Nyon, Switzerland). After four washes in this buffer, the P81 papers were dried before counting incorporated radioactivity using a Beckman Liquid Scintillation Counter (Beckman Coulter, Nyon, Switzerland).
Site-directed mutagenesis
Point mutations and amino-acid changes were performed using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, Basel, Switzerland), according to the manufacturer's instructions. Briefly, a pLenti6 lentiviral destination vector containing the CMV-FLAG-tagged wild-type hRFPL1 sequences was used as a template and mutant strand synthesis was obtained by PCR by using the appropriate primers. The methylated template DNA was then digested using DpnI for 1 h at 37°C. The mutated DNA was transformed into Stbl3 bacteria (Invitrogen). Clones were sequenced (Macrogen) to verify the mutation. The stability of each mutant was controlled by immunoblotting from transduced HeLa cells using a FLAG antibody (data not shown).
Cell-cycle analysis
DNA content of HeLa cells was determined by propidium iodide (PI) staining of DNA and FACS analysis. The cells were synchronized in G1 phase by adding, for 20 h, 500 μM -mimosine (Sigma). The cells were then washed twice in PBS and were transduced with eGFP- or hRFPL1-expressing lentivectors. They were then collected by trypsinisation 3 days later, extensively washed in PBS to detach apoptotic cells and thus avoid interference with the cell-death process and resuspended in ice-cold 70% ethanol. After incubating on ice for 15 min, they were centrifuged (2000 × g, 10 min). The cell pellet was resuspended in 62.5 U ribonuclease A, incubated in a 37°C water bath for 15 min, and subsequently exposed to 25 μg/ml PI for 30 min at 4°C. A total of 10 000 events were analyzed on a FACSCalibur four-color flow cytometer (Becton Dickinson), equipped with both a 488 nm argon laser and a 635 nm diode laser. Cell-cycle analysis was carried out with CellQuest software (Becton Dickinson) and histograms were generated by WinMDI 2.9 software (Joe Trotter; Scripps Research Institute, La Jolla, CA, USA).
Western blotting
HeLa cells were plated on a 10-cm dish and transduced the next day with lentiviral vectors expressing eGFP or the different RFPLs. At 2 days after transduction, the cells were lyzed on ice in 1% NP-40 in 25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EGTA supplemented with a complete protease inhibitor cocktail (Roche) and centrifuged at 14 000 × g for 15 min at 4°C. Proteins from the supernatant were quantified using Bradford protein assay (Bio-Rad). Proteins (25–50 μg) per lane were diluted in loading buffer, denatured at 70°C for 10 min before being separated by SDS-PAGE and then transferred to PVDF membrane (Millipor, Zug, Switzerland). Membranes were blocked with PBS containing 0.05% Tween 20 and 5% non-fat dried milk and incubated overnight at 4°C with cyclin B1 (60 kDa, 1 : 300; Sigma), Cdc2 (34 kDa, 1 : 500; Cell Signaling), cyclin D1 (36 kDa, 1 : 200; Thermo Scientific, Wohlen, Switzerland), cyclin A (55 kDa, 1 : 1000; Cell Signaling), p53 (53 kDa, 1 : 500; Cell Signaling), p21WAF1/CIP1 (21 kDa, 1 : 500; Cell Signaling), Cdc25C (60 kDa, 1 : 1000; Cell Signaling), PLK1 (62 kDa, 1 : 1000; Cell Signaling), Emi1 (56 kDa, 1 : 200; Santa Cruz Biotechnology) or α-actin (42 kDa, 1 : 400; NeoMarkers, Wohlen, Switzerland) antibodies. Membranes were washed three times with PBS/0.05% Tween 20 buffer and incubated for 1 h at room temperature with appropriate secondary antibody with PBS containing 0.05% Tween 20 and 5% non-fat dried milk. Membranes were then washed twice in PBS/0.05% Tween 20 buffer and once in PBS buffer. Blots were revealed using ECL Western Blotting Detection Reagents (Amersham) and quantified using the ImageJ software (http://rsbweb.nih.gov/ij/).
Impact of hRFPL1 siRNA on Pax6-mediated cyclin B1 and Cdc2 downregulations
HeLa cells were plated in six-well culture dishes and transfected right away in suspension using 30 ng of either control or hRFPL1 siRNAs (Qiagen) with Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. Medium was replaced 6 h later and cells were transduced the next day either with eGFP- or Pax6-expressing lentiviral vectors prepared as described above. Cells were then collected 30 h after transduction for protein or mRNA extraction. For each condition, cyclin B1 and Cdc2 protein levels were assessed by western blotting and the results were quantified using the ImageJ software, whereas hRFPL1 transcript levels were determined by RT–PCR according to the above-described protocols.
Immunocytochemistry
At 3 days after transduction, cells were washed in PBS and fixed with 2% paraformaldehyde (w/v) for 30 min at room temperature. After two washes in PBS/0.05% Tween 20 buffer, cells were permeabilized in PBS containing 0.5% (v/v) Triton X-100, for 30 min at room temperature and washed twice more in PBS/0.05% Tween 20 buffer. The fixed cells were then treated with PBS containing 1% fetal calf serum (blocking buffer) for 30 min and incubated with an RFPL (1 : 100; Abnova, Heidelberg, Germany), cleaved caspase-3 (1 : 1000; Cell Signaling), FLAG M1 (1 : 100; Sigma) or cyclin B1 (1 : 50; Sigma) primary antibody for 1 h at room temperature. After two washes in blocking buffer, cells were incubated with appropriate secondary antibody (Alexa Fluor 488 or 555 conjugates, 1 : 1000; Molecular Probes, Basel, Switzerland) for 1 h at room temperature. The cells were finally washed twice in PBS/0.05% Tween 20 buffer, once in PBS buffer and once in distilled water before mounting using FluorSave (Calbiochem). Visualization analysis was performed using an AxioSkop 2 Plus microscope equipped for epifluorescence and recorded using an AxioCam HR CCD camera and the AxioVision 4 software (Zeiss).
Subcellular fractionation
Subcellular localization of hRFPL1 was assessed using the Qproteome Cell Compartment Kit (Qiagen). Briefly, HeLa cells were transduced with FLAG-tagged hRFPL1 and collected the following day. Subcellular fractionation was performed according to the manufacturer's instructions. At the end, proteins of each fraction were quantified using Bradford protein assay (Bio-Rad) and the same amount in each condition was concentrated by incubation with four volumes of ice-cold acetone for 20 min at −20°C and centrifugation at 12 000 × g for 20 min at 4°C. Presence of FLAG-tagged hRFPL1 in the pellets was then analyzed by western blotting as indicated above using FLAG (1 : 200; Sigma), EGFR (120 kDa, 1 : 1000; Cell Signaling), GM130 (140 kDa, 1 : 2000; Abcam, Cambridge, UK) and Histone H3 (17 kDa, 1 : 5000; Sigma) antibodies.
Kinetics of cyclin B1 and Cdc2 accumulation
HeLa cells were plated in 6-well dishes (for western blotting analysis) or on coverslips in 12-well dishes (for immunofluorescence analysis) and transduced the next day with eGFP- or hRFPL1-expressing lentivectors. After 1 day, the cells were synchronized in G1 phase by adding 500 μM -mimosine (Sigma). On the third day, cells were extensively washed in PBS to remove -mimosine and were collected or fixed at 0, 3, 6, 9, 12, and 15 h following cell-cycle block release. Western blotting and immunofluorescence were then performed and analyzed as above described.
EdU cumulative labeling
GFP-positive HeLa cells were plated on coverslips in six-well culture dishes and transduced the following day with eGFP- or hRFPL1-expressing lentivectors. After 24 h, 10 μM EdU (Invitrogen) was added and after 2, 3, 4, 5 and 6 h cells were fixed with 2% paraformaldehyde. Similar to the immunohistochemistry protocol described above, cells were washed, permeabilized and incubated in blocking buffer. EdU labeling was then revealed using the Click-iT EdU Alexa Fluor 594 imaging kit, according to the manufacturer's instructions (Invitrogen). Coverslips were then mounted on slides using FluorSave (Calbiochem). Number of EdU-positive cells over the total number of cells was counted under a Zeiss Axioplan microscope equipped for epifluorescence in five randomly selected fields per experiment.
Percentage of labeled mitosis
HeLa cells were plated on coverslips in six-well culture dishes and transduced the following day with an empty vector or hRFPL1-expressing lentivector. After 24 h, 10 μM EdU (Invitrogen) was added for 30 min. The cells were then rinsed twice in PBS buffer before adding new DMEM medium, and fixed at 30, 60, 90, 120, 150, 180, 210, 240, 300 and 360 min following the single EdU pulse with 2% paraformaldehyde. The cells were then washed, permeabilized and incubated in blocking buffer. The cells in mitosis were determined using a phospho-Histone H3 antibody (1 : 2000; Upstate) and an anti-rabbit secondary antibody (Alexa Fluor 488 conjugates, 1 : 800; Molecular Probes). EdU labeling was revealed as described above. Coverslips were then mounted on slides using FluorSave (Calbiochem). Number of EdU-positive cells among the population of phospho-Histone H3-positive cells was counted under a Zeiss Axioplan microscope equipped for epifluorescence in five randomly selected fields per experiment.
Statistical analysis
Results are shown as mean±standard error (S.E.M.). Student's unpaired t-test for two group comparisons was used to analyze cell-cycle analysis, measurement of cell-cycle length, real-time RT-PCR, western blots. Analyses of multiple groups (cell number at a single time point, kinase assays, real-time RT-PCR, quantitative immunocytochemistry, western blots) were performed by a one-way analysis of variance (ANOVA) followed by post hoc multiple comparisons using Tukey's test. Analyses of cell number (time course) and kinetics of cyclin B1 and Cdc2 accumulations were carried out by two-way ANOVA followed by post hoc multiple comparisons using Tukey's test. For all tests, a P-value <0.05 was taken as statistically significant.
Acknowledgments
JB was supported by the Auvergne Regional Council, the Geneva Department of Public Education and FP7-2007 ICT-216593 (SECO). CD was supported by FP7-2007 ICT-216593 (SECO) and ANR-06-NEUR-010. KHK was supported by the Swiss National Science Foundation.
Glossary
- cPFTα
cyclic-Pifithrin-α
- RDM
RFPL-defining motif
- RFPL
Ret finger protein-like
- RSH
RFPL1,2,3-specifying helix
- SNP
single nucleotide polymorphism
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer G, Bonné S, Luco RF, Van De Casteele M, Lenne-Samuel N, Collombat P, et al. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25:1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefont J, Nikolaev SI, Perrier AL, Guo S, Cartier L, Sorce S, et al. Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. Am J Hum Genet. 2008;83:208–218. doi: 10.1016/j.ajhg.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Lee JH, Yan C, Matzuk MM. The ret finger protein-like 4 gene, Rfpl4, encodes a putative E3 ubiquitin-protein ligase expressed in adult germ cells. Mech Dev. 2002;112:173–177. doi: 10.1016/s0925-4773(01)00635-9. [DOI] [PubMed] [Google Scholar]
- Suzumori N, Burns KH, Yan W, Matzuk MM. RFPL4 interacts with oocyte proteins of the ubiquitin–proteasome degradation pathway. Proc Natl Acad Sci USA. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledan E, Polanski Z, Terret ME, Maro B. Meiotic maturation of the mouse oocyte requires an equilibrium between cyclin B synthesis and degradation. Dev Biol. 2001;232:400–413. doi: 10.1006/dbio.2001.0188. [DOI] [PubMed] [Google Scholar]
- Bassermann F, Peschel C, Duyster J. Mitotic entry: a matter of oscillating destruction. Cell Cycle. 2005;4:1515–1517. doi: 10.4161/cc.4.11.2192. [DOI] [PubMed] [Google Scholar]
- Cartier L, Laforge T, Feki A, Arnaudeau S, Dubois-Dauphin M, Krause KH. Pax6-induced alteration of cell fate: shape changes, expression of neuronal alpha tubulin, postmitotic phenotype, and cell migration. J Neurobiol. 2006;66:421–436. doi: 10.1002/neu.20225. [DOI] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Dunand-Sauthier I, Wiznerowicz M, Clarkson SG. UV-induced apoptosis in XPG-deficient fibroblasts involves activation of CD95 and caspases but not p53. DNA Repair (Amst) 2007;6:602–614. doi: 10.1016/j.dnarep.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Pim D, Banks L. The role of the E6–p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18:7690–7700. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- Yang J, Bardes ES, Moore JD, Brennan J, Powers MA, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Hsieh YW, Yang XJ. Dynamic Pax6 expression during the neurogenic cell cycle influences proliferation and cell fate choices of retinal progenitors. Neural Dev. 2009;4:32. doi: 10.1186/1749-8104-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Suter DM, Tirefort D, Julien S, Krause KH. A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells. 2009;27:49–58. doi: 10.1634/stemcells.2008-0319. [DOI] [PubMed] [Google Scholar]
- Innocente SA, Abrahamson JL, Cogswell JP, Lee JM. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H, Kim M, Yoshikawa Y, Ashida H, Ogawa M, Fujita Y, et al. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell. 2007;130:611–623. doi: 10.1016/j.cell.2007.06.043. [DOI] [PubMed] [Google Scholar]
- Yuan J, Krämer A, Matthess Y, Yan R, Spänkuch B, Gätje R, et al. Stable gene silencing of cyclin B1 in tumor cells increases susceptibility to taxol and leads to growth arrest in vivo. Oncogene. 2006;25:1753–1762. doi: 10.1038/sj.onc.1209202. [DOI] [PubMed] [Google Scholar]
- Xie XH, An HJ, Kang S, Hong S, Choi YP, Kim YT, et al. Loss of cyclin B1 followed by downregulation of cyclin A/Cdk2, apoptosis and antiproliferation in HeLa cell line. Int J Cancer. 2005;116:520–525. doi: 10.1002/ijc.21056. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Santamaria D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- Listovsky T, Zor A, Laronne A, Brandeis M. Cdk1 is essential for mammalian cyclosome/APC regulation. Exp Cell Res. 2000;255:184–191. doi: 10.1006/excr.1999.4788. [DOI] [PubMed] [Google Scholar]
- Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schönfelder G. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci USA. 2006;103:1406–1411. doi: 10.1073/pnas.0508103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Inga A, Snipe J, Krysiak O, Schönfelder G, Resnick MA. A single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol Cell Biol. 2007;27:2590–2600. doi: 10.1128/MCB.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal MK, Ruhul Amin AR, Agarwal ML. DNA replication licensing factor minichromosome maintenance deficient 5 rescues p53-mediated growth arrest. Cancer Res. 2007;67:116–121. doi: 10.1158/0008-5472.CAN-06-2835. [DOI] [PubMed] [Google Scholar]
- Pandit B, Halasi M, Gartel AL. p53 negatively regulates expression of FoxM1. Cell Cycle. 2009;8:3425–3427. doi: 10.4161/cc.8.20.9628. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Reed NA, Zhang W, Duncan MK, Kilimann MW, Cvekl A. Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. J Biol Chem. 2002;277:11539–11548. doi: 10.1074/jbc.M110531200. [DOI] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, et al. Identification of pax6-dependent gene regulatory networks in the mouse lens. PLoS One. 2009;4:e4159. doi: 10.1371/journal.pone.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Zhang G, Wang G, Wang S, Li Q, Ouyang G, Peng X. Applying proteomic methodologies to analyze the effect of hexamethylene bisacetamide (HMBA) on proliferation and differentiation of human gastric carcinoma BGC-823 cells. Int J Biochem Cell Biol. 2004;36:1613–1623. doi: 10.1016/j.biocel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Kuan CY, Roth KA, Flavell RA, Rakic P. Mechanisms of programmed cell death in the developing brain. Trends Neurosci. 2000;23:291–297. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Cartier L, Bettiol E, Tirefort D, Jaconi ME, Dubois-Dauphin M, et al. Rapid generation of stable transgenic embryonic stem cell lines using modular lentivectors. Stem Cells. 2006;24:615–623. doi: 10.1634/stemcells.2005-0226. [DOI] [PubMed] [Google Scholar]
- Bonnefont J, Daulhac L, Etienne M, Chapuy E, Mallet C, Ouchchane L, et al. Acetaminophen recruits spinal p42/p44 MAPKs and GH/IGF-1 receptors to produce analgesia via the serotonergic system. Mol Pharmacol. 2007;71:407–415. doi: 10.1124/mol.106.025775. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.