Abstract
As the number of protein folds is quite limited, a mode of analysis that will be increasingly common in the future, especially with the advent of structural genomics, is to survey and re-survey the finite parts list of folds from an expanding number of perspectives. We have developed a new resource, called PartsList, that lets one dynamically perform these comparative fold surveys. It is available on the web at http://bioinfo.mbb.yale.edu/partslist and http://www.partslist.org. The system is based on the existing fold classifications and functions as a form of companion annotation for them, providing ‘global views’ of many already completed fold surveys. The central idea in the system is that of comparison through ranking; PartsList will rank the approximately 420 folds based on more than 180 attributes. These include: (i) occurrence in a number of completely sequenced genomes (e.g. it will show the most common folds in the worm versus yeast); (ii) occurrence in the structure databank (e.g. most common folds in the PDB); (iii) both absolute and relative gene expression information (e.g. most changing folds in expression over the cell cycle); (iv) protein–protein interactions, based on experimental data in yeast and comprehensive PDB surveys (e.g. most interacting fold); (v) sensitivity to inserted transposons; (vi) the number of functions associated with the fold (e.g. most multi-functional folds); (vii) amino acid composition (e.g. most Cys-rich folds); (viii) protein motions (e.g. most mobile folds); and (ix) the level of similarity based on a comprehensive set of structural alignments (e.g. most structurally variable folds). The integration of whole-genome expression and protein–protein interaction data with structural information is a particularly novel feature of our system. We provide three ways of visualizing the rankings: a profiler emphasizing the progression of high and low ranks across many pre-selected attributes, a dynamic comparer for custom comparisons and a numerical rankings correlator. These allow one to directly compare very different attributes of a fold (e.g. expression level, genome occurrence and maximum motion) in the uniform numerical format of ranks. This uniform framework, in turn, highlights the way that the frequency of many of the attributes falls off with approximate power-law behavior (i.e. according to V–b, for attribute value V and constant exponent b), with a few folds having large values and most having small values.
INTRODUCTION
Protein folds can be considered the most basic molecular parts. There are a very limited number of them in biology. Currently, about 500 are known, and it is believed that there may be no more than a few thousand in total (1–3). This number is considerably less than the number of genes in complex, multicellular organisms (>10 000 for multicellular organisms; 4). Consequently, folds provide a valuable way of simplifying and making manageable complex genomic information. In addition, folds are useful for studying the relationships between evolutionarily distant organisms since, in making comparisons, structure is more conserved than sequence or function.
In a general sense, how should one approach the analysis of molecular parts? A simple analogy to mechanical parts may be useful in this regard. Given the ‘parts’ from a number of devices (e.g. a car, a bicycle, and a plane) one might like to know which ones are shared by all and which are unique (say, wings for a plane). Furthermore, one might want to know which are common, generic parts and which are more specialized. Finally, one might like to organize the parts by a number of standardized attributes (e.g. the most flexible parts, the parts with the most functions, and the biggest parts). PartsList aims to provide answers to simple questions such as these for the domain of protein folds.
Properties related to protein folds can be divided into those that are ‘intrinsic’ versus ‘extrinsic’. Intrinsic information concerns an individual fold itself, e.g. its sequence, 3D structure and function, while ‘extrinsic’ information relates to a fold in the context of all other folds, e.g. its occurrence in many genomes and expression level in relation to that for other folds. Web-based search tools already provide intrinsic information about protein structures in the form of reports about individual structures. Valuable examples include the PDB Structure Explorer (5), PDBsum (6) and the MMDB (7). However, current resources lack the ability to fully present extrinsic information.
Likewise, while there are many databases storing information related to individual organisms (e.g. SGD, MIPS and FlyBase; 8–10), comparative genomics (PEDANT and COGs; 9,11), gene expression (GEO, the Gene Expression Omnibus at the NCBI, and ExpressDB; 12) and protein–protein interactions (DIP and BIND; 13,14), none of these integrates gene sequences, protein interactions, expression levels and other attributes with structure. (However, it should be mentioned that the Sacc3D module of SGD and PEDANT do tabulate the occurrence of folds in genomes.)
PartsList is arranged somewhat differently from most other biological resources. In a usual database (e.g. GenBank; 15) the number of entries increases as the database develops, while each entry has a fairly fixed number of attributes to describe it. In contrast, PartsList is envisioned to have a relatively stable number of entries, i.e. the finite list of protein folds, while the attributes that describe each entry are expected to increase considerably. In the current version of PartsList the properties for a protein fold include: amino acid composition, alignment information, fold occurrences in various genomes, statistics related to motions, absolute expression levels of yeast in different experiments, relative expression ratios for yeast, worm and Escherichia coli in various conditions, information on protein–protein interactions (based on whole-genome yeast interaction data and databank surveys) and sensitivity of the genes associated with the fold to inserted transposons.
One reason to build the database is to compare protein folds in a rich context and in a unified way. This was achieved through ranking. This allows users to directly compare very different attributes of a fold in a uniform numerical format. The rankings can be visualized in three ways: a profiler emphasizing the progression of high and low ranks across many pre-selected attributes, a rankings comparer for custom comparisons and a numerical rankings correlator. This can help users gain insight into the functions of protein folds in the context of the whole genome. Our system makes it very easy to answer questions like: ‘What is the most common fold in the worm as compared to E.coli?’, ‘What is the most highly expressed fold in yeast and how does this compare to the fold that changes most in expression level during the cell-cycle?’ and ‘Which fold has the most protein–protein interactions in the PDB and is it highly ranked in terms of protein motions?’
One of the strengths of the uniform numerical system of ranks in PartsList is that it puts everything into a common framework so that one can see hidden similarities in the occurrence of parts ordered according to many different attributes. In particular, as we describe below, we found that the frequency of many of the attributes falls off according to a power-law distribution (i.e. according to V–b, for attribute value V and a constant b), with a few folds having large attribute values and most having small values. For instance, there are only a few folds that occur many times in the yeast genome and most only occur once or twice. Likewise, most folds only have a few functions associated with them, but there are a few ‘Swiss-army-knife’ folds that are associated with many distinct functions. Similar power-law-like expressions have been found to apply in a variety of other situations relating to proteins, for instance, in the occurrence of oligo-peptide words (16–18), in the frequency of transmembrane helices (19) and sequence families with given size (20), and in the structure of biological networks, with a few nodes having many connections and most have only a few (21,22).
PartsList is built on top of the Structural Classification of Proteins (SCOP) (23) fold classification and acts as an accompanying annotation to this system. SCOP is divided into a hierarchy of five levels: class, fold, superfamily, family and protein. The ‘parts’ in our system can be either SCOP folds or superfamilies. However, sometimes for ease of expression we will just refer to ‘folds’ when we really mean ‘folds and/or superfamilies’. We currently use 420 folds and 610 superfamilies in PartsList. Each is represented by a representative domain, which is also the key for each entry of protein fold.
While we chose to use the SCOP classification, we could equally well have based the system on the other existing fold classifications, e.g. CATH (24), FSSP (25) or VAST (26,27). Moreover, for most attributes, we could also have developed our system around non-structural classifications of protein parts, e.g. Pfam (28), Blocks (29) or SMART (30). However, basing it around actual structural folds has the advantage that each part is more precisely and physically defined.
ATTRIBUTES THAT CAN BE RANKED: INFORMATION IN THE SYSTEM
Currently the attributes for each entry (i.e. protein fold) can be separated into several main categories: statistical information from a comprehensive set of structural alignments, amino acid composition information, fold occurrences in various genomes, expression levels in different experiments, protein interactions, macromolecular motion, transposon sensitivity and miscellaneous.
We have developed a formalism for expressing each of the attributes, which is described in Table 1. In the table, the term PART refers to either fold or superfamily, depending on which of these is being ranked. Essentially, we have a database of attributes where each attribute is given a standardized description and associated with a precise reference. In the following, we describe some main categories of attributes.
Table 1. All the attributes ranked by PartsList.
| A | ||||
| Category |
Symbol |
Definition of symbol |
Attributes in category |
Reference |
| Genome Occurrence | G(x) | Number of times a particular PART occurs in genome x. (These are based on PSI-BLAST comparisons between PDB and the genomes with an e-value cutoff in these comparisons of 0.0001.) | 20 | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) |
| Expression | L(e) | Average expression level of a particular PART. This is the average expression level over all genes that contain this PART. | 8 | (46) |
| C(e) | PART composition of the yeast transcriptome in expression level experiment e. This refers to the fraction of the mRNA population with this PART as opposed to all other parts. (This is only applicable to expression experiments, such as SAGE and GeneChips, that measure absolute mRNA levels in copies per cell.) | 8 | (46) | |
| E(e) | Transcriptome enrichment compared to genome in experiment e. [Transcriptome enrichment is defined as percentage difference of PART composition in the transcriptome and the genome. In symbols: E(e) = [C(e)-G(Scer)] / G(Scer).] | 8 | (46 | |
| F(r) | Expression level fluctuation in experiment r. [This is the standard deviation in the expression ratio measurement R(i,t) over a timecourse, for example, <(R(i,t)–<R(i,t)>)2> where one averages over all times t and genes i that have a particular PART.] | 7 | (67) | |
| Alignments | V(f) | The number of aligned pairs in pair-set f. | 2 | (39) |
| U(f) | RMS deviation in Cα atoms averaged over all alignments in pair-set f | 2 | (39) | |
| R(f) | Similar to U(f) for pair-set f but only the best fitting half of the atoms are included in the calculation | 2 | (39) | |
| S(f) | Average percentage identity between pairs of aligned proteins in pair-set f | 2 | (39) | |
| P(f) | Average sequence P value for pair-set f | 2 | (39) | |
| Q(f) | Average structural P value for pair-set f | 2 | (39) | |
| Compositions | N(p) | The number of structures associated with a particular PART in dataset p. | 2 | |
| B(a,p) | Composition of amino acid a in a particular PART where one averages over all structures in dataset p associated with the PART | 40 | ||
| Motion | M(s,d) | The maximum value of statistic s derived from surveying set of motions d in the Macromolecular Motions Database for a particular PART, where s is only calculated from the entries in the database that are associated with the PART. | 7 | (56,57) |
| A(s,d) | Similar to M(s,d) but now we take the average instead of the maximum. | 7 | (56,57) | |
| Interaction | I(y,c) | For a given PART, the number of types of protein–protein interactions in interaction dataset y subject to the restriction c regarding whether or not the proteins are on the same chain. The number of interaction types is the number of distinctly different PARTs that interacts with a given PART. | 24 | (51,68) |
| J(y,c) | For a given PART, the total number of types of interactions in interaction dataset y subject to the restriction c regarding whether or not the proteins are on the same chain. Here we show all interactions observed not just the number of distinct PART-PART interactions tabulated in I(y,c). | 24 | (52,68) | |
| Transposon | T(b) | The sensitivity of the cell to a transposon inserted into genes containing a particular PART under different growth condition b. The sensitivity was indicated by negative logarithm of a P value, which measures the degree to which the observations for one particular gene could have resulted from wild-type cells that randomly change their phenotype. | 20 | (58) |
| Miscelleneous | X(q) | Various miscellaneous ranks. | 5 | |
| Total |
|
|
182 |
|
| B | ||||
| Attributes |
Value |
Description |
Reference |
|
| Genome x = | aful | Archaeoglobus fulgidus | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | |
| mjan | Methanococcus jannaschii | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| mthe | Methanobacterium thermoautotrophicum | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| phor | Pyrococcus horikoshii | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| scer | Saccharomyces cerevisiae | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| cele | Caenorhabditis elegans | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| aaeo | Aquifex aeolicus | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| syne | Synechocystis sp. | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| ecol | Escherichia coli | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| bsub | Bacillus subtilis | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| mtub | Mycobacterium tuberculosis | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| hinf | Haemophilus influenza Rd | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| hpyl | Helicobacter pylor | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| mgen | Mycoplasma genitalium | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| mpne | Mycoplasma pneumoniae | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| bbur | Borrelia burgdorferi | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| tpal | Treponema pallidum | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| ctra | Chlamydia trachomatis | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| cpne | Chlamydia pneumoniae | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| rpro | Rickettsia prowazekii | (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35) | ||
| Absolute Expression Experimente = | vegsam | GeneChip mRNA expression analysis of 6200 yeast ORFs under vegetative growth conditions. | (48) | |
| vegyou | GeneChip mRNA expression analysis of 5455 yeast ORFs under vegetative growth conditions. | (49) | ||
| sage | mRNA expression analysis of 3788 yeast ORFs determined by SAGE. | (43) | ||
| matea | GeneChip mRNA expression analysis of yeast mating type a strain grown on glucose. | (50) | ||
| mateal | GeneChip mRNA expression analysis of yeast mating type α strain grown on glucose. | (50) | ||
| gal | GeneChip mRNA expression analysis of yeast mating type a strain grown on galactose. | (50) | ||
| heat | GeneChip mRNA analysis of yeast mating type a strain grown on glucose at 30°C before a 39°C heat shock. | (50) | ||
| ref | Reference transcriptome. This is a scaling and merging of the above experiments. | (46) | ||
| Microarray Experimentr = | cdc28 | cDNA microarray genome-wide characterization of mRNA transcript levels for CDC28 synchronized yeast cells during the cell cycle. | (69) | |
| cdc15 | cDNA microarray genome-wide characterization of mRNA transcript levels for CDC15 synchronized yeast cells during the cell cycle. | (69) | ||
| alpha | Analysis using cDNA microarrays of yeast mRNA levels after synchronization of cell cycle via α arrest factor. | (69) | ||
| diaux | Genome-wide cDNA microarray analysis of the temporal program of yeast mRNA expression accompanying the metabolic shift from fermentation to respiration. | (70) | ||
| spor | cDNA microarray genome-wide analysis to assay changes in gene expression during sporulation. | (71) | ||
| heatec | cDNA microarray experiment and analysis on 4290 E.coli ORFs after exposure of the bacteria to heat shock. | (72) | ||
| deve | Analysis of genome wide changes during successive larval stages using cDNA microarrays of ∼12 000 C.elegans ORFs. | (73) | ||
| Pair-setf = | all | All pairs within a PART included in the calculations in Wilson et al. (For example, for fold rankings this would be the total number of pairs within a fold.) | (39) | |
| foldonly | A subset of the pair-set ‘all’ that only includes pairs between structures that are in the same PART but different sub-PART. (If PART is fold, then sub-PART is superfamily; If PART is superfamily, then sub-PART is family.) | (39) | ||
| Amino acid a= | Ala, Cys, Asp, Glu, Phe, Gly, His, Ile, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Val, Trp, Tyr. | (31) | ||
| Datasetp = | pdb100 | All structures within the fold (as defined by SCOP pdb100d). | (31) | |
| pdb40 | Similar to pdb100 but now using a version of the PDB clustered at 40% similarity (as defined by SCOP pdb40d) | (31) | ||
| Interaction typey = | pdball | Interactions for a PART are computed with all other PARTS in the PDB databank based on the distances between atoms in the coordinate files. Five or more contacts between atoms separated by <5 Å was considered a valid PART–PART contact. | (9,51,55) | |
| pdba | A subset of ‘pdball’. Interactions for a PART are computed just with all-α proteins (SCOP class 1) in the PDB. | (9,51,55) | ||
| pdbb | Similar to ‘pdba’ but now just with all-β proteins (SCOP class 2). | (9,51,55) | ||
| pdbab | Similar to ‘pdba’ but now just with mixed helix-sheet proteins (SCOP class 3 and 4) | (9,51,55) | ||
| scerall | Interactions for a PART are computed with all other PARTS based on the yeast two-hybrid experimental data. In particular, interactions between structural domains in the yeast genome were obtained by assigning protein structures to the yeast proteins. Structural domains contained within the same ORF that were within 30 amino acids were assumed to interact in an intramolecular fashion. To derive intermolecular interactions, we combined three sets of protein–protein interactions: (i) the MIPS web pages on complexes and pairwise interactions (February 2000) (9), (ii) the global yeast two-hybrid experiments by Uetz et al. (51) and (iii) large-scale yeast two-hybrid experiments by Ito et al. (52). Out of all these pairwise interactions known for yeast ORFs, there is a limited set in which both partners are completely covered by one structural domain (to within 100 residues). | (9,51,55) | ||
| scera | A subset of ‘scerall’. Interactions for a PART are computed just with all-α proteins (SCOP class 1) in the yeast experiment. | (9,51,55) | ||
| scerb | Similar to ‘scera’ but now just with all-β proteins (SCOP class 2). | (9,51,55) | ||
| scerab | Similar to ‘scera’ but now just with mixed helix-sheet proteins (SCOP class 3 and 4). | (9,51,55) | ||
| Interaction restrictionc = | inter | The interaction must occur between PARTS in different chains | (9,51,55) | |
| intra | The interaction must occur between PARTS in the same chain. | (9,51,55) | ||
| none | The union of ‘inter’ and ‘intra’. Interactions can occur in PARTS on the same or different chains. | (9,51,55) | ||
| Motion statistics = | nresidue | Number of residues. | (56,57) | |
| maxcadev | Maximal displacement of a Cα atom, in Å, of any residue during the motion (after fitting on the first core). | (56,57) | ||
| rmsoverall | Overall RMS of two structures after they are superimposed by a sieve-fit technique. Note that they are larger than traditionally used RMS. | (56,57) | ||
| nhinges | Number of hinges involved in the motion. | (56,57) | ||
| kappa | The rotation (in degrees) around the screw axis necessary to superimpose two domains of motion. | (56,57) | ||
| transe | Transition energy of the motion (maximum energy less minimum energy over the motion) (in kcal/mol). | (56,57) | ||
| deltae | Absolute value of energy difference between the ‘starting’ and ‘ending’ conformations of a motion (in kcal/mol). | (56,57) | ||
| Motion datasetd = | goldstd | List of approximately 220 ‘gold-standard’ manually curated motions | (56,57) | |
| auto | List of approximately 4000 conformational different proteins based on analyzing the SCOP database for similar proteins with large conformational differences (as measured by RMS) but close sequence similarity. | (56,57) | ||
| Transposon conditionsb = | caff | YPD + 8mM caffeine. | (58) | |
| cyss | Cyclohexmide hypersensitivity: YPD + 0.08 µg/ml cycloheximide at 30°C. | (58) | ||
| wr | White/red colour on YPD. | (58) | ||
| ypg | YPGlycerol. | (58) | ||
| calcs | Calcofluor hypersensitivity: YPD+12µg/ml calcoluor at 30°C. | (58) | ||
| hyg | YPD + 46µg/ml hygromycin at 30°C. | (58) | ||
| sds | YPD + 0.003% SDS. | (58) | ||
| bens | Benomyl hypersensitivity: YPD + 10 µg/ml benomyl. | (58) | ||
| bcip | YPD + 5-bromo-4-chloro-3-indolyl phosphate at 37°C | (58) | ||
| mb | YPD + 0.001% methylene blue at 30°C. | (58) | ||
| benr | Benomyl resistance: YPD + 20 µg/ml benomyl. | (58) | ||
| ypd37 | YPD at 37°C. | (58) | ||
| egta | YPD + 2mM EGTA | (58) | ||
| mms | YPD + 0.008% MMS. | (58) | ||
| hu | YPD + 75mM hydroxyurea. | (58) | ||
| ypd11 | YPD at 11°C. | (58) | ||
| calcr | Calcofluor resistance: YPD + 0.3 µg/ml calcofluor at 30°C. | (58) | ||
| cycr | Cyclohexmide resistance: YPD + 0.3 µg/ml cycloheximide. | (58) | ||
| hhig | Hyperhaploid invasive growth mutants. | (58) | ||
| nacl | YPD + 0.9 M NaCl. | (58) | ||
| Misc. quantitiesq = | pseu | Number of pseudogenes in worm genome matching a particular PART. | (59) | |
| func | Total number of functions associated with this PART. (In this survey all non-enzyme functions were lumped into a single category.) | (60) | ||
| enz | Total number of enzymatic functions associated with this PART. | (60) | ||
| size | Average length of a PART in the pdb40d clustering of the PDB. | |||
| age | The year of the first structure that is part of the PART was determined. |
The formalism for specifying an attribute has two parts: an overall category, denoted by a single uppercase symbol, and some parameter choices, which are denoted by lower-case arguments to the first symbol. Some examples for folds will suffice to make this clear: G(aful) is genome occurrence of a particular fold in A.fulgidus; M(nhinges,goldstd) is the maximum value of the number of hinges statistic from surveying a set of motions in the gold-standard subset of the Macromolecular Motions Database, where this statistic is only calculated for the entries in the motions database that are associated with a particular fold; and I(pdball,inter) is the number of distinct types of protein-protein interactions found in a survey of the PDB, subject to the restriction that the interactions must be between folds on different chains.
Genome occurrence
The data in this category reveal fold occurrences in 20 different genomes, including four archaea, two eukaryotes and 14 bacteria (additional details online).
The data were obtained in the following fashion: Once a library of folds has been constructed, representative sequences can be extracted (31). Then one can use these to search genomes by comparing each representative sequence against the genomes using the standard pairwise comparison programs, FASTA (32) and BLAST (33) and well-established thresholds (34).
Alternatively, one can build up profiles by running each representative sequence against PDB with PSI-BLAST and then comparing these profiles against each of the genomes. This latter procedure is more sensitive than pairwise comparison and relatively efficient once the profiles are made up. However, in doing large-scale surveys one has to be conscious of the potential biases introduced due to the profiles being more sensitive for larger families, which often results in the big families getting even bigger.
After the structure assignment, it becomes easy to enumerate how often a fold or structure feature occurs in a given genome or organism. Detailed information can be found in previous reports (H.Hegyi, J.Lin and M.Gerstein, manuscript submitted; 19,35,36). This pools assignments from previous work (37,38).
Alignment
Number of structures. We did a comprehensive set of structural alignments of structures in the PDB structure databank (39–41). The number of structures and aligned pairs used in these comparisons, which are based around Astral (31), give approximate measures of the occurrence of folds in the PDB. Comparison of these values to those for genome occurrence provides a measure of how biased the composition of the PDB is (42).
Sequence diversity. The scores from the alignments indicate the sequence diversity between the related structures within folds or superfamilies, in terms of percentage sequence identity and a sequence-based P value. P values are useful measures of statistical significance of the similarity calculation. A P value is the probability that one can obtain the same or better alignment score from a randomly composed alignment. A smaller P value is less likely to have been obtained by chance than a larger P value. Large P values close to 1.0 indicate that the similarity is characteristically random and thus insignificant.
Structural diversity. We also give analogous measures of the diversity of the structures with a given fold, allowing one to rank folds by their degree of variability. We tabulate untrimmed and trimmed RMS, along with the structural P value. RMS, root-mean-squared deviation in α carbon positions, has been the traditional statistic that gauges the divergence between two related structures. Smaller RMS scores indicate more closely related structures. However, sometimes a few ill-fitting atoms may significantly increase the RMS of structures known to be similar. To compensate for this we also report a ‘trimmed’ RMS for a conserved core structure, which is based on the better fitting half of the aligned α carbons, and structural P value, which compensates for other effects such as structure size. For details, see Wilson et al. (39).
Composition
This allows us to see which folds are most biased in composition of particular amino acids. We use various levels of the Astral clustering of the SCOP sequences to arrive at the composition (31).
Expression
Three techniques are frequently used to obtain genome-wide gene expression data. They are Affymetrix oligonucleotide gene chips, Serial Analysis of Gene Expression (SAGE) and cDNA microarrays (43–45). SAGE and, to some degree, gene chips measure the absolute expression levels (in units of mRNA transcripts per cell), while microarrays are used to obtain the expression level changes of a given open reading frame (ORF) as the ratio to a reference state.
A main motivation for expression experiments is often to study protein function and to characterize the functions of unannotated genes. However, this does not preclude relating other attributes of proteins, such as their structure, to expression data. For instance, it may be that highly expressed protein folds share a number of characteristics, such as a particularly stable architecture or a composition biased in a certain way. Relating expression and structure involved matching the PDB structure database against the genome and then summing the expression levels of all ORFs containing the same fold. However, if one is trying to find genes expressed in a particular metabolic state, PartsList is not the right place to look.
Absolute. The absolute expression level data gives a good representation of highly expressed genes. All the experiments currently indexed by PartsList are for yeast. For each experiment, in addition to ranking based on the average expression level for a fold, we also consider the composition in the transcriptome and the enrichment of this value relative to its composition in the genome. Transcriptome composition is the fractional composition of a fold (relative to that for other folds) in the mRNA population. In other words, it is the composition of a fold in the genome weighted by the expression levels of each of the genes. The enrichment is the relative change between the composition of a fold in the genome and the transcriptome. Further details are provided in previous reports (46,47). We report values for experiments from a number of different labs (43,48–50) and a single reference set that merges and scales all the expression sets together.
Ratio. The expression ratio data shows the most actively changing genes over a period of time (e.g. cell cycle) or based on a change in states (e.g. healthy versus diseased). Source data for expression ratios are the fluctuations in expression of a certain fold over a period of time (e.g. the cell cycle). These are measured in terms of standard deviations for a particular fold, which is calculated from the average of the expression ratio standard deviations for each gene that matches the fold structure.
Interactions
Information on protein–protein interactions is derived from surveys of the contacts in the PDB and the experiments in yeast.
PDB. To determine which domains interact with one another in the PDB entries indexed by SCOP (9580 at the time of the analysis), the coordinates of each domain were parsed to check whether there are five or more contacts within 5 Å to another domain, as described by Park et al. (51). The distance of 5 Å was chosen, as this is a conservative threshold for interaction between two atoms, where the atoms are either Cαs or atoms in side chains. The five-contact threshold was chosen to make sure the contact between the domains was reasonably extensive. (In fact, the number of domains identified as contacting each other hardly changed for thresholds between one and 10 contacts and 3 and 6 Å distances.)
Yeast. The interactions between structural domains in the yeast genome were obtained by assigning protein structures to the yeast proteins using PSI-BLAST and PDB-ISL as described by Teichmann et al. (52,53). Assigned structural domains contained within the same ORF that were adjacent within 30 amino acids were assumed to interact. (This is generally true of the domains in the PDB, with a few exceptions, such as domains in transcription factors like adjacent zinc fingers or variable and constant immunoglobulin domains.) To derive intermolecular interactions in the yeast genome we combined three sets of protein–protein interactions: (i) the MIPS web pages on complexes and pairwise interactions (February 2000) (9), (ii) the global yeast two-hybrid experiments by Uetz et al. (54) and (iii) large-scale yeast two-hybrid experiments by Ito et al. (55). Out of all these pairwise interactions known for yeast ORFs, there is a limited set in which both partners are completely covered by one structural domain (to within 100 residues). This set of protein pairs was used to derive a further set of domain contacts in the yeast genome as described by Park et al. (51).
Motions
Information on motions is from the Macromolecular Motions Database (56,57). We consider a set of approximately 4400 motions automatically identified by examining the PDB and a smaller, manually curated set of motions. For each fold we determine the number of entries in the motions database that are associated with it. Then, over this set of motions we either average or take the maximum value of a number of relevant statistics describing the motion, i.e. the maximum Cα displacement in the motion, the overall rotation of the motion and the energy difference between the start and endpoints of structures involved in the motion.
Transposon sensitivity
Ross-Macdonald et al. (58) developed a procedure for randomly inserting transposons throughout the yeast genome. They investigated the phenotypes resulting from each insertion in 20 different growth conditions in comparison to wild-type growth. The experiment for each insertion in each condition was repeated several times. If the observed phenotype of the mutant deviates from the average wild-type phenotype, this could be either because of a real effect of the mutation on the cell or it could just be a typical variation of the phenotype of wild-type cells. We developed a P value score that measures the degree of confidence that the observed phenotype results from randomly changing wild-type cells. The negative logarithm of this P value rises with the significance of the phenotype measurements and can be understood as the sensitivity of the cell to mutations in a particular gene. We calculated a value for the transposon sensitivity for protein folds by geometrically averaging the P values of the associated genes.
Miscellaneous
The miscellaneous section includes any information that does not fit into a major category. It includes: number of pseudogenes in worm associated with a fold (59), total number of functions and number of enzymatic functions associated with a fold (60), the average length of the sequence, and the year the domain structure was originally determined.
Errors
The above data, of course, have systematic and statistical errors. For some attributes we expect considerably smaller errors than others. For instance, we expect the numbers related to the sequence composition of different folds (e.g. the Ala composition) to be particularly accurate, since the only factors affecting these are errors in the underlying sequence of the protein and in the SCOP fold classification itself. In contrast, there is a considerable known rate of false positives associated with the global protein interaction experiments using the two-hybrid method (54,61), and this suggests statistics based on yeast interactions may be somewhat less accurate. Furthermore, the precise values for the rankings in PartsList are also contingent on the evolving contents of various databanks. Thus, over time as more structures are determined, one should expect statistics such as the most common folds in a particular genome to change somewhat. A very detailed discussion of the expected errors in the various quantities in PartsList is available on the web from the help section.
RANKING ALL THE FOLDS BASED ON EXTRINSIC INFORMATION
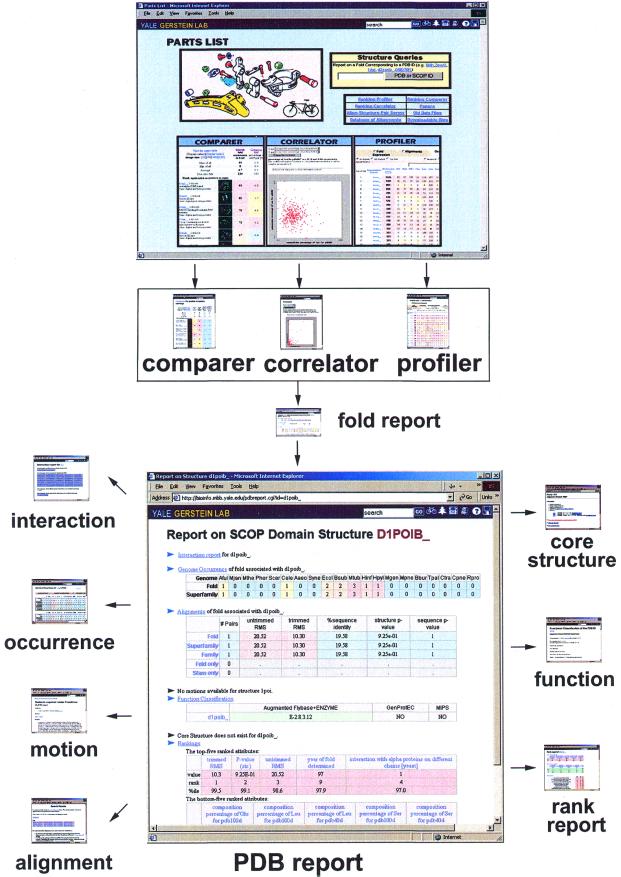
The PartsList resource facilitates exploring extrinsic information by dynamically ranking protein folds in different contexts, such as genome and expression levels. We provide three tools for visualizing the rankings: Comparer, Correlator and Profiler. The overall structure of PartsList is schematically shown in Figure 1.
Figure 1.
The overall structure of PartsList. Three tools (Profiler, Comparer and Correlator) provide an easy way to access and manipulate the display of the dataset. With these tools, users can isolate interesting folds and obtain fold reports about them. Further clicks take one to PDB report, which gives detailed information about an individual structural domain, including its genome occurrence, alignment information, molecular motions, functional annotation, interactions and core structure.
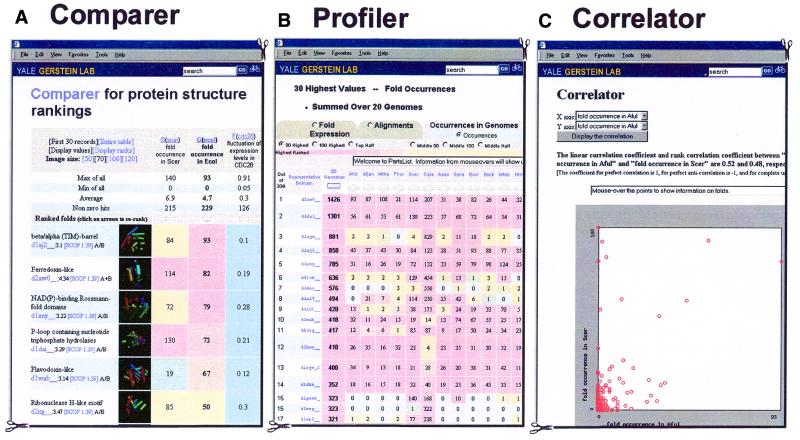
Comparer
The motivation behind Comparer is to allow one to rank folds according to a given attribute and then see the ranks associated with other attributes. The ranking attribute and the additional attributes are selected by the user. Figure 2A shows an example. The most common folds in E.coli are shown alongside three other attributes: fold occurrence in yeast, fluctuation in expression level during the yeast cell cycle and fluctuation in expression level in E.coli during heat shock. Which displayed attribute is used to rank the folds can be easily changed; in the example in Figure 2A the report can be re-sorted based on the other three attributes by clicking on arrows.
Figure 2.
Sample displays. (A) A sample Comparer display: the four selected attributes are the fold genome occurrence in yeast, the analogous quantity for E.coli, fluctuation of expression level for CDC28 synchronized yeast cell during the cell cycle, and the corresponding values for E.coli to heat shock. (Using the nomenclature in Table 1 these quantities are G(scer), G(ecol), F(cdc28) and F(heatec).) The folds are ranked in terms of fold occurrence in E.coli and the most common fold here is the TIM-barrel (represented by the SCOP domain d1aj2__). If one clicks the ‘Display ranks’ button, the values in the cells will be replaced by the ranks in their respective columns. By clicking the ‘re-rank’ arrows, one can also obtain other views by sorting on other attributes. (B) Shows the occurrences of folds in 20 genomes in Profiler. (C) Shows the correlation between the fold occurrences in the A.fulgidus and S.cerevisiae genomes [G(aful) and G(scer)]. Both linear and rank correlation coefficients are calculated. The linear correlation coefficient is defined as: R = [1/(N–1)]X·Y, where X and Y are two vectors with N elements. Each element of the X vector is normalized thus: Xi = (Xi′ – X)/σx, where X and σx are the average and standard deviation of the values of the original data vector X′, respectively. Y is normalized in a similar fashion. For two perfectly correlated datasets, R = 1, while for two completely uncorrelated datasets, R = 0. If we replace Xi by its rank among all the other Xi in the sample (i.e., 1,2,3 … N), then we get the rank correlation coefficient. A scatter plot is also shown to help in visualizing this correlation.
Profiler
In principle, Profiler presents the same information as Comparer. However, it shows the progressing pattern for several pre-selected categories and is intended to give people an easy-to-use interface that gives some simple views of the data. Figure 2B shows an example that highlights the phylogenetic pattern of fold occurrence in 20 genomes.
Correlator
Correlator uses linear and rank correlation coefficients to measure the association between two selected attributes. The difference between these two types of correlation coefficients is that the former relates to the actual values while the latter relates to the ranks among the samples. The interpretation of the linear correlation coefficient can be completely meaningless if the joint probability distribution of the variables is too different from a binormal distribution. This is the reason for introducing the rank correlation coefficient. Correlator provides both coefficients for the selected quantities. In most cases, they are close. For example, the linear correlation coefficient and rank correlation coefficient for fold occurrence in genomes Archaeoglobus fulgidus and Methanococcus jannaschii (Aful and Mjan) are 0.88 and 0.77, respectively, while the corresponding coefficients for fold occurrence in A.fulgidus and Saccharomyces cerevisiae (Scer) are 0.52 and 0.48, respectively. This is not surprising, as the first two genomes are both Archaeal, while in the second comparison one genome belongs to Archaea (Aful) and another to Eucarya (Scer). As one would expect, the fold occurrences for the more closely related genomes have a higher correlation.
In addition to the coefficients, Correlator displays a scatter plot to aid in visualizing the correlation between the selected fold attributes. Figure 2C shows the scatter plot for the second example above: the correlation between occurrences in the A.fulgidus and S.cerevisiae genomes. One can easily observe that some folds appear frequently in Scer but seldom or never in A.fulgidus. By clicking on a point on the plot, one obtains detailed information about the corresponding fold. This kind of plot can reveal interesting folds with certain relationships between attributes even though in some cases the overall correlation coefficients between the two attributes are almost zero (i.e. no correlation).
POWER-LAW BEHAVIOR OF MANY DISPARATE ATTRIBUTES
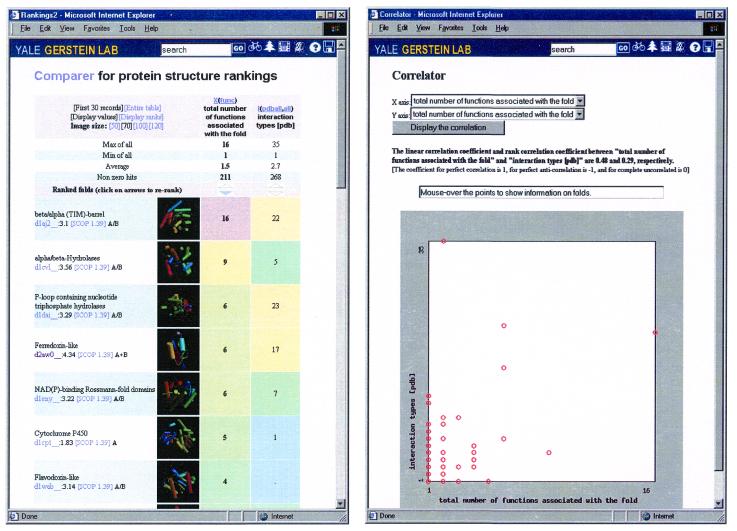
Going back and forth between Correlator and Comparer allows one to see interesting relationships between disparate attributes of proteins. Figure 3 illustrates a comparison of two attributes, functions and interactions. It shows a ranking of the folds that have the most interactions in the PDB in comparison to those that have the most functions. It is immediately apparent that there are only a few folds with large values of either attribute, i.e. many functions or interactions. Moreover, the most multi-functional folds also have the most distinct interactions with other folds, suggesting that a few a folds may function as general-purpose parts.
Figure 3.
The relation between the number of functions associated with a protein fold and the number of distinct protein–protein interactions it has (based on a survey of the PDB databank). These are X(func) and I(pdball,none) using the nomenclature in Table 1. This relationship can be displayed both in Comparer (left) and Correlator (right).
In fact, the uniform system of ranks in PartsList shows that ‘only a few folds having large values for an attribute’ is a generally true statement for many of the disparate attributes catalogued by the system. Moreover, the falloff from high to low values for a given attribute often follows a power-law distribution. That is, the normalized frequency F that a number of distinct folds have a particular attribute value V follows a functional form like:
F(V) = aV–b
where a and b are constants. Note that F(V) is just the number of folds with an attribute value V divided by the total number of folds and that on a log–log plot this function becomes a straight line with slope –b. Often the attribute value V itself reflects the ‘occurrence’ of a fold in a particular context, e.g. V could be the number of times a given fold occurs in a particular genome. Quantities that follow a power-law-like behavior are often said to have a form like that of Zipf’s law, which often occurs in the analysis of word frequency in documents (62).
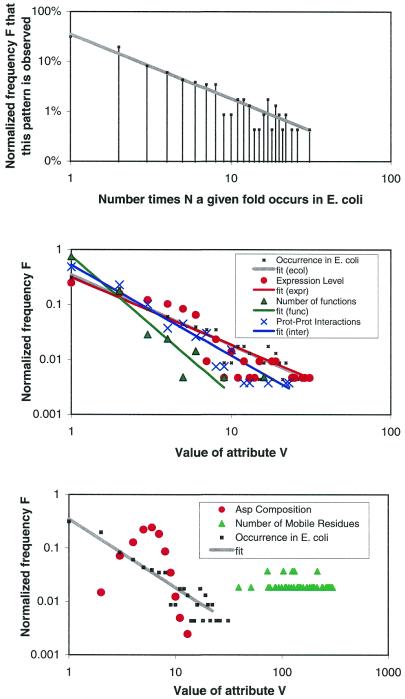
Thus far, this general conclusion is described in language sufficiently abstract to accommodate the many different types of attributes in PartsList. A few concrete examples will make the conclusion clearer. For instance, we find that in genomes most folds occur only once while there are only a very few folds that occur many times. An illustration is shown in the upper panel of Figure 5 for E.coli. The x-axis is the number of times a particular fold occurs in the E.coli genome and the y-axis shows the number of distinct folds that have same occurrence. (This is normalized by dividing by the total number of folds so that the maximum value on y-axis is 100%.) From the log–log format of the plot, one can immediately see that the falloff obeys a power-law, with a few folds occurring many times and most only once or twice. The middle panel shows other attributes that display similar power-law-like behavior, including expression level in yeast, number of functions associated with a fold, and number of protein–protein interactions found in the PDB. Of course, not all attributes follow a power-law. The lower panel shows two of these less typical attributes: Asp composition in a fold and average number of residues involved in a motion.
Figure 5.
Some novel relationships that are highlighted by the PartsList system. (Upper panel) The occurrence of folds in the E.coli genome plotted on a log–log scale, i.e. G(ecol) using the nomenclature in Table 1. The x-axis is the fold occurrence in the genome, while the y-axis is the number of folds with a particular occurrence. The fit of the points to a straight line shows that the falloff obeys a power-law with constants a = 0.35 and b = 1.3 (see text). (Middle panel) Other attributes that also follow power-law behavior: the average expression level according to our merged and scaled set [L(ref) with a = 0.3 and b = 1.2), the number of protein–protein interactions [I(pdball,none) with a = 0.52 and b = 1.6], and the number of functions [X(func) with a = 0.76 and b = 2.5]. (Lower panel) Some attributes that do not follow power-law behavior: the Asp composition of the fold [B(Ala,pdb100)] and the number of mobile residues during a motion [M(nresidue,auto)]. The fold occurrence in E.coli is plotted as a reference.
One of the strengths of the uniform numerical system of ranks in PartsList is that it puts everything into a common framework so that one can see similarities across disparate attributes. We believe it would be difficult to see a common power-law behavior for many aspects of protein structure without PartsList.
TRADITIONAL SINGLE-STRUCTURE REPORTS
In addition to the tools that compare and relate the extrinsic properties of protein folds, we provide traditional reports that are more focused on an individual structure.
Occurrence report. This allows users to see the number of times that a fold corresponding to the queried protein structure occurs in various genomes. This gives a phylogenetic profile of the occurrence of a particular fold in 20 genomes, similar in spirit to the fold patterns discussed earlier (19).
Function report. This summarizes the functional classification of the queried PDB structure. It merges a number of functional classifications, including FlyBase (10), ENZYME (63), GenProtEC (64) and MIPS (9). Our approach to functional classification is described in a number of previous publications (e.g. 39,60). In short, we used pairwise comparison to cross-reference the PDB domains against SWISS-PROT. Depending on whether they had an Enzyme Commission (EC) number, we were able to divide all entries into enzymes and non-enzymes, a division that represents the highest level in our classification. (For the enzyme category, we only transferred EC numbers to those SCOP domains with a one-to-one match to a SWISS-PROT enzyme.) In the absence of an EC-type classification for non-enzymes, we assigned functions to non-enzymatic SCOP domains according to Ashburner’s original classification of Drosophila protein functions. This classification is derived from a controlled vocabulary of fly terms, is available on the web and is loosely connected with the FlyBase database (10). It has recently been superceded by the GO functional classification (65). MIPS and GenProtEC classifications to SCOP domains were assigned based on sequence comparisons to classified yeast and E.coli ORFs, respectively. The SCOP domain most closely matching each ORF classified in MIPS or GenProtEC was assigned the corresponding MIPS or GenProtEC function number. Only matches of ≥80% sequence identity were considered.
Alignment report. This gives detailed information on structural alignments available between pairs of protein domains associated with a fold. A pair viewer is provided, which gives many key statistics about the alignment (e.g. RMS, sequence identity, number of fit atoms, etc.), in addition to a listing of the actual aligned residues. Both HTML and parseable text views are available.
Interaction report. This shows all the pairs of protein–protein interactions associated with a fold based on either the PDB survey or yeast genome data.
Rank report. This highlights the top-five and bottom-five ranked attributes associated with a fold. It also shows all attributes ordered by the rank they are given in that fold. Thus, it highlights for a particular fold the attributes with respect to which it most stands out. That is, it highlights the ‘outlier attributes’ of each fold, the way each fold is most unique. The rank report could be used, for example, by a protein engineer interested in determining the unique properties of a structure he is working on.
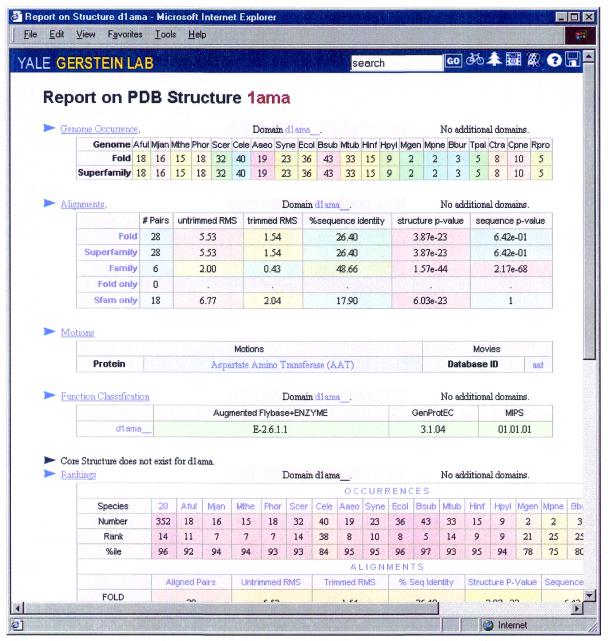
PDB report. This summarizes all the information concerning a domain or a representative PDB structure. It includes: (i) a summary of the occurrence report; (ii) a summary of the alignments available for structures in the same superfamily and fold; (iii) a description of motions and motion-movies associated with the structure in the Macromolecular Motions database (56,57); (iv) a summary of the merged functional classification; (v) a core structure, if available (66); (vi) ranking tables of the queried structure in various datasets; and (vii) a summary of the interactions report. Figure 4 shows a sample PDB report for structure 1AMA.
Figure 4.
A sample PDB report for structure 1AMA. The report summarizes the relevant information for this domain, including genome occurrences, alignment, motions, function classification, core structure and rankings. By clicking on the headers, one can get the detailed reports for these quantities.
Fold report. This lists all the SCOP domains associated with the queried fold and provides information (similar to that in the PDB report) that is common to all, i.e. genome occurrence, alignment report and rankings.
DISCUSSION
We developed a web-based system for dynamically ranking protein folds based on disparate attributes, including fold occurrence in various genomes, expression level, alignment statistics, protein–protein interactions, motion statistics and transposon sensitivity. Three ranking tools are provided, Comparer, Profiler and Correlator, which can help users to place one fold in context of all other ones. The uniform system of ranks employed by PartsList provides a good framework for comparing different experiments and gaining a broad perspective on the complexity of genomes.
We anticipate that PartsList will have a relatively stable number of entries (i.e. folds), while for each entry the attributes that describe it will increase over time. In the future, as experiments yield new information, PartsList will include more and more attributes. In particular, we anticipate that much new expression information will be incorporated. We also plan to develop a form to allow automatic submission of new ranking attributes and to encourage people to submit any ranking information.
Acknowledgments
ACKNOWLEDGEMENTS
We thank NIH Structural Genomics Program and the Keck Foundation for support.
References
- 1.Chothia C. (1992) Proteins. One thousand families for the molecular biologist. Nature, 357, 543–544. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S.E., Hubbard,T., Murzin,A. and Chothia,C. (1995) Gene duplications in H. influenzae. Nature, 378, 140. [DOI] [PubMed] [Google Scholar]
- 3.Wolf Y.I., Grishin,N.V. and Koonin,E.V. (2000) Estimating the number of protein folds and families from complete genome data. J. Mol. Biol., 299, 897–905. [DOI] [PubMed] [Google Scholar]
- 4. The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 5.Berman H.,M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski R.A., Hutchinson,E.G., Michie. A.D., Wallace,A.C., Jones,M.L. and Thornton,J.M. (1997) PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci., 22, 488–490. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Addess,K.J., Geer,L., Madej,T., Marchler-Bauer,A., Zimmernan,D. and Bryant,S.H. (2000) MMDB: 3D structure data in Entrez. Nucleic Acids Res., 28, 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Issel-Tarver,L., Kasarskis,A., Scafe,C.R., Sherlock,G., Binkley,G., Jin,H., Kaloper,M., Orr,S.D., Schroeder,M., Weng,S., Zhu,Y., Botstein,D. and Cherry,J.M. (2000) Integrating functional genomic information into the Saccharomyces genome database. Nucleic Acids Res., 28, 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frishman D., Heumann,K., Lesk,A. and Mewes,H.W. (1998) Comprehensive, comprehensible, distributed and intelligent databases: current status. Bioinformatics, 14, 551–561. [DOI] [PubMed] [Google Scholar]
- 10.The FlyBase Consortium. (1999) The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res., 27, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aach J., Rindone,W. and Church,G.M. (2000) Systematic management and analysis of yeast gene expression data. Genome Res., 10, 431–445. [DOI] [PubMed] [Google Scholar]
- 13.Bader G.D. and Hogue,C.W. (2000) BIND—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics, 16, 465–477. [DOI] [PubMed] [Google Scholar]
- 14.Xenarios I., Rice,D.W., Salwinski,L., Baron,M.K., Marcotte,E.M. and Eisenberg,D. (2000) DIP: the database of interacting proteins. Nucleic Acids Res., 28, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konopka A.K. and Martindale,C. (1995) Noncoding DNA, Zipf’s law, and language. Science, 268, 789. [DOI] [PubMed] [Google Scholar]
- 17.Flam F. (1994) Hints of a language in junk DNA. Science, 266, 1320. [DOI] [PubMed] [Google Scholar]
- 18.Bornberg-Bauer E. (1997) How are model protein structures distributed in sequence space? Biophys. J., 73, 2393–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerstein M. (1998) Patterns of protein-fold usage in eight microbial genomes: a comprehensive structural census. Proteins, 33, 518–534. [DOI] [PubMed] [Google Scholar]
- 20.Gerstein M. (1997) A structural census of genomes: comparing eukaryotic, bacterial and archaeal genomes in terms of protein structure. J. Mol. Biol., 274, 562–576. [DOI] [PubMed] [Google Scholar]
- 21.Jeong H., Tombor,B., Albert,R., Oltvai,Z.N. and Barabasi,A.L. (2000) The large-scale organization of metabolic networks. Nature, 407, 651–654. [DOI] [PubMed] [Google Scholar]
- 22.Amaral L.A.N., Scala,A., Barthelemy,M. and Stanley,H.E. (2000) Classes of small-world networks Proc. Natl Acad. Sci. USA, 97, 11149–11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 24.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH—a hierarchic classification of protein domain structures. Structures, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 25.Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- 26.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 337–385. [DOI] [PubMed] [Google Scholar]
- 27.Madej T., Gibrat,J.-F. and Bryant,S.H. (1995) Threading a database of protein cores. Proteins, 23, 356–369. [DOI] [PubMed] [Google Scholar]
- 28.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L.L. (1999) The Pfam protein families database. Nucleic Acids Res., 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner S.E., Koehl,P. and Levitt,M. (2000) The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res., 28, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipman D.J. and Pearson,W.R. (1985) Rapid and sensitive protein similarity searches. Science, 227, 1435–1441. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 34.Brenner S., Chothia,C. and Hubbard,T. (1998) Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl Acad. Sci. USA, 95, 6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerstein M. and Levitt,M. (1997) A structural census of the current population of protein sequences. Proc. Natl Acad. Sci. USA, 94, 11911–11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teichmann S., Chothia,C. and Gerstein,M. (1999) Advances in structural genomics. Curr. Opin. Struct. Biol., 9, 390–399. [DOI] [PubMed] [Google Scholar]
- 37.Gerstein M., Lin,J. and Hegyi,H. (2000) Protein folds in the worm genome. Pac. Symp. Biocomput., 5, 30–42. [DOI] [PubMed] [Google Scholar]
- 38.Lin J. and Gerstein,M. (2000) Whole-genome trees based on the occurrence of folds and orthologs: implications for comparing genomes on different levels. Genome Res., 10, 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson C.A., Kreychman,J. and Gerstein,M. (2000) Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol., 297, 233–249. [DOI] [PubMed] [Google Scholar]
- 40.Levitt M. and Gerstein,M. (1998) A unified statistical framework for sequence comparison and structure comparison. Proc. Natl Acad. Sci. USA, 95, 5913–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerstein M. and Levitt,M. (1998) Comprehensive assessment of automatic structural alignment against a manual sandard, the Scop classification of proteins. Protein Sci., 7, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerstein M. (1998) How representative are the known structures of the proteins in a complete genome? A comprehensive structural census. Fold. Des., 3, 497–512. [DOI] [PubMed] [Google Scholar]
- 43.Velculescu V.E., Zhang,L., Zhou,W., Vogelstein,J., Basrai,M.A., Bassett,D.E.,Jr, Hieter,P., Vogelstein,B. and Kinzler,K.W. (1997) Characterization of the yeast transcriptome. Cell, 88, 243–251. [DOI] [PubMed] [Google Scholar]
- 44.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nat. Genet., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 45.Lipshutz R.J., Fodor,S.P., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nat. Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 46.Jansen R. and Gerstein,M. (2000) Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res., 28, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerstein M. and Jansen,R. (2000) The current excitement in bioinformatics-analysis of whole-genome expression data: how does it relate to protein structure and function Curr. Opin. Struct. Biol., 10, 574–584. [DOI] [PubMed] [Google Scholar]
- 48.Jelinsky S.A. and Samson,L.D. (1999) Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl Acad. Sci. USA., 96, 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 50.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 51.Park J., Lappe,M. and Teichmann,S.A. (2001) Mapping protein family interactions: intra- and intermolecular interactions repertoires are distinct. J. Mol. Biol., 307, 929–939. [DOI] [PubMed] [Google Scholar]
- 52.Teichmann S., Chothia,C., Church,G. and Park,J. (2000) Fast assignment of protein structures to sequences using the intermediate sequence library PDB-ISL. Bioinformatics, 16, 117–124. [DOI] [PubMed] [Google Scholar]
- 53.Teichmann S.A., Park,J. and Chothia,C. (1998) Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements. Proc. Natl Acad. Sci. USA, 95, 14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P., Qureshi-Emili,A., Li,Y., Godwin,B., Conover,D., Kalbfleisch,T., Vijayadamodar,G., Yang,M., Johnston,M., Fields,S. and Rothberg,J.M. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 55.Ito T., Tashiro,K., Muta,S., Ozawa,R., Chiba,T., Nishizawa,M., Yamamoto,K., Kuhara,S. and Sakaki,Y. (2000) Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl Acad. Sci. USA, 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerstein M. and Krebs,W. (1998) A database of macromolecular motions. Nucleic Acids Res., 26, 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krebs W. and Gerstein,M. (2000) The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res., 28, 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ross-Macdonald P., Coelho,P.S., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K., Sheehan,A., Symoniatis,D., Umansky,L., Heidtman,M., Nelson,F.K., Iwasaki,H., Hager,K., Gerstein,M., Miller,P., Roeder,G.S. and Snyder,M. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 59.Harrison P., Echols,N. and Gerstein,M. (2001) Digging for dead genes: an analysis of the characteristics of the pseudogene population in the C. elegans genome. Nucleic Acids Res., 29, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hegyi H. and Gerstein,M. (1999) The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J. Mol. Biol., 228, 147–164. [DOI] [PubMed] [Google Scholar]
- 61.Schwikowski B., Uetz,P. and Fields,S. (2000) A network of protein–protein interactions in yeast. Nat. Biotechnol., 18, 1257–1261. [DOI] [PubMed] [Google Scholar]
- 62.Knuth D. (1973) The Art of Computer Programming 3. Addison-Wesley, Reading, MA.
- 63.Bairoch A. (1993) The ENZYME data bank. Nucleic Acids Res., 21, 3155–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riley M. and Labedan,B. (1996) E. coli gene products: physiological functions and common ancestries. In Neidhardt,F., Curtiss,R.,III, Lin,E.C.C., Ingraham,J., Low,K.B., Magasanik,B., Reznikoff,W., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 2118–2202.
- 65.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock,G. (2000) Gene ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt R.B., Gerstein,M. and Altman,R.B. (1997) LPFC: an internet library of protein family core structures. Protein Sci., 6, 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drawid A., Jansen,R. and Gerstein,M. (2000) Genome-wide analysis relating expression level with protein subcellular localization. Trends Genet., 16, 426–429. [DOI] [PubMed] [Google Scholar]
- 68.Park J., Karplus,K., Barrett,C., Hughey,R., Haussler,D., Hubbard,T. and Chothia,C. (1998) Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol., 284, 1201–1210. [DOI] [PubMed] [Google Scholar]
- 69.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K. Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeRisi J.L., Iyer,V.R. and Brown P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 71.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 72.Richmond C.S., Glasner,J.D., Mau,R., Jin,H. and Blattner,F.R. (1999) Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wixon J., Blaxter,M., Hope,I., Barstead,R. and Kim,S. (2000) Caenorhabditis elegans. Yeast, 17, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]