Abstract
Collaborator of ARF (CARF) was cloned as an ARF-interacting protein and shown to regulate the p53–p21WAF1–HDM2 pathway, which is central to tumor suppression via senescence and apoptosis. We had previously reported that CARF inhibition in cancer cells led to polyploidy and caspase-dependent apoptosis, however, the mechanisms governing this phenomenon remained unknown. Thus, we examined various cell death and survival pathways including the mitochondrial stress, ataxia telangiectasia mutated (ATM)–ATR, Ras–MAP kinase and retinoblastoma cascades. We found that CARF is a pleiotropic regulator with widespread effects; its suppression affected all investigated pathways. Most remarkably, it protected the cells against genotoxicity; CARF knockdown elicited DNA damage response as evidenced by increased levels of phosphorylated ATM and γH2AX, leading to induction of mitotic arrest and eventual apoptosis. We also show that the CARF-silencing-induced apoptosis in vitro translates to in vivo. In a human tumor xenograft mouse model, treatment of developing tumors with short hairpin RNA (shRNA) against CARF via an adenovirus carrier induced complete suppression of tumor growth, suggesting that CARF shRNA is a strong candidate for an anticancer reagent. We demonstrate that CARF has a vital role in genome preservation and tumor suppression and CARF siRNA is an effective novel cancer therapeutic agent.
Keywords: cell death, mechanisms, stress pathways, ATM/ATR, mitotic catastrophe
Tumor suppression largely function via the p53–p21WAF1–HDM2 pathway for promotion of cell growth arrest, senescence, or programmed cell death to eliminate damaged cells or to halt neoplastic activities.1 We had previously isolated an alternative reading frame (ARF)-binding protein, Collaborator of ARF (CARF), which functions in regulation of p53-mediated tumor suppression.2 Further, we discovered that the CARF protein is a vital regulator of p53-dependent senescence and apoptosis, and the determinant of cell fate is reliant on the dosage or level of CARF.3 CARF is critical for cell cycle progression, and CARF-compromised cells display hallmarks of mitotic catastrophe (MC) including hypercondensed chromatin, polyploidy, as well as deranged spindle fiber and centrosome assemblies. In this study, we sought to elucidate the mechanisms responsible for CARF-suppression-induced cell death and its in vivo effects.
MC is characterized by mitotic arrest accompanied by increases in histone H3, cyclin B1 and cyclin-dependent kinase 1 and deregulation of the cell cycle checkpoints, culminating into cell death.4 Although a clear definition of MC is lacking, it is widely considered as a mode of apoptosis owing to accompanying changes in mitochondrial membrane potential and caspase activation.5 Regulation of the cell survival and death processes has been largely attributed to p53-dependent and p53-independent pathways involving retinoblastoma (RB), E2F1, p21WAF1, Ras–mitogen-activated protein kinases (MAPK) and ataxia telangiectasia mutated (ATM)/ATM- and RAD3-related (ATR) functions.6, 7, 8, 9 The latter serve as prime mediators of the DNA damage response, instigating apoptosis through Ras–MAPK, RB–E2F1 and ARF–p53–p21WAF1 or mitotic DNA damage checkpoint mediated by the BRCA1 and CHK1 pathways.10, 11, 12, 13, 14
In the present study, we report that the suppression of CARF induces MC accompanied by activation of the mitochondrial stress and caspase-dependent pathways via induction of DNA damage and disruption of the cell cycle checkpoint regulation, culminating into apoptosis of cancer cells. Furthermore, in an in vivo tumor model using adeno-oncolytic virus armed with CARF siRNA, complete suppression of tumors was observed, suggesting that CARF siRNA is a strong candidate for antitumor therapy.
Results
CARF-silencing-induced cell death is p53-independent and involves the mitochondrial stress pathway
We previously showed that the suppression of CARF compromised p53 function causing reduction in the level of p21WAF1 expression.15 However, it remained unclear whether functional compromise of p53 was critically involved in the cell death phenotype. CARF-silencing induced cell death in HeLa (compromised p53 function owing to the presence of human papilloma virus; Figure 1a), DLD-1 and C33A (mutant p53; data not shown), as well as in HCT116 p53−/− cells (Figure 1b). These data indicated that p53 is not a crucial factor for CARF-silencing-induced cell death and hence, other factors and pathways warranted further investigations.
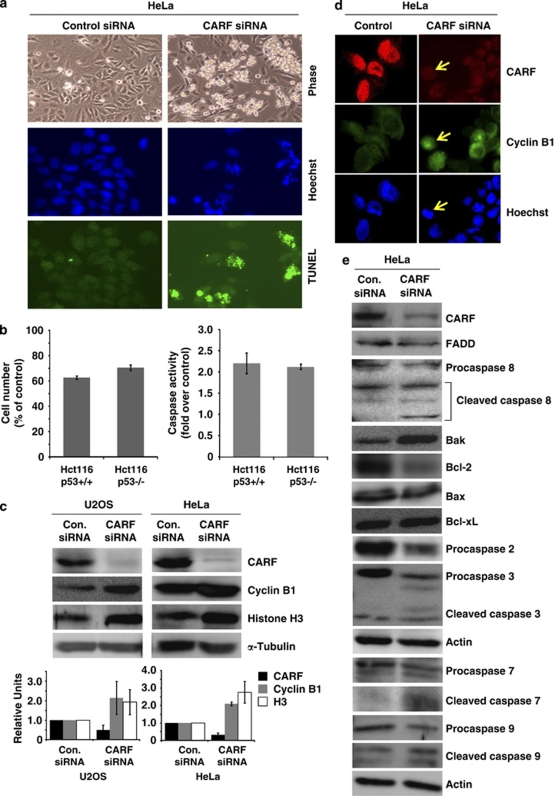
Figure 1.
Cell death induced by CARF suppression occurs after mitotic arrest through the mitochondrial stress and caspase-dependent pathway. TUNEL staining of HeLa cells transfected with CARF-targeting siRNA shows increased cell death following CARF suppression (a). Syngeneic p53 +/+ and p53 −/− HCT116 cells showed comparable apoptosis (decreased cell viability, left and increase in caspase 3 activity, right) after CARF suppression (b). CARF-compromised cells underwent mitotic arrest as evidenced by accumulation of cyclin B1 and histone H3 with densitometric quantitation of representative blots from at least three experiments, in which the CARF-suppressed group is shown as fold change over control, which was set as 1 (c). Immunofluorescence shows that cells with reduced CARF levels had increased cyclin B1 nuclear accumulation, as shown by the arrows (d). CARF-compromised HeLa cells were subjected to immunoblotting analyses for proteins constituting the mitochondrial stress and caspase pathways (e). Actin and α-Tubulin were used as loading controls. Graphs are represented as average mean±S.D. (f) Schematic diagram of the hypothetical pathways that lead to cell death following CARF inhibition. We considered that apoptosis induced by CARF suppression might activate multiple pathways, including the ATM/ATR/CHK1/CHK2, Ras-MAPK and/or RB/E2F1 networks
We utilized CARF siRNA in U2OS (functional wild-type p53) and HeLa cells and examined the expression of cyclin B1 and histone H3 (crucial regulators of mitosis and major markers of MC).5 As shown in Figure 1c, cyclin B1 and histone H3 were increased following CARF suppression. In contrast to the predominantly pancytoplasmic cyclin B1 in normal cycling cells, cyclin B1 accumulated in the nucleus of CARF-compromised cells (Figure1d, arrows), which exhibited compact and condensed chromosomes as in prophase and metaphase cells, suggesting that the CARF-compromised cells were arrested at mitosis owing to inhibition of cyclin B1 degradation that normally occurs for mitotic exit and may have thus undergone MC before cell death.16, 17 The fact that there was no change in FADD expression after CARF suppression (Figure 1e) was suggestive that the CARF suppression was not recognized as an external stress. On the other hand, specific activation of mediators of the internal stress response-apoptosis pathway, such as upregulation of Bak (pro-apoptotic protein) and downregulation of Bcl-2 (antiapoptotic protein), were observed (Figure 1e) suggesting that CARF suppression was recognized as an internal stress response leading to cleavage and activation of caspases 2, 3, 7 and 9. The data suggested that the CARF-silencing-induced apoptosis was mediated predominantly by the mitochondrial–internal stress pathway.18, 19 To elucidate the mechanistic processes involved in this phenomenon, we next investigated three major cell stress pathways, including the Ras–MAPK, RB–E2F1 and ATM–ATR–CHK cascades, involved in p53-independent growth arrest and cell death (Figure 1f).
Ras pathways are activated, but not essential in CARF-silencing-induced cell death
We earlier showed that normal cells undergo stress-induced premature senescence by overexpression of CARF, which is mediated by upregulation of Ras, a small GTPase proto-oncogene activated by receptor tyrosine kinases that regulates cell survival and death pathways.3 We first sought to determine whether the Ras-MAPK pathway is involved in CARF-silencing-induced apoptosis. As shown in Figure 2a, CARF suppression led to downregulation of Ras and inactivation of its downstream effector MAP kinases, extracellular regulated kinases (ERK)1/2; the level of phosphorylated ERK1/2 was decreased in CARF-compromised cells. In order to address whether this pathway is critical for CARF-silencing-induced apoptosis, we investigated whether exogenous expression of ERK1/2 could reverse the apoptosis caused by CARF inhibition. As ERK1 and ERK2 are functionally comparable in the regulation of apoptosis, only ERK1-overexpressing U2OS cells were generated. Cells expressing control GFP and GFP-ERK1 proteins were compared for CARF-silencing-induced apoptosis (Figure 2b). As shown in Figure 2c and d, cell viability and cleavage of caspase 3 were observed at a similar level in control and ERK1-overexpressing cells, implying that downregulation of the Ras-MAPK signaling was not essential in cell death induced by CARF-suppression. This was further verified by an independent experiment in which we suppressed CARF in HT1080 cells, which harbor Ras overexpression and constitutively active MAPK signaling. HT1080 cells showed a high rate of spontaneous apoptosis, as supported by caspase 3 cleavage, but suppression of CARF did not cause any further increase in cell death (Figure 2e).
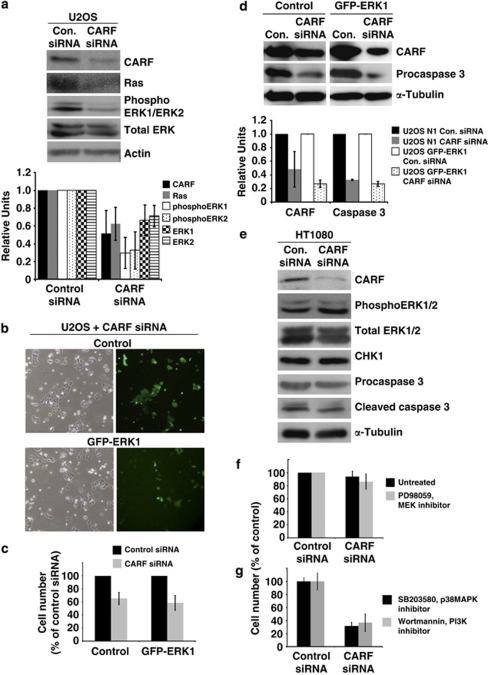
Figure 2.
The Ras-associated pathways are not required for cell death induced by CARF suppression. Ras, total ERK and phosphoERK1/2 were evaluated by immunoblotting in CARF-compromised U2OS cells with densitometric quantitation of representative blots from at least three experiments (a). U2OS cells with overexpression of GFP-ERK1 was transfected with CARF siRNA (b), and cell viability was measured by trypan blue exclusion assay (c) and immunoblotting for procaspase 3 with densitometric quantitation of representative blots from at least three experiments (d). CARF-compromised HT1080 cells were analyzed for total ERK, phosphoERK1/2, CHK1 and caspase cleavage by immunoblotting (e). ERK1/2 was inhibited in CARF-suppressed HT1080 cells by treatment with PD98059, and cell viability was measured using the trypan blue exclusion method (f). p38MAPK and PI3K were inhibited in CARF-compromised U2OS cells by treatment with SB203580 and wortmannin, respectively, and cell viability was measured as above (g). Actin and α-Tubulin were used as loading controls. Densitometric quantitations were performed wherein, the CARF-suppressed group is shown as fold change over control siRNA, which was set as 1. Graphs are represented as average mean±S.D. Cell viability was measured as pecentage of surviving CARF-targeted cells to control siRNA-transfected cells, which was considered as 100%
In order to further investigate and verify the crucial involvement of the MAPK cascade, we recruited MAPK inhibitors. As shown in Figure 2f, HT1080 cells transfected with CARF siRNA were treated with PD98059, a potent MEK1/2 inhibitor. Cell viability of control and treated cells was the same indicating that the MAPK pathway was not critically involved in CARF-suppression induced apoptosis. Treatment with another MEK1/2 inhibitor, U0126, generated similar results (data not shown).
To determine whether the suppression of CARF also involves the other Ras-regulated pathways mediated by PI3K–Akt and p38MAPK, we treated cells transfected with CARF siRNA cells with wortmannin (PI3K inhibitor) or SB203580 (p38MAPK inhibitor). Cell death was induced in an identical manner after both treatments in the CARF-compromised cells (Figure 2g), indicating that the various Ras pathways are not critically involved in this cell death process.
RB–E2F1 pathway is involved, but not required for CARF silencing-induced cell death
We had earlier shown that premature senescence induced in normal cells by CARF overexpression was associated with increase in RB.3 As the RB–E2F1 pathway modulates apoptosis by p53-independent mechanisms, we investigated if RB has a role in the CARF cell death pathway. As shown in Figure 3a, although total RB protein was not significantly altered after CARF suppression, there were proportional reductions in all phosphorylated forms of RB, including serines 780, 795 and 807/811 and in E2F1 (Figure 3a), signifying growth arrest, which may have ultimately resulted in apoptosis via cleavage of caspase 3. To determine whether RB is essential for CARF-silencing-induced cell death, we suppressed CARF in RB−/− Saos-2 cells. In contrast to U2OS and HeLa, Saos-2 cells were resistant to CARF-silencing-induced cell death (Figure 3b) and did not show caspase 3 cleavage or changes in E2F1, p21WAF1 and CHK1 expression (Figure 3c). CARF inhibition was thus, conducted in RB-restored Saos-2 cells to determine whether the cell death phenotype could be reversed. As shown in Figure 3d, HA-tagged RB was restored in Saos-2 cells, however, after CARF suppression, caspase activation was still comparable to non-restored CARF-compromised cells, and no apoptosis was observed (Figure 3d and e). Thus, although CARF siRNA-transfected cells were compromised for RB phosphorylation leading to growth arrest, it was not critical for the cell death phenotype.
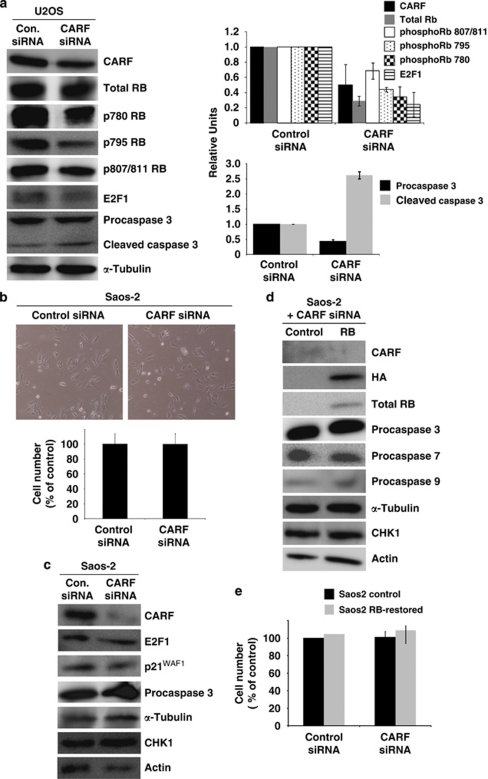
Figure 3.
Cell death induced by CARF inhibition is not critically dependent on RB. Total RB, phosphorylated RB, E2F1 and caspase 3 were analyzed by immunoblotting in CARF-compromised U2OS cells with densitometric quantitation of representative blots from at least three experiments, in which the CARF-suppressed group is shown as fold change over control, which was set as 1 (a). Saos-2 cells were transfected with CARF-targeting siRNA and cell viability was measured by the trypan blue exclusion assay (b). CARF, E2F1, p21CIP1/WAF1, caspase 3 and CHK1 were evaluated by western blotting in CARF-compromised Saos-2 cells (c). Control and RB-restored Saos-2 cells were subjected to CARF inhibition and immunoblotting for CARF, HA tag, RB, CHK1 and caspases 3, 7 and 9 was performed (d). Cell viability of control and RB-restored Saos-2 cells following CARF suppression was also conducted (e). Actin and α-Tubulin were used as loading controls. Graphs are represented as average mean±S.D. Cell viability was measured as percentage of surviving CARF-targeted cells to control siRNA-transfected cells, which was considered as 100%
ATM is involved, but not essential for CARF silencing-induced cell death
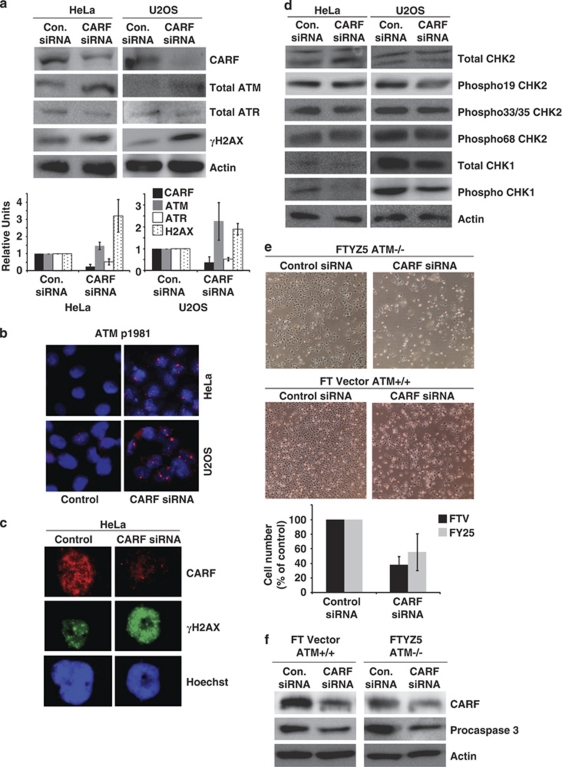
As CARF was upregulated by DNA damage-inducing stress signals, such as treatment with doxorubicin, etoposide and camptothecin, we considered whether CARF suppression causes genotoxicity leading to apoptotic cell death and examined the effect of CARF suppression on the ATM–ATR pathways, critical regulators of the DNA damage response. As shown in Figure 4a, b and c, CARF-silencing was associated with increases in total and phosphorylated ATM (at serine 1981), as well as γH2AX in both HeLa and U2OS cells suggesting that CARF suppression initiated a DNA damage response. However, total expression of CHK2, a downstream effector of ATM, did not show any changes and analysis of the phosphorylated forms of CHK2, including phosphorylation at serine 19, serines 33/35 and threonine 68, also did not reveal any alterations between control and CARF-suppressed cells, suggesting that CARF apoptosis is not critically regulated by the ATM pathway (Figure 4d). This was further supported using ATM-deficient cells, FTYZ5 and AT5 BIVA (data not shown for the latter). We found that the ATM−/− cells underwent cell death following CARF suppression very similar to ATM+/+ cells (Figure 4e). Furthermore, cell death in both ATM−/− and ATM+/+ cells showed cleavage of caspase 3 (Figure 4f) demonstrating that ATM is not essential for CARF-silencing-induced apoptosis.
Figure 4.
CARF suppression activates the ATM pathway, but it is not required for cell death. Total ATM, total ATR and γH2AX, as well as CARF levels were analyzed by immunoblotting with densitometric quantitation of representative blots from at least three experiments, in which the CARF-suppressed group is shown as fold change over control, which was set as 1 (a). Phosphorylated ATM at serine 1981 (b) and γH2AX (c) were detected by immunofluorescent staining, wherein, blue stain denotes nuclei. The phosphorylated forms of CHK2, including phosphorylation at serine 19, serines 33/35 and threonine 68, total CHK1 and phosphorylated CHK1 were examined by western blotting (d). ATM +/+ and null cells were transfected with CARF siRNA, in which apoptosis is shown as rounded, floating cells in the images (e), and cleavage of procaspase 3 was detected by immunoblotting (f). Actin was used as loading control and Hoechst 33 258 was used for nuclear staining. Graphs are represented as average mean±S.D.
Suppression of the ATR–CHK1 pathway is a critical factor in CARF-silencing-induced cell death
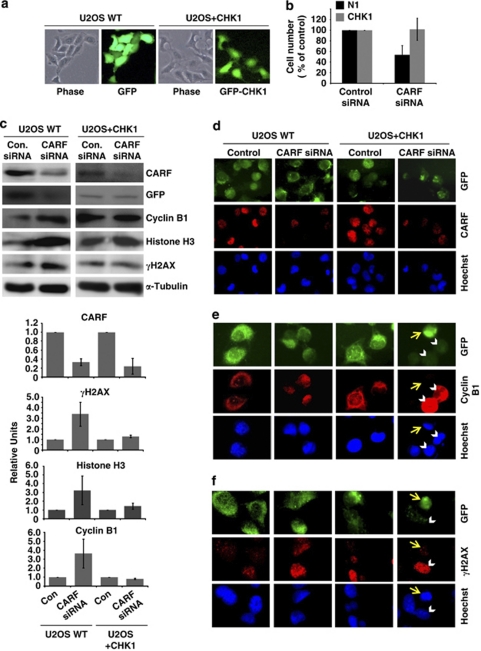
In contrast to ATM and CHK2, ATR was suppressed after CARF inhibition in both HeLa and U2OS cells (Figure 4a), which was accompanied by decreases in total CHK1 and phospho-CHK1 (Figure 4d). To verify that the suppression of this pathway is a critical factor in CARF-inhibition-induced cell death, GFP-CHK1 was transiently overexpressed (Figure 5a) and then subjected to CARF suppression. As shown in Figure 5b, although cell death was still observed in the control vector cells following CARF suppression, cell viability was maintained in the cells with CHK1 overexpression. Further, cyclin B1 and histone H3 (the major markers of MC), as well as γH2AX, which showed increases in control CARF-compromised cells (Figures 1c and 5c), were normalized in the CHK1-overexpressing, CARF-compromised cells (Figure 5c and d). Immunofluorescent staining showed that CARF siRNA transfection effectively decreased CARF expression (Figure 5d). Cyclin B1 was found predominately in the cytoplasm of cells and γH2AX foci were very rare in the control cells (Figure 5e and f, 1st and 3rd columns). In the CARF-compromised control vector cells, nuclear cyclin B1 and γH2AX showed accumulation (Figure 5e and f, 2nd column). In contrast, the CHK1-overexpressing cells did not accumulate cyclin B1 and γH2AX after CARF suppression (Figure 5e and f, 4th column). Noticeably, cells that lacked CHK1 expression (as evidenced by lack of GFP signal, white arrowheads), accumulated nuclear cyclin B1 and γH2AX after CARF suppression while CHK1-overexpressing cells did not (yellow arrows). These results were also seen in HeLa cells (data not shown).
Figure 5.
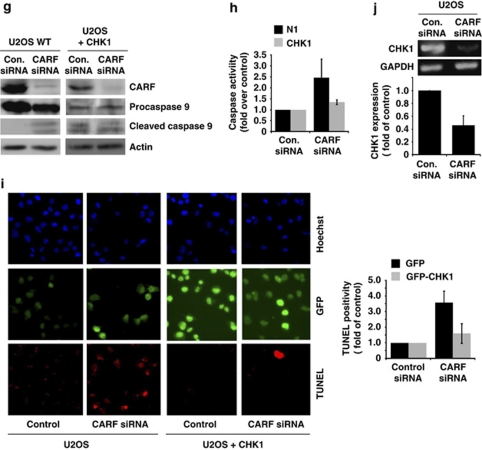
ATR–CHK1 are required for CARF-suppression-induced cell death. U2OS cells were transiently transfected with GFP-CHK1 or control vector (a). After CARF suppression, cell viability was measured by the trypan blue exclusion method, wherein, the percentage of surviving CARF-targeted cells was compared with control siRNA-transfected cells, which was considered as 100% (b). Immunoblots for CARF, GFP, γH2AX, histone H3 and cyclin B1 were performed in control and CHK-1 overexpressing cells following CARF inhibition with densitometric quantitation of representative blots from at least three experiments, in which the CARF-suppressed group is shown as fold change over control, which was set as 1 (c). Immunofluorescent staining was also performed for CARF, cyclin B1 and γH2AX in methanol/acetone-fixed cells (CARF siRNA-transfected control and CHK1 overexpressing) (d–f). For characterization of the apoptotic phenotype, caspase activation was examined by immunoblotting (g) and fluorescence-based assay (h) in control and CHK1-overexpressing cells following CARF knockdown. Further, TUNEL staining was conducted and quantitated by counting a total of 500–2000 cells from two independent experiments (i). Lastly, using reverse-transcription PCR for CHK1 performed in control and CARF-suppressed cells, we demonstrated that CHK1 transcripts were reduced following CARF knockdown. The PCR products were quantitated from two independent experiments, and the CARF siRNA sample is shown as fold change over control. α-Tubulin was used as loading control and Hoechst 33 258 was used for nuclear staining. Graphs are represented as average mean±S.D.
We further confirmed that CHK1 overexpression reverted the apoptosis in CARF-compromised cells by investigating the activation of caspases, as well as by TUNEL staining (Figure 5g and h). We observed increased cleavage of caspase 9 in the control U2OS cells following CARF knockdown, as shown previously, however, this cleavage was normalized in the CHK1-overexpressing cells (Figure 5g). Consistent with apoptosis, caspase 3 activity was substantially increased in control U2OS, but not in CHK1-overexpressing cells after CARF knockdown (Figure 5h). TUNEL staining also revealed increased apoptosis in the control U2OS cells following CARF knockdown and a reversion of this phenotype in the CHK1-overexpressing cells (Figure 5i).
In order to determine the mechanism by which CARF regulates CHK1, we performed reverse-transcription PCR to observe the transcriptional status of CHK1 following CARF knockdown. We found that CHK1 was significantly downregulated in the CARF-compromised cells, suggesting that CARF is a transcriptional regulator of CHK1.
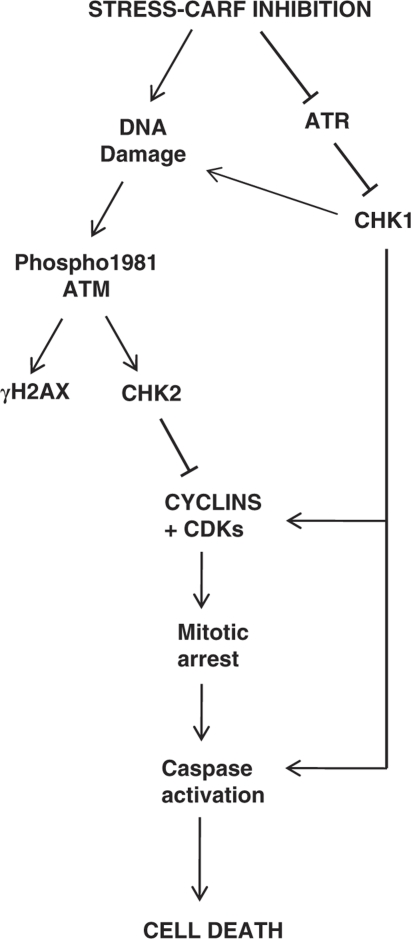
Thus, we conclude here that suppression of CARF resulted in downregulation of the ATR–CHK1 pathway via transcriptional repression of CHK1 expression, leading to MC and apoptosis, and exogenous upregulation of CHK1 reverted the cell death phenotype (Figure 6).
Figure 6.
Schematic diagram of the hypothetical pathways that lead to CARF-suppression induced apoptosis. Our results excluded the crucial involvement of Ras-MAPK and RB–E2F1 pathways and demonstrated that mitotic arrest and cell death induced by CARF inhibition progresses via the ATR–CHK1 pathway
CARF targeting causes tumor suppression in vivo
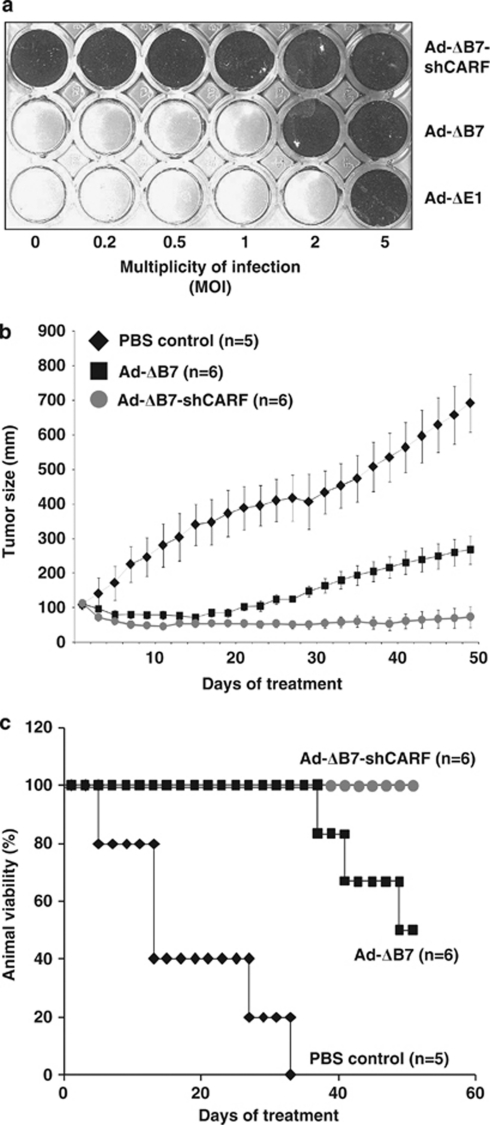
As described above, siRNA-mediated CARF suppression induced apoptosis in cultured cancer cells implicating its antitumor potential. To verify whether the suppression of CARF is antitumoral in vivo, we used A549 cells (a lung carcinoma with wildtype p53, RB and ATR–CHK1 pathways) in a human tumor xenograft nude mouse model. In order to achieve stable suppression of CARF expression, we used adenovirus as a carrier. We introduced CARF shRNA (shCARF) under the control of a U6 promoter in the E3 region of an E1A-mutated and E1B-deleted adenovirus, Ad-ΔB7, generating Ad-ΔB7-shCARF. Relative efficacy of the virus infection and cytotoxicity were compared with the control vectors in in vitro cytotoxicity assays (Figure 7a). We found that the CARF-targeting adenovirus evoked a cytotoxic response in A549 cells even at multiplicities of infection (MOI) 0.2; it was about threefold more potent than the control carrier ΔB7. Oncolytic control and CARF-targeting adenoviruses were examined for their ability to suppress the growth of A549 xenografts established in nude mice. When the subcutaneously implanted A549 tumors reached approximately 100 mm3 in volume, tumors were randomized and treated with phosphate-buffered saline (PBS), Ad-ΔB7 or Ad-ΔB7-shCARF thrice daily on alternate days. Daily observations on tumor volume, body weight and general activity were continued until the experiment was terminated at 50 days. As early as 3 days of treatment, there was a regression in the size of the tumors in the shCARF group (Figure 7b, gray circles). Although there was a cytotoxic effect from the adenovirus carrier, beginning from day 20 of treatment, tumors of the shCARF group continued to regress while tumors of the Ad-ΔB7 control group began to enlarge in size. By day 50, in comparison to the PBS-treated control tumors (average tumor volume; 692.6 mm3), Ad-ΔB7 treated group showed ∼61% reduction in tumor size (average tumor volume; 267.9 mm3). The cytotoxic effect was much more pronounced (average tumor volume 73.3 mm3 implicating ∼89% reduction in tumor growth) with CARF-targeting adenoviruses, Ad-ΔB7-shCARF. Furthermore, CARF-targeting adenovirus significantly averted mortality related to large tumor size. Although all the PBS control mice died by day 35 and 50% of the Ad-ΔB7 control group died by the end of the experiment, all the Ad-ΔB7-shCARF mice were alive with no or very small tumors by day 50 (Figure 7c). We observed no marked treatment-related toxicities affecting feeding, activity or body weight of mice. These data confirmed that our CARF-targeting strategy caused tumor suppression in vivo and, hence, shCARF is a potential antitumor reagent either by itself or in concert with adeno-oncolytic viruses.
Figure 7.
CARF suppression in vivo induces tumor regression. A549 cells were infected with MOI from 0.2–5 of shCARF-carrying adenovirus to determine the optimal dose (a). Nude mice (n=5–6 per group) were injected with 1 × 107 A549 cells, and when tumors reached 100 mm3 in volume, either 3 × 108 plaque forming unit of Ad-ΔB7 (squares) or Ad-ΔB7-shCARF (gray circles) were injected intratumorally three times every 2 days, at which time the tumor size was also measured (b). Survival of the animals was also recorded as percentage (%) viability (c). The mice were killed on day 50
Discussion
We had previously found that CARF, a novel p53-regulatory protein, can dose-dependently induce senescence (overexpression) and apoptosis (downregulation).20 In this report, we elucidated the mechanisms that regulated the apoptotic process following CARF inhibition. We show here that, CARF is a pleiotropic protein that not only mediates p53 and its related pathways, but also synergizes with multiple signaling cascades including the ATM–ATR, Ras–MAPK and RB pathways responsible for cell survival and cell death. As shown in Figure 1, we found that the apoptotic program is, through MC, mediated by activation of the mitochondrial–internal stress and caspase pathways including upregulation of Bak (pro-apoptotic protein), downregulation of Bcl-2 (antiapoptotic protein) and activation of caspases 9, 7, 2 and 3.
Genome integrity is the basis for proper cell growth, proliferation and survival, and loss of genome integrity may result either in cell death or arrest to prevent growth of abnormal cells or tumorigenesis.21 It has been established that genotoxic stress or checkpoint abrogation through CHK1 inhibition can lead to MC and caspase-dependent apoptosis for which cyclin B1 accumulation is a biomarker.22, 23 We found that CARF suppression downregulated the essential genome safeguards, ATR and CHK1, which led to DNA damage as evidenced by increased γH2AX (Figure 5a and c). This appeared to be the primary cause of DNA damage in CARF-compromised cells, as overexpression of CHK1 in CARF-compromised cells reverted not only the cell death phenotype, but also abolished the induction of γH2AX (Figure 5). The significance of ATR and CHK1 in maintaining genome integrity has been shown by other studies, in which exogenous suppression of this pathway leads to formation of single-stranded DNA, DNA breaks and telomere instability, and complete knockout of ATR or CHK1 is embryonic lethal.24, 25 CHK1 may also function downstream of ATM, but after CARF inhibition, ATM was upregulated and activated as evidenced by phosphorylation at serine 1981, although CHK2 was not activated and CARF knockdown in ATM deficient cells did not alter the cell fate (Figures 5 and 6). The data suggest that CARF knockdown induced genotoxicity and severely stunted the DNA damage response. As CHK1 inhibitors are currently being developed as adjuvants to enhance the efficacy of genotoxic antitumor agents, our results suggest that CARF may also be useful as a therapeutic reagent.
CARF suppression and the subsequent DNA damage also affected the Ras–MAPK and RB–E2F1 pathways seemingly to induce cell cycle arrest, but the reconstitution of these factors following CARF inhibition did not protect the cells against CARF-silencing-induced apoptosis. The quintessential MAPK pathway is crucial in organismal development, having pivotal roles in cell survival, proliferation, differentiation and death pathways.8 This pathway consists of three major groups: the ERKs, stress-activated protein kinase/c-Jun NH2-terminal kinase and p38MAPK. Ras, a key upstream player in this pathway, can invoke both cellular proliferation, as observed by neoplastic activity via oncogenic Ras activation and subsequent MAPK or phosphoinositide 3-kinases (PI3-K) transduction and cell death. Further, MAPK are known to participate in the maintenance of G2/M arrest and they are required for exit from growth arrest and transition through mitosis.26 In our previous study, we had found that Ras can induce CARF, and we show here that CARF inhibition reciprocally decreases Ras, leading to downregulation of the Ras-mediated MAPK pathway, possibly as a measure to halt the cell cycle. Nonetheless, using two independent assays including ERK-overexpressing and HT1080 cells, we found that the Ras–MAPK pathway is not essential for CARF-inhibition-induced apoptosis. In contrast, we show that CHK1 levels remained the same in control and CARF-siRNA treated HT1080 cells (Figure 2e), which suggested that CHK1 has a role in preventing cell death in these cells.
Similarly, RB–E2F1 pathway, another important pathway in regulation of cell cycle arrest and apoptosis was not critically involved in CARF siRNA-involved cell death. Cell cycle progression requires phosphorylation of RB by cyclin-dependent kinases and their partner cyclins to free E2F proteins and subsequent transactivation of a plethora of cell cycle regulating genes.6 Hypophosphorylated RB binds to and inhibits the activity of E2F proteins, leading to suppression of a variety of genes including those required for cell cycle progression. Further, RB and E2F proteins have been implicated in regulation of apoptosis, wherein, hypophosphorylated RB binds to E2F1, leading to growth arrest followed by apoptosis.27, 28 Proper RB function is required for maintenance of G2/M arrest following DNA damage and disruption of RB accelerated G2/M progression in the presence of DNA damage by elevating E2F activity and the expression of a mitotic regulatory genes.29 Our results demonstrated that CARF suppression led to hypophosphorylated RB and downregulation of E2F1, suggesting that CARF inhibition may also activate this pathway to bring about growth arrest. However, although Saos-2 cells, which lack RB, were resistant to CARF-inhibition induced cell death, restoration of RB to Saos-2 did not revert the phenotype, which indicated that RB is not sufficient to revert the apoptosis induced by CARF suppression. On the other hand, CHK1 expression was identical between control and CARF siRNA-treated Saos-2 cells (Figure 3c and d), suggesting that in Saos-2, similar to those in HT1080 cells, the retention of CHK1 levels prevented the apoptotic phenotype as seen in the other cell lines with downregulated CHK1.
The data suggested that the primary effect of CARF suppression is loss of DNA protection and induction of DNA damage owing to downregulation of the ATR–CHK1 pathway through transcriptional repression of the CHK1 gene, leading to induction of mitotic arrest and cell death. We show that CARF is an essential genome safeguard that critically regulates the ATR–CHK1 pathway and its inhibition induces cell death via ATR–CHK1 dysregulation that also involves the Ras-MAPK, ATM–CHK2 and RB–E2F1 pathways (Figure 6). These results highlight the pleiotropic effects of CARF as its knockdown impinges upon multiple pathways and its potential as a novel therapeutic reagent.
Furthermore, in vivo suppression of tumoral CARF completely abrogated tumor growth leading to 100% animal survival, providing evidence that CARF is a strong anticancer therapeutic target (Figure 7). Our in vivo strategy involved using oncolytic adenovirus to carry shCARF directly to tumors, which caused complete tumor regression, and viability of the animals was preserved as compared with the vehicle controls. Infection of tumor cells with adenoviruses induces an immune response which enhances the recognition of tumor antigens.30 However, a high infective dose may cause acute cytotoxicity. To circumvent this, generation of mutant adenoviruses with enhanced specificity to tumor cells to decrease side effects and increase efficacy has been developed. Adenovirus carrying the E1B7 deletion (Ad-ΔB7) has been shown to markedly increase oncolysis and apoptosis, potentiating its development as a novel anticancer therapy, but its tumor-targeting effect is still considered mild, thus, it may be more effective as an adjuvant treatment.31 Previously, adenovirus with E1 deletions exhibited enhanced efficacy and tumorilytic activities when administered in conjunction with radiotherapy or chemotherapeutic drugs.32, 33 Alternatively, using the mutant adenovirus as anticancer gene carriers had also demonstrated increased oncolytic efficacy.34 Thus, in our study, we utilized Ad-ΔB7 as a gene carrier for shCARF. Although Ad-ΔB7 alone showed a moderate degree of oncolysis and inhibition of tumor growth, by 21 days of treatment, tumor growth resumed followed by animal mortality presumably owing to the increase in tumor burden (Figure 7). In contrast, Ad-ΔB7-shCARF showed tumor regression throughout the experimental period and complete preservation of animal viability. This demonstrated that CARF suppression might be a safe and effective antitumor reagent in conjunction with oncolytic adenovirus administration with no observed side effects such as cytotoxicity.
Materials and Methods
Cell culture
All the cell lines were obtained from the America Type Culture Collection (Manassas, VA, USA) unless otherwise specified. The HCT116 human colon cancer cell lines (p53−/− and p53+/+) were a gift from Dr. Bert Vogelstein (The Johns Hopkins Kimmel Cancer Center, Baltimore, MD, USA) and the ATM-deficient cells FT/pEBS7 (hereby referred to as FT vector or FTV) and control cell line FT/pEBS7-YZ5 (FTYZ5) were derived from the AT22IJE-T line, an immortalized fibroblast line, and generously provided by Dr. KumKum Khanna (Queensland Institute of Medical Research, Herston, Australia).35 All the cell lines were cultured in Dulbecco's modified Eagle's minimal essential medium (Invitrogen, Carlsbad, CA, USA) supplemented with 5–10% fetal bovine serum (Invitrogen) at 37°C with 95% O2 and 5% CO2 in a humidified chamber.
CARF siRNA and plasmid transfections
The synthesis and sequences of CARF siRNAs are described elsewhere.2 Briefly, for annealing of siRNAs, 20 μM of each control or target sense and antisense strand were incubated in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4 and 2 mM magnesium acetate) for 1 min at 90°C followed by 1 h at 37°C. Transfection of siRNA duplexes was carried out using Oligofectamine reagent (Invitrogen) as previously described. For plasmid transfections, the cDNA encoding full-length ERK1 tagged with GFP (Addgene, Cambridge, MA, USA; plasmid #14 747) generously provided by Dr. Rony Seger from the Weizmann Institute of Science (Rehouot, Israel), RB tagged with HA (Addgene plasmid #10 720) provided by Dr. William Sellers from the Dana Farber Cancer Institute (Boston, MA, USA), and GFP-tagged CHK1 which was a gift from Dr. Aziz Sancar from the University of North Carolina School of Medicine (Addgene plasmid #22 888) were transiently transfected into cells using Fugene 6 (Roche, Basel, Switzerland) following the manufacturer's protocol. Briefly, cells were plated into a 6-well plate, 2 μg of each vector was transfected into cells at a ratio of 6 : 1 of transfection reagent to DNA in antibiotic-free media with 10% fetal bovine serum, and after 48 h, cells were washed and subsequently transfected with CARF siRNA.
TUNEL staining
TUNEL staining was performed as before.3 Briefly, cells were plated onto coverslips, transfected with CARF or control siRNA, and then subjected to TUNEL staining using the DeadEnd Fluorometric TUNEL System (Promega, Madison, WI, USA) or the ApopTag Red In Situ Apoptosis Detection (Millipore, Billerica, MA, USA) kits as described previously.
Cell culture treatments
All chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. Cells were treated with 20 μM each of wortmannin (PI3K/Akt inhibitor), PD98059 (MEK inhibitor) and SB203580 (p38MAK inhibitor) for inhibition of the various Ras-associated pathways and with 50 μM of zVAD.fmk for inhibition of caspases. The treatments were performed simultaneously with CARF siRNA transfection complexes to the cells, using dimethyl sulfoxide as the vehicle control and cells were harvested after 48 h. All the experiments were performed in triplicate at least three times.
Western blot analysis
The protein samples (10–20 μg) were harvested using Nonidet-P40 lysis buffer (20 mM Tris, 100 mM EDTA, 100 mM EGTA, 100 μM PMSF, 150 mM NaCl and 1% NP-40) or RIPA buffer (Thermo Scientific Corp., Waltham, MA, USA), separated in SDS-polyacrylamide gels, and electroblotted onto PVDF membranes (Millipore) using a semidry transfer blotter (Biometra, Tokyo, Japan). Immunoblotting was performed with antibodies against: Bak, Bax, caspase 3,7, 8 and caspase 9, ATM, ATR, E2F1, p21WAF1, GFP and HA purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); Bcl-xL, Bcl-2, total CHK1, phospho-CHK1, total CHK2, phospho-CHK2, total RB and phospho-RB from Cell Signaling (Danvers, MA, USA); total ERK1/2 from Abcam (Cambridge, MA, USA); and Ras, phospho-ERK1/2, cyclin B1, histone H3 and caspase 2 obtained from BD Biosciences (Franklin Lakes, NJ, USA). The monoclonal anti-actin (Millipore), monoclonal anti-αtubulin (Sigma-Aldrich), anti-phospho1981 ATM (Genetex, Irvine, CA, USA), anti-γH2AX (Millipore), anti-FADD (MBL, Nagoya, Japan) and polyclonal anti-CARF2 antibodies were also used. The immunoblots were incubated with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies (Santa Cruz Biotechnology) and detected using ECL substrate (Amersham Pharmacia Biotech/GE Healthcare, Piscataway, NJ, USA). Densitometric quantitation of the representative immunoblots was carried out using the ImageJ software (National Institute of Health). The data are shown as relative units wherein, control siRNA bands are given a value of 1 and CARF siRNA bands are calculated as fold change over control. All the experiments were performed in triplicate at least three times.
Immunofluorescent staining
For immunocytostaining, cells were grown on glass coverslips or trypsinized after treatments and cytospun onto coated glass slides using the Cytospin 4 instrument (Thermo Scientific), and then fixed with equal volume of cold methanol and acetone for 10 min, followed by permeabilization using PBS with 2% Triton 0X-100 for 10 min at room temperature. The cells were incubated with anti-phospho1981 ATM, anti-cyclin B1, anti-γH2AX and/or anti-CARF antibodies at room temperature for 1 h or at 4°C overnight, probed with Alexa Fluor-conjugated secondary antibodies (Molecular Probes, Invitrogen) and finally counterstained with Hoechst 33 258 (Sigma-Aldrich). The slides were viewed using a Zeiss Axioplan 2 microscope and images were taken using a AxioCam HRc camera (Carl Zeiss, Tokyo, Japan).
Caspase 3 activity assay
Caspase 3 activity was measured using the BD Biosciences kit as per the manufacturer's instructions. Briefly, cells were treated with control or CARF siRNA and 48 h following transfection, cells were lysed and subjected to the assay. Fluorescence was measured using a plate reader with an excitation wavelength of 380 nm and an emission wavelength range of 420–460 nm. The fluorescence intensity is plotted as a ratio of fold change over control siRNA.
Cell viability assay
Cells were trypsinized, stained with trypan blue and counted using a hemocytometer. The data is shown as percentage of control cells, which is represented as 100%. All the experiments were performed in triplicate at least three times.
Reverse-transcription PCR
Total RNA was isolated from control and CARF-siRNA transfected cells at 48 h using the Rneasy Mini Kit from Qiagen (Germantown, MD, USA). The primer sequences were as follows: (sense) 5′ GATGCAGACAAATCTTATCAATGC 3′ and (antisense) 5′ AGTTTGCAGGACAGGATAATCTTC 3′ for CHK1 and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was used as an internal control. The PCR cycle included an initial 10 min denaturation step at 95°C followed by 25 cycles of 95°C for 45 s, 60 °C for 45 s and 72 °C for 45 s, with a final annealing step at 72 °C for 10 min. The PCR products were separated in 0.8% agarose gel and quantitated using the ImageJ program. The CHK1 product was normalized to GAPDH and the CARF-compromised group was plotted as fold change over control, which was set as 1. All samples were performed in triplicate from at least two independent experiments.
Generation of CARF-carrying adenovirus (Ad-ΔB7-shCARF)
Adenoviruses were chosen to carry shRNA against CARF as an in vivo carrier because of their enhanced oncolytic ability and safety.33 We introduced shCARF, the sequences of which are identical to the CARF siRNA described above, under the control of an U6 promoter in the E3 region of an E1A-mutated and E1B-deleted adenovirus, Ad-ΔB7, generating Ad-ΔB7-shCARF.
Cytopathic effect assay
A549 lung adenocarcinoma cells were plated into 48-well plates at approximately 30–70% confluency and then infected with Ad-ΔE1, Ad-ΔB7 or Ad-ΔB7-shCARF at an MOI of 0.2–5. After 5–7 days of incubation at 37°C, the plates were gently washed to remove non-viable cells and the living cells on the plate were then stained with 0.5% crystal violet in 50% methanol.
Antitumor effects in human xenograft model
Human lung tumor xenografts were established in 6-to-8-week-old male athymic nu/nu mice (Charles River Japan Inc., Yokohama, Japan). Mice were subcutaneously implanted with 1 × 107 A549 human lung cancer cells in the abdominal region. When tumors grew to about 100 mm3 in volume, they were injected thrice daily every 2 days with either 3 × 108 plaque forming unit of Ad-ΔB7 or Ad-ΔB7-shCARF. Tumor growth was monitored thrice weekly by measuring the length and width of the tumor until the end of the study. Tumor volume was estimated on the basis of the following formula: volume=0.523Lw2 (L, length and w, width). All the animal studies were conducted at the Yonsei University College of Medicine, an Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility, according to institutional regulations.
Acknowledgments
This study was supported by grants from the Japan Society for Promotion of Science, New Energy and Industrial Technology Development Organization (NEDO) of Japan, and the Ministry of Economy, Trade and Industry (METI) of Japan and the National Research Foundation of Korea (2010-0029220, 2009K001644).
Glossary
- CARF
collaborator of alternative reading frame
- RB
retinoblastoma
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia mutated- and Rad3-related
- MAPK
mitogen-activated protein kinase
The authors declare no conflict of interest.
Footnotes
Edited by B Zhivotovsky
References
- Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Hasan MK, Yaguchi T, Sugihara T, Kumar PK, Taira K, Reddel RR, et al. CARF is a novel protein that cooperates with mouse p19ARF (human p14ARF) in activating p53. J Biol Chem. 2002;277:37765–37770. doi: 10.1074/jbc.M204177200. [DOI] [PubMed] [Google Scholar]
- Hasan K, Cheung C, Kaul Z, Shah N, Sakaushi S, Sugimoto K, et al. CARF is a vital dual regulator of cellular senescence and apoptosis. J Biol Chem. 2009;284:1664–1672. doi: 10.1074/jbc.M805778200. [DOI] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: A molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: Mitotic catastrophe. Cell Death Differ. 2008;15:1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- Buchmann AM, Swaminathan S, Thimmapaya B. Regulation of cellular genes in a chromosomal context by the retinoblastoma tumor suppressor protein. Mol Cell Biol. 1998;18:4565–4576. doi: 10.1128/mcb.18.8.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: A function for each cell compartment. Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Karreth FA, Tuveson DA. Modelling oncogenic ras/raf signalling in the mouse. Curr Opin Genet Dev. 2009;19:4–11. doi: 10.1016/j.gde.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Rajasekaran B, Brown KD. Mlh1- and ATM-dependent MAPK signaling is activated through c-abl in response to the alkylator n-methyl-n'-nitro-n'-nitrosoguanidine. J Biol Chem. 2007;282:32021–32031. doi: 10.1074/jbc.M701451200. [DOI] [PubMed] [Google Scholar]
- Carcagno AL, Ogara MF, Sonzogni SV, Marazita MC, Sirkin PF, Ceruti JM, et al. E2F1 transcription is induced by genotoxic stress through ATM/ATR activation. IUBMB Life. 2009;61:537–543. doi: 10.1002/iub.197. [DOI] [PubMed] [Google Scholar]
- Pauklin S, Kristjuhan A, Maimets T, Jaks V. ARF and ATM/ATR cooperate in p53-mediated apoptosis upon oncogenic stress. Biochem Biophys Res Commun. 2005;334:386–394. doi: 10.1016/j.bbrc.2005.06.097. [DOI] [PubMed] [Google Scholar]
- Niida H, Tsuge S, Katsuno Y, Konishi A, Takeda N, Nakanishi M. Depletion of CHK1 leads to premature activation of CDC2-cyclin B and mitotic catastrophe. J Biol Chem. 2005;280:39246–39252. doi: 10.1074/jbc.M505009200. [DOI] [PubMed] [Google Scholar]
- Huang X, Tran T, Zhang L, Hatcher R, Zhang P. DNA damage-induced mitotic catastrophe is mediated by the CHK1-dependent mitotic exit DNA damage checkpoint. Proc Natl Acad Sci USA. 2005;102:1065–1070. doi: 10.1073/pnas.0409130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MK, Yaguchi T, Harada JI, Hirano T, Wadhwa R, Kaul SC. CARF (collaborator of ARF) interacts with HDM2: Evidence for a novel regulatory feedback regulation of CARF-p53-HDM2-p21waf1 pathway. Int J Oncol. 2008;32:663–671. [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang X, Jiang Q, Clarke PR, Zhang C. Cyclin B1 is localized to unattached kinetochores and contributes to efficient microtubule attachment and proper chromosome alignment during mitosis. Cell Res. 2008;18:268–280. doi: 10.1038/cr.2008.11. [DOI] [PubMed] [Google Scholar]
- Shin S, Lee Y, Kim W, Ko H, Choi H, Kim K. Caspase-2 primes cancer cells for TRAIL-mediated apoptosis by processing procaspase-8. EMBO J. 2005;24:3532–3542. doi: 10.1038/sj.emboj.7600827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CF, Chen CL, Chang WT, Jan MS, Hsu LJ, Wu RH, et al. Sequential caspase-2 and caspase-8 activation upstream of mitochondria during ceramide and etoposide-induced apoptosis. J Biol Chem. 2004;279:40755–40761. doi: 10.1074/jbc.M404726200. [DOI] [PubMed] [Google Scholar]
- Cheung CT, Kaul SC, Wadhwa R. Molecular bridging of aging and cancer: A CARF link. Ann N Y Acad Sci. 2010;1197:129–133. doi: 10.1111/j.1749-6632.2009.05392.x. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Xue J, Sowin TJ, Rosenberg SH, Zhang H. A novel mechanism of checkpoint abrogation conferred by CHK1 downregulation. Oncogene. 2005;24:1403–1411. doi: 10.1038/sj.onc.1208309. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Xue J, Gu WZ, Bui M, Li G, Tao ZF, et al. Cyclin B1 is an efficacy-predicting biomarker for CHK1 inhibitors. Biomarkers. 2008;13:579–596. doi: 10.1080/13547500802063240. [DOI] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human CHK1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun G, Hoffschir F, Revaud D, Granotier C, Gauthier LR, Mailliet P, et al. ATR contributes to telomere maintenance in human cells. Nucleic Acids Res. 2010;38:2955–2963. doi: 10.1093/nar/gkp1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf JA, Ouyang B, Knudsen ES, Fukasawa K, Babcock G, Fagin JA. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J Biol Chem. 2006;281:3800–3809. doi: 10.1074/jbc.M511690200. [DOI] [PubMed] [Google Scholar]
- Chinni SR, Li Y, Upadhyay S, Koppolu PK, Sarkar FH. Indole-3-carbinol (I3C) induced cell growth inhibition, g1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene. 2001;20:2927–2936. doi: 10.1038/sj.onc.1204365. [DOI] [PubMed] [Google Scholar]
- Singh RP, Agarwal C, Agarwal R. Inositol hexaphosphate inhibits growth, and induces G1 arrest and apoptotic death of prostate carcinoma DU145 cells: Modulation of CDKI-CDK-cyclin and PRB-related protein-E2F complexes. Carcinogenesis. 2003;24:555–563. doi: 10.1093/carcin/24.3.555. [DOI] [PubMed] [Google Scholar]
- Eguchi T, Takaki T, Itadani H, Kotani H. RB silencing compromises the DNA damage-induced G2/M checkpoint and causes deregulated expression of the ECT2 oncogene. Oncogene. 2007;26:509–520. doi: 10.1038/sj.onc.1209810. [DOI] [PubMed] [Google Scholar]
- Rein DT, Breidenbach M, Curiel DT. Current developments in adenovirus-based cancer gene therapy. Future Oncol. 2006;2:137–143. doi: 10.2217/14796694.2.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JH, Choi KJ, Kim PH, Yun CO. E1A- and E1B-double mutant replicating adenovirus elicits enhanced oncolytic and antitumor effects. Hum Gene Ther. 2007;18:773–786. doi: 10.1089/hum.2006.167. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim PH, Yoo JY, Yoon AR, Choi HJ, Seong J, et al. Double E1B 19 kDa- and E1B 55 kDa-deleted oncolytic adenovirus in combination with radiotherapy elicits an enhanced anti-tumor effect. Gene Ther. 2009;16:1111–1121. doi: 10.1038/gt.2009.72. [DOI] [PubMed] [Google Scholar]
- Yoon AR, Kim JH, Lee YS, Kim H, Yoo JY, Sohn JH, et al. Markedly enhanced cytolysis by E1B-19 kDa-deleted oncolytic adenovirus in combination with cisplatin. Hum Gene Ther. 2006;17:379–390. doi: 10.1089/hum.2006.17.379. [DOI] [PubMed] [Google Scholar]
- Choi KJ, Kim JH, Lee YS, Kim J, Suh BS, Kim H, et al. Concurrent delivery of GM-CSF and B7-1 using an oncolytic adenovirus elicits potent antitumor effect. Gene Ther. 2006;13:1010–1020. doi: 10.1038/sj.gt.3302759. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Jaspers NG, Etkin S, Danieli T, Trakhtenbrot L, Amiel A, et al. Cellular and molecular characteristics of an immortalized ataxia-telangiectasia (group AB) cell line. Cancer Res. 1989;49:2495–2501. [PubMed] [Google Scholar]