Abstract
Beclin 1, the mammalian orthologue of yeast Atg6, has a central role in autophagy, a process of programmed cell survival, which is increased during periods of cell stress and extinguished during the cell cycle. It interacts with several cofactors (Atg14L, UVRAG, Bif-1, Rubicon, Ambra1, HMGB1, nPIST, VMP1, SLAM, IP3R, PINK and survivin) to regulate the lipid kinase Vps-34 protein and promote formation of Beclin 1-Vps34-Vps15 core complexes, thereby inducing autophagy. In contrast, the BH3 domain of Beclin 1 is bound to, and inhibited by Bcl-2 or Bcl-XL. This interaction can be disrupted by phosphorylation of Bcl-2 and Beclin 1, or ubiquitination of Beclin 1. Interestingly, caspase-mediated cleavage of Beclin 1 promotes crosstalk between apoptosis and autophagy. Beclin 1 dysfunction has been implicated in many disorders, including cancer and neurodegeneration. Here, we summarize new findings regarding the organization and function of the Beclin 1 network in cellular homeostasis, focusing on the cross-regulation between apoptosis and autophagy.
Keywords: Beclin 1, PI3K, Bcl-2, autophagy, apoptosis, signal transduction
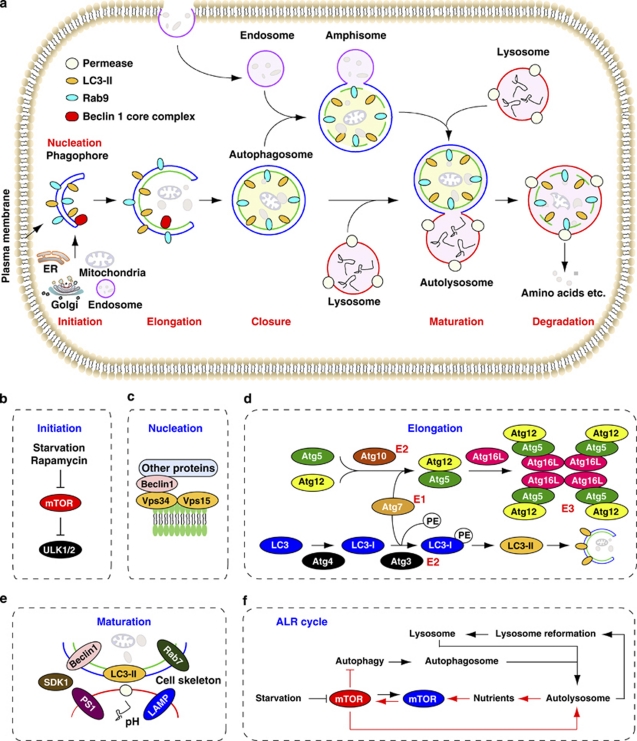
Autophagy is an essential process that consists of selective degradation of cellular components. There are at least three different types of autophagy described and possibly more. These autophagy types include macroautophagy (hereafter referred to as autophagy), microautophagy and chaperone-mediated autophagy.1 The initial step of autophagy is the surrounding and sequestering of cytoplasmic organelles and proteins within an isolation membrane (phagophore). Potential sources for the membrane to generate the phagophore include the Golgi complex, endosomes, the endoplasmic reticulum (ER), mitochondria and the plasma membrane.2 The nascent membranes are fused at their edges to form double-membrane vesicles, called autophagosomes. Autophagosomes undergo a stepwise maturation process, including fusion with acidified endosomal and/or lysosomal vesicles, eventually leading to the delivery of cytoplasmic contents to lysosomal components, where they fuse, then degrade and are recycled (Figure 1a). The process of mammalian autophagy is divided into six principal steps: initiation (Figure 1b), nucleation (Figure 1c), elongation (Figure 1d), closure, maturation (Figure 1e) and degradation or extrusion.
Figure 1.
Stages of autophagy. (a) Different types of autophagy. LC3-II is a marker of Atg5/Atg7-dependent autophagy, whereas Rab-9 is a marker of Atg5/Atg7-independent autophagy. (b) The initiation is sustained by activation of ULK1 and ULK2 complexes, which are inhibited by mTOR. (c) The nucleation depends on Beclin 1-Vps34-Vps15 core complexes and other proteins. (d) The elongation of the phagophore is mediated by two ubiquitin-like conjugation systems that together promote the assembly of the ATG16L complex and the processing of LC3. PE, phosphatidylethanolamine. (e) The maturation is promoted by LC3, Beclin 1, the lysosomal membrane proteins LAMP-1 and LAMP-2, the GTP-binding protein RAB7, the ATPase SKD1, the cell skeleton, the pH of lysosomes and possibly presenilin 1 (PS1). (f) Autophagic lysosome reformation (ALR) cycle. mTOR signaling is inhibited during initiation of autophagy, but reactivated by prolonged starvation. Reactivation of mTOR is autophagy-dependent and requires the degradation of autolysosomal products. Increased mTOR activity attenuates autophagy and generates proto-lysosomal tubules and vesicles that extrude from autolysosomes and ultimately mature into functional lysosomes, thereby restoring the full complement of lysosomes in the cell (figure modified from 1, 6, 73, 74)
It has been well demonstrated that autophagy depends on Atg5/Atg7, is associated with microtubule-associated protein light chain 3 (LC3) truncation and lipidation, and may originate directly from the ER membrane and other membrane organelles. Furthermore, recent study has identified a Atg5/Atg7-independent pathway of autophagy.3 This pathway of autophagy was not associated with LC3 processing but appeared to involve autophagosome formation from late endosomes and the trans-Golgi.3 Atg7-independent autophagy had been implicated in mitochondrial clearance from reticulocytes.4 The exact molecular basis of Atg5/Atg7-independent autophagy remains to be elucidated. Interestingly, Beclin 1 is required for Atg5/Atg7-dependent and -independent autophagy.3 However, the presence of Beclin 1-independent autophagy is evaluated in various experimental setting,5 further complicates discernment of the role of these individual pathways.
As a central element signaling cell growth and enhancing protein translation, the mammalian target of rapamycin (mTOR), when inhibited, induces autophagy. Moreover, as a critical feedback mechanism, reactivation of mTOR terminates autophagy and initiates lysosome reformation (Figure 1f).6 Autophagy principally serves an adaptive, or as we have termed it, a ‘programmed cell survival' role to protect organisms during periods of enhanced cellular distress. In rare settings, however, the self-cannibalistic functions, or paradoxically, the pro-survival functions of autophagy may be deleterious and lead to cell death.
The mammalian autophagy gene Beclin 1, an ortholog of the Atg6/ vacuolar protein sorting (Vps)-30 protein in yeast, was cloned through a yeast two-hybrid screen in 1998 by Beth Levine's group.7 Beclin 1 is important for localization of autophagic proteins to a pre-autophagosomal structure (PAS), depending on interaction with the class III type phosphoinositide 3-kinase (PI3KC3)/Vps34. Together they form the Beclin 1-Vps34-Vps15 core complex.8 Our studies, and those from other laboratories, suggest that Beclin 1 coordinately regulates the autophagy and membrane trafficking involved in several physiological and pathological processes. This review focuses on the Beclin 1 network of associating proteins and its regulation of several critical cellular processes.
Beclin 1 is a BH3-only domain autophagy protein
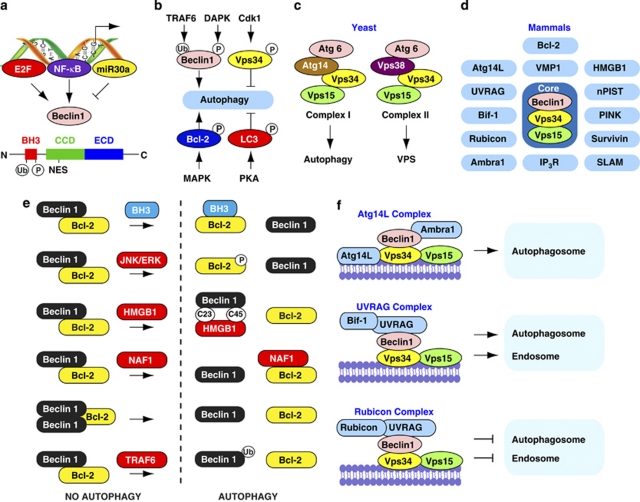
Beclin 1 is a novel Bcl-2-homology (BH)-3 domain only protein.9 The embryonic phenotype of Beclin 1 null mice is even more severe than that of other autophagy gene-deficient mice, which die in early embryonic development (E7.5 or earlier) with defects in proamniotic canal closure.10 Beclin 1 is expressed in many human and murine tissues, and is localized primarily within cytoplasmic structures, including the ER, mitochondria and the perinuclear membrane. In human colon cancer tissue observed via immunohistochemistry, Beclin 1 is distributed within the plasma-membrane, the cytoplasm and the nucleus.11 Beclin 1 contains three identified structural domains: a BH3 domain (amino acids 114–123) at the N-terminus, a central coiled-coil domain (CCD, amino acids 144–269) and an evolutionarily conserved domain (ECD, amino acids 244–337) (Figure 2a). The ECD is essential for Beclin 1's ability to mediate autophagy and to inhibit tumorigenesis. Beclin 1 also contains a short leucine-rich amino acid sequence that is responsible for its efficient nuclear export signal (NES) (Figure 2a). Mutations of the Beclin 1 NES interfere with its abilities to promote nutrient deprivation-induced autophagy and suppress tumorigenesis.12 Anti-apoptotic Bcl-2 family members interact with the BH3 domain of Beclin 1. The activating molecule in Beclin 1-regulated autophagy (Ambra1)/UV radiation resistance-associated gene (UVRAG)/Atg14L interact with the CCD domain, and PI3KC3/Vps34 interacts with the ECD and CCD domains.
Figure 2.
The complex Beclin 1 network. (a) NF-κB and E2F positively regulates Beclin 1 expression, whereas miR30a negatively regulates Beclin 1 expression. Conversely, miR-9 increases autophagy by regulating histone deacetylases activity in lymphoma cells (not shown). Structurally, Beclin 1 has a BH3 domain, a central CCD and an ECD. (b) Phosphorylation and ubiquitination modification regulates autophagy. (c) In yeast, Atg6-Vps34-Vps15 complexes I and II regulate autophagy and Vps, respectively. (d) In mammals, Beclin 1-Vps34-Vps15 core complexes and Beclin 1's binding proteins. (e) Models of Beclin 1–Bcl-2 complex dissociation in autophagy. (f) Model of the role of the three Beclin 1-Vps34-Vps15 complexes. The Atg14L complex functions positively in autophagosome formation. The UVRAG complex functions positively in autophagosome and endosome maturation. The Rubicon complex functions negatively in autophagosome and endosome maturation (figure modified from 49, 74)
As a BH3-only domain protein, Beclin 1 also has the capability to self-associate. Oligomerized Beclin 1 is proposed to serve as a platform enabling rapid nucleation of its associating proteins. The CCD, a universal oligomerization domain, mediates Beclin 1 self-interaction and dimer formation in vivo and in vitro.13, 14 The amino terminus binds less effectively than the CCD to contribute to Beclin 1 self-assembly.13 More specifically, the Bcl-2-binding domain of Beclin 1 is not essential for its self-interaction.13 Early studies suggested that Bcl-XL and UVRAG cause a monomer-dimer switch in Beclin 1 (Noble et al.14). The viral Bcl-2 homologue M11 partially disrupts the Beclin 1 homo-oligomer.15 Interestingly, viral Bcl-2 binds independently to two sites on the Beclin 1 dimer, one with high affinity and one with lower affinity, whereas human Bcl-XL binds to both sites equally with relatively low affinity by sedimentation equilibrium and velocity analysis.14 Beclin 1 self-interaction is not affected by amino acid deprivation, rapamycin-induced autophagy, PI3KC3/Vps34, Bcl-XL or UVRAG overexpression in live cells.13 Therefore, regulation of Beclin 1 oligomerization in vivo is more complex. We do not yet know whether other Bcl-2 family proteins or cofactors influence Beclin 1 self-association.
Transcriptional regulation of Beclin 1 in autophagy
Increasing evidence suggests that nuclear factor (NF)-κB, E2F transcription factors (E2F) and microRNAs (miRNAs) are involved in regulation of Beclin 1 expression in autophagy (Figure 2a).
p65, one of the canonical NF-κB pathway components, directly binds the Beclin 1 promoter and upregulates its mRNA and protein levels, leading to autophagy in T cells.16 However, the relationship between NF-κB and regulation of autophagy seems puzzling and complex. NF-κB has emerged as a negative regulator of autophagy, as induced by tumor necrosis factor, reactive oxygen species (ROS) and starvation in some cell lines.17 On the other hand, other studies suggest NF-κB and its counter-regulator IKK have an activating role in autophagy.18 Constitutively, active IKK subunits stimulate autophagy.18 Remarkably, NF-κB p65 positively modulates Beclin 1 transcription and autophagy.16 Further study, likely evaluating regulatable systems in various cellular targets in vivo, will be required to sort out the complexity.
Recent work suggests that E2F1 binds Atg1/ULK1, LC3, DRAM1, BNIP3, ULK2, Atg4, Atg7, GABARAPL2, Atg9A, Atg10, Atg12, p73, Apaf1 and caspase promoters, increasing their expression and regulating autophagy and apoptosis.19 A ChIP-on-chip study suggested that Beclin 1 could be an E2F target.20 Indeed, E2Fs can directly transactivate the Beclin 1 promoter (Figure 2a).21 When E2F-1 or -2 or -3 is depleted, Beclin 1 mRNA and protein levels are significantly reduced in the U2OS cell line.21
miRNAs are post-transcriptional regulators that bind to complementary sequences in the three prime untranslated regions (3′ UTRs) of target mRNAs, usually resulting in gene silencing. The miR-30a-binding sequences in the 3′-UTR of Beclin 1 contributes to the modulation of Beclin 1 expression by miR-30a (Figure 2a). Transfection with a miR-30a mimic decreased Beclin 1 mRNA and protein expression; whereas treatment with a miR-30a antagomir resulted in upregulation of Beclin 1 (Zhu et al.22). It is not known whether miR-30a directly mediated the activity of NF-κB and E2F during Beclin 1 expression. Consistent treatment of tumor cells with a miR-30a mimic results in decreased autophagic activity in HeLa cells.22
Modification of Beclin 1 by phosphorylation and ubiquitination in autophagy
Beclin 1 is a new phosphorylation substrate of the death-associated protein kinase (DAPK),23 a tumor suppressor lost in many cancer types (Figure 2b). In addition to stimulating the functions of cell death and growth, DAPK can stimulate autophagy and membrane blebbing by binding to LC3. Notably, DAPK phosphorylates Beclin 1 on Thr 119 at the BH3 domain, thus promoting the dissociation of Beclin 1 from Bcl-2-like proteins, which in turn induces autophagy.23 The relationship between Beclin 1 and other substrates of DAPK in autophagy is unknown.
Phosphorylation of Bcl-2 by the c-Jun N-terminal kinase 1 (JNK1), located within the nonstructural loop of Bcl-2, is another mechanism that reduces the interaction between Beclin 1 and its inhibitor.24 We find that high mobility group box 1 (HMGB1) promotes the phosphorylation of Bcl-2 by extracellular signal-regulated kinase (ERK), the subsequent dissociation of Beclin 1 from Bcl-2, and the induction of autophagy.25 Thus, the interaction between Beclin 1 and its inhibitors is dynamic and subject to regulation by phosphorylation of either one of the two partners in this complex. Unfortunately, we know almost nothing about how these mitogen-activated protein kinases (MAPKs, e.g., JNK1 and ERK) might induce conformational changes between Beclin 1 and its binding partners, preventing their interaction with one another. Autophagy is negatively regulated by the mTOR and Ras-cAMP-PKA pathways. Interestingly, LC3 is a direct substrate of PKA, and its phosphorylation prevents recruitment of LC3 to autophagosomes (Figure 2b).26 Thus, DAPK and PKA target individual substrates, such as Beclin 1 and LC3 in autophagy.
The tumor necrosis factor receptor-associated factor 6 (TRAF6) and the deubiquitinating enzyme A20 control K63-linked ubiquitination of Beclin 1 to trigger autophagy.27 The K63-linked ubiquitination of Beclin 1 facilitates oligomerization of Beclin 1 and the activation of PI3KC3. This helps trigger the formation of autophagosomes. It is important that future studies assess the state of phosphorylation and ubiquitination of Beclin 1 in the setting of autophagy.
Induction of autophagy by Beclin 1-Vps34-Vps15 core complexes
Although more and more Beclin 1-binding proteins and complexes are being identified in mammals, each of the individual complexes seems to recycle various elements derived from other cellular processes. Beclin 1 (Atg6/Vps30), PI3KC3/Vps34 and Vps15 (Figure 2d) have been predicted to regulate autophagy in a similar manner to yeast (Figure 2c).
The phosphoinositol-3-kinase (PI3K) family is divided into three different classes in mammals – class I, class II and class III. Yeast cells have only a solitary class III PI3K and Vps34. Vps34 forms at least two distinct PI3K complexes in yeast (Figure 2c): complex I consisting of Vps34, Vps15, Atg6/Vps30 and Atg14, and complex II containing Vps34, Vps15, Atg6/Vps30 and Vps38 (Itakura et al.28). Complexes I and II function in autophagy and the Vps pathway, respectively. Complex II is more abundant than complex I, suggesting that Atg6/Vps30-Vps34-Vps15 is primarily involved in the Vps pathways under normal conditions and that yeast autophagy is strictly regulated at a basal level in yeast. Interestingly, recent studies show that Vps34 is phosphorylated on Thr159 by cyclin-dependent kinase 1 (Cdk1), which negatively regulates its interaction with Beclin 1 during mitosis in mammalian cells (Figure 2b).29 Vps34 also appears to have a role in other membrane-trafficking processes distinct from autophagy. It would be interesting to know if Vps34 phosphorylation affects these processes, and if cell cycle proteins are required for autophagy.
Inhibition of autophagy by Beclin 1–Bcl-2 /Bcl-XL complexes
Mutations of either the BH3-only domain within Beclin 1, or the BH3 receptor domain within Bcl-2 or Bcl-XL, disrupted the Beclin 1–Bcl-2 complex, resulting in the stimulation of autophagy. The relationship between Beclin 1 and Bcl-2/Bcl-XL is complicated. Beclin 1 cannot neutralize the anti-apoptotic function of Bcl-2, which is exerted at the mitochondrial membrane.30 In contrast, Bcl-2 or Bcl-XL reduces the pro-autophagic activity of Beclin 1 (Pattingre et al.31; Maiuri et al.32). Interestingly, ER-localized Bcl-2, but not mitochondrial-localized Bcl-2, inhibits autophagy,31 which is consistent with the older notion that ER-associated class III PI3K activity may be crucial in the nucleation of autophagosome formation. Beclin 1 can colocalize with Bcl-XL within mitochondria via its BH3 domain,14 suggesting a differential role of Bcl-XL in Beclin 1 complex when compared with Bcl-2.
Currently, there are several different means to regulate dissociation of Beclin 1 and Bcl-2/Bcl-XL during autophagy in mammalian cells (Figure 2e). These include: (1) competitive displacement of the Beclin 1 BH3 domain by other Bcl-2 family proteins. The interaction between Beclin 1 and the anti-apoptotic proteins is inhibited by tBid, Bad and BNIP3, but not by Bax and Bak.33 Moreover, the pro-apoptotic BH3-only proteins such as BNIP3, Bad, Noxa, Puma, BimEL and Bik all induce autophagy.33 (2) JNK1 or ERK-mediated phosphorylation of Bcl-2 (Wei et al.24; Tang et al.25) or DAPK-mediated phosphorylation of Beclin 1 (Zalckvar et al.23). Phosphorylation of DAPK at Ser 735 by ERK increases the catalytic activity of DAPK both in vitro and in vivo.34 In contrast, DAPK promotes the cytoplasmic retention of ERK, thereby inhibiting ERK signaling in the nucleus.34 Furthermore, DAPK regulates JNK signaling by binding and activating protein kinase D under conditions of oxidative stress.35 Future studies are required to define the crosstalk between the MAP kinases (e.g., JNK, ERK) and DAPK to regulate phosphorylation/dephosphorylation events in autophagy. (3) Competitive displacement of Bcl-2 by other Beclin 1-binding proteins such as HMGB1, UVRAG or Atg14L/Barkor; this likely initiates a program of heightened anti-apoptotic state, promotes autophagy, and ultimately protects the cell during cell stress. (4) NAF-1 (nutrient-deprivation autophagy factor-1) dysfunction. NAF-1 is a component of the inositol-1,4,5 trisphosphate (IP3) receptor complex, which contributes to Beclin 1–Bcl-2 interaction within the ER. NAF-1 binds Bcl-2 and this interaction is independent of a BH3 domain, but depends at least in part on its redox sensitive CDGSH iron/ sulphur-binding domain.36 ROS are important signaling molecules that initiate autophagy. Beclin 1 is also a cysteine-rich protein. Whether redox regulates the structure and function of Beclin 1 is unknown. (5) Beclin 1 self-interaction: Beclin 1 can form large homo-oligomers, which may provide a platform for further protein–protein interactions and displacement of Bcl-2 or Bcl-XL. In general, the anti-autophagic binding of Bcl-2 promotes Beclin 1 homodimerization, and this prevents heterodimerization with Atg14L or UVRAG. It is not clear whether the disulfide bond is involved in the formation of Beclin 1 homo-oligomers. (6) TRAF6-mediated ubiquitination of Beclin 1. The deubiquitinating enzyme A20 reduces ubiquitination of Beclin 1 and limits the induction of autophagy in response to Toll-like receptor (TLR) signaling, increasing the interaction of Bcl-2 with Beclin 1 (Shi and Kehrl27).
Other Beclin 1-binding proteins in autophagy
Atg14L/Barkor
Atg14L, also known as Barkor,37 contains two CCD that are required for binding the CCD regions of Beclin 1 and Vps34. Atg14L shares an 18% sequence identity and a 32% sequence similarity with yeast Atg14. In yeast, Atg14 acts as an adaptor molecule promoting interaction of Vps30 and the rest of the complex. It is responsible for targeting the entire complex to the PAS (Figure 2c). Similar to the role of Atg14 in yeast, human Atg14L is required for autophagosome formation.38 In mammals, Atg14L localizes to the ER.39 Furthermore, Atg14L colocalizes with Atg5 and Atg16L1 on the isolation membranes during autophagy.28 Notably, Atg14L and UVRAG form two distinct complexes with Beclin 1-Vps34 in autophagy (Figure 2f). The interaction between Beclin 1 and Vps34 is not affected by Atg14L. In contrast, Beclin 1 is required for the interaction between Vps34 and Atg14L.37 Loss of Beclin 1 or Vps34 greatly reduces Atg14L protein levels, suggesting that these proteins have some non-autophagic functions or altered stability in the absence of forming a complex with Beclin 1/Vps34. Interestingly, PI3K activity is required for starvation-induced Atg5 and ATG16L punctae formation, but not for Atg14L punctae, suggesting that precursor structures of Atg14L form independent of Vps34 and Beclin 1 (Itakura et al.28).
UVRAG
UVRAG shows tumor-suppressor activity in cancer cells through autophagy-dependent40 or -independent mechanisms.41 Remarkably, UVRAG promotes autophagosome formation by activation of the Beclin 1 complex40 and autophagosome maturation by recruitment of the fusion machinery to the late endosome in mammalian cells42 (Figure 2f). Suppressing UVRAG expression in cells does not affect autophagic flux or autophagosome formation.28, 41 UVRAG promoted autophagosome maturation, and endocytic trafficking is independent of Beclin 1. These findings suggest that UVRAG has some unknown role(s) in regulating autophagy dynamics and membrane trafficking.
UVRAG directly binds Beclin 1 through the CCD.40 UVRAG can disrupt the Beclin 1-dimer interface, forming a heterodimer with Beclin 1 (Noble et al.14). However, other study shows that overexpression of UVRAG has no effect on Beclin 1 self-interaction.13 Loss of Beclin 1 or Vps34 reduces UVRAG protein levels in starvation-induced autophagy.28 However, UVRAG and Atg5/Atg16L1 do not colocalize during heightened autophagy. Moreover, endogenous UVRAG punctae were also not colocalized with LC3 under starvation conditions, suggesting that UVRAG punctae do not represent autophagy-related structures. Indeed, UVRAG primarily resides in endocytic compartments such as Rab9-positive late endosomes, as well as partially with Rab5 and Rab7 (Itakura et al.28; Liang et al.42), raising the possibility that it has an additional non-autophagic role.
Bif-1
Endophilin B1/Bax-interacting factor 1 (Bif-1) is localized in the cytosol and regulates the membrane dynamics of organelles, such as the Golgi complex and mitochondria, as well as autophagosomes. In autophagy, Bif-1 interacts with Beclin 1 through UVRAG and functions as a positive mediator of PI3KC3 and the induction of autophagy in mammalian cells.43 Loss of Bif-1 significantly suppresses PI3KC3 activation and the formation of autophagosomes. As in Beclin 1- and UVRAG-deficient mice, knockout of Bif-1 promotes spontaneous tumorigenesis in mice, although embryonic Bif-1−/− mice develop normally.43 Bif-1 accumulates in punctate structures, along with Atg5 and LC3, suggesting that Bif-1 is involved in the early stages of autophagosome formation and may have a role in the biogenesis and/or expansion of phagophores. Moreover, Bif-1 interacts with Beclin 1 through UVRAG (Figure 2f).
Rubicon
RUN domain protein as Beclin 1 interacting and cysteine-rich containing (Rubicon) was recently identified as another novel Beclin 1-binding partner.38, 44 Rubicon contains a RUN domain, an asparagine-rich region, a CCD, and a cysteine-rich region.38, 44 Rubicon localizes to the late endosome/lysosome, but not the autophagosome. In contrast to Atg14L, Rubicon negatively regulates autophagy, likely through inhibition of autophagosome maturation (Figure 2f). Overexpression of Rubicon remarkably reduces PI3KC3/Vps34 activity and impairs the acidification of LC3-associated vacuoles. Knockdown of Rubicon by shRNA leads to a dramatic increase in LC3 punctae formation, even under nutrient-rich conditions in mammalian cells. Overexpression of Rubicon also results in the formation of many abnormal structures that are positive for the late endosome/lysosome marker lysosomal-associated membrane protein 1 (LAMP-1) and the multivesicular body marker lysobisphosphatidic acid. However, Beclin 1 does not appear to be involved in the formation of aberrant endosomal structures resulting from Rubicon overexpression that apparently inhibit autophagosome maturation. At present, Beclin 1's participation in Rubicon-mediated endocytic trafficking is unclear. In addition, careful analysis of the previous study38 Beth Levine's group unpublished data have confirmed that Rubicon likely also inhibits autophagosome initiation.
Ambra1
Ambra1 has a role in the development of the nervous system.45 Ambra1 functional deficiency in mouse embryos leads to severe neural tube defects associated with autophagy impairment, accumulation of ubiquitinated proteins, unbalanced cell proliferation, and excessive apoptotic cell death. As a Beclin 1-binding protein (Figure 2d), downregulation of Ambra1 results in a remarkable decrease in rapamycin and nutrient deprivation-induced autophagy, whereas Ambra1 overexpression increases basal and rapamycin-induced autophagy. In addition, recent studies demonstrate that DLC1, a component of the microtubule-based molecular dynein motor complex, is a negative regulator of Ambra1 by yeast two-hybrid assays.46 Ambra1 is required for Beclin 1 translocation to the ER following induction of autophagy.46 These findings provide an understanding at the molecular level detailing the relationship between cytoskeleton dynamics and autophagosome formation.
HMGB1
HMGB1 is a chromatin-associated nuclear protein and extracellular damage-associated molecular pattern molecule.47, 48 Recently, we have shown that HMGB1 is a novel Beclin 1-binding protein important in sustaining autophagy.25, 49 Deletion, depletion or inhibition of HMGB1 in mouse embryonic fibroblasts or tumor cells markedly diminishes autophagy. Endogenous HMGB1 interacts with Beclin 1 in normal and tumor cells.49 Structurally, HMGB1 is composed of three domains: two positively charged domains (A box and B box) and a negatively charged carboxyl terminus (the acidic tail). The three cysteines of human HMGB1 are encoded at positions 23, 45 (A box) and 106 (B box). C106S mutants have much higher cytosolic levels of HMGB1 and demonstrate enhanced binding to Beclin 1, leading to the subsequent dissociation of Bcl-2 from Beclin 1 in mammalian cells. Meanwhile, C23S and C45S mutants lose their ability to mediate autophagy as they are unable to bind Beclin 1 and therefore cannot disrupt Bcl-2–Beclin 1 interactions (Figures 2d and e). These findings demonstrate that oxidation of HMGB1 regulates its localization and ability to sustain autophagy. Moreover, cytosolic translocation of HMGB1 is necessary, but may not be sufficient, to enhance autophagy.
In addition, HMGB1 is released with sustained autophagy, late apoptosis and necrosis in mammalian cells.50, 51, 52, 53 Delivery of exogenous HMGB1 protein to cells triggers autophagy or apoptosis in cancer cells, depending on its redox status and receptors. Reducible HMGB1 (i.e., where the cysteines are not terminally oxidized) binds to the receptor for advanced glycation end products (RAGE), induces Beclin 1-dependent autophagy, and promotes pancreatic or colon tumor cell line resistance to chemotherapeutic agents and ionizing radiation. Moreover, RAGE sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival in vivo and in vitro.54, 55 In contrast, oxidized HMGB1 increases the cytotoxicity of these agents and induces apoptosis via the mitochondrial pathway. We propose a new function for HMGB1 within the tumor microenvironment: as a regulator of cell death and survival, suggesting that HMGB1 has an important functional role in cross-regulating apoptosis and autophagy.
nPIST
Neuronal isoform of protein-interaction, specifically with TC10 (nPIST), was originally discovered as glutamate receptor δ2 (GluRδ2) and a Beclin 1-binding protein by yeast two-hybrid studies.56 nPIST can regulate the ability of Beclin 1 to induce autophagy.56 nPIST binds GluRδ2 by a PDZ domain, whereas nPIST binds Beclin 1 by a CCD (Figure 2d). These interactions provide a physical linkage between the GluRδ2 receptor and Beclin 1.
VMP1
The pancreatitis-associated protein, vacuole membrane protein 1 (VMP1) is an ER transmembrane protein and candidate marker of autophagosomal membranes with no known homologues in yeast (Figure 2d). VMP1 is induced by starvation and rapamycin treatment, and its overexpression triggers conversion of LC3-I to LC3-II and autophagosome formation in mammalian cells.57, 58 Moreover, VMP1 hydrophilic C-terminal region (Atg domain) has proved to be essential for Beclin 1 interaction and autophagy induction. Atg mutant VMP1 expression fails to induce LC3 recruitment, and the triple colocalization as well as interaction of VMP1 and Beclin 1 is abolished. Interestingly, tumor protein 53-induced nuclear protein 2 (TP53INP2) can translocate from the nucleus to the autophagosome structures and interact with LC3 and VMP1, following activation of autophagy by rapamycin or starvation.59 It is unknown, however, whether Beclin 1 mediates interaction between TP53INP2 and LC3/VMP1.
SLAM
Recent studies demonstrated that SLAM (SLAMF1; CD150), a microbial/measles virus sensor, is a novel Beclin 1-binding protein involved in immune cell killing of Gram-negative bacteria through the phagosome and autophagy (Figure 2d).60 Loss of SLAM in macrophages decreased the activity of the cell membrane nicotinamide adenine dinucleotide phosphate oxidase NOX2 complex, one of the key mechanisms by which phagocytes kill bacteria.60
IP3R
IP3R is a membrane glycoprotein complex acting as Ca2+ channel, activated by IP3. In addition to apoptosis, the IP3R also regulates autophagy. IP3R antagonists or knockdown of IP3R by RNAi induces autophagy.61 In contrast, IP3R agonists inhibit autophagy. Notably, IP3R antagonists induce autophagy by disrupting a molecular complex formed by interaction of IP3R and Beclin 1 (Figure 2d), which is increased or inhibited by overexpression or knockdown of Bcl-2.61 In cells, IP3R-mediated Ca2+ fluxes are irrelevant to the regulation of autophagy. Knockdown of IP3R impairs the activity of mTORC1 but not the Beclin 1 core complex in chicken DT40 B-lymphocyte cells.62
PINK1
PTEN-induced putative kinase 1 (PINK1) is a serine/threonine protein kinase that localizes to mitochondria. It has been implicated in protecting cells from stress-induced mitochondrial dysfunction by regulation of mitophagy.63 Moreover, the full-length PINK1, but not its cleaved isoforms, or a mutant protein truncated at the C-terminus, interacts with the pro-autophagic protein Beclin 1 and enhances autophagy (Figure 2d).64
Survivin
Survivin is an anti-apoptotic protein highly expressed during fetal development and malignancy. Recent study indicates that interaction of Beclin 1 with survivin regulates sensitivity of human glioma cells to TRAIL (a death receptor ligand) – induced apoptosis (Figure 2d).65 This study presents a possible mechanism regulating the cross-talk between apoptosis and autophagy by Beclin 1-mediated degradation of survivin.
Pathogen-derived Beclin 1 interaction partners
In recent years, a number of pathogen-derived proteins that also bind to Beclin 1 have been characterized. These include vBcl-2 (KSHV and murine γHV68 M11), ICP34.5 (HSV), M2 (influenza) and Nef (HIV). vBcl-2 of γ-herpesviruses including KSHV and M11 can inhibit autophagy via a direct interaction with Beclin 1 (Pattingre et al.31). The M11 has higher affinity to Beclin 1 when compared with other vBcl-2 (Ku et al.15; Sinha et al.66). The herpes simplex virus protein ICP34.5 interacts with Beclin 1 to block double-stranded RNA-activated protein kinase R-dependent macroautophagy induction, which limits neurovirulence by HSV-1 (Orvedahl et al.67). Recent studies also provide evidence that HIV-1 Nef and the influenza M2 proteins interact with Beclin 1 and block autophagosome maturation.68, 69 These studies suggest that Beclin 1 complexes not only promote autophagosome formation but also maturation in different pathogens, which regulates host response in immunologic defense.
Crosstalk between apoptosis and autophagy by Beclin 1
In general, the ‘BH3-only members' of Bcl-2 family can bind to and antagonize the pro-survival proteins leading to increased apoptosis. However, unlike other known BH3-only proteins, Beclin 1 does not function as a pro-apoptotic molecule, even if it is overexpressed. Bcl-2, as it interacts with Beclin 1, does not lose its anti-apoptotic potential.30 In contrast, Beclin 1 has an anti-apoptotic role in several settings including TRAIL, chemotherapy, irradiation, immunotherapy, nutrient deprivation, angiogenesis inhibitors and hypoxia. In C. elegans, depletion of Beclin 1 triggers CED-3/caspase-dependent programmed cell death. Bid knockdown protects cancer cells against apoptosis and induces autophagy, manifest by increased expression of Beclin 1. The precise mechanism by which Beclin 1 inhibits apoptosis is not yet clear, but may be related to unregulated autophagy as an adaptive or anti-injury mechanism, clearing apoptotic cells.
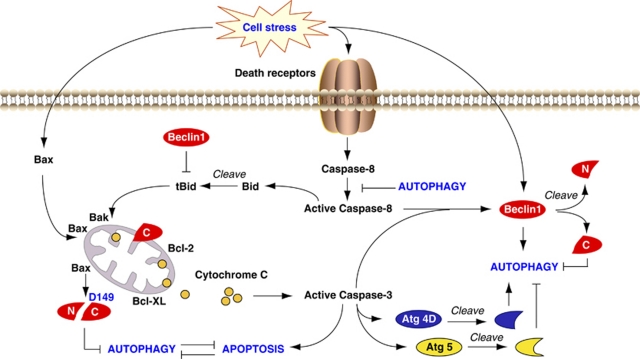
Caspases are cysteine aspartyl proteases that have a cardinal role in apoptotic cell death. Interestingly, caspases can cleave Beclin 1 in apoptosis, thereby destroying its pro-autophagic activity.70, 71 For example, caspase-3-, 7- and 8-mediated cleavage of Beclin 1 generates N- and C-terminal fragments that lose their ability to induce autophagy (Figure 3). The C-terminal fragments translocate to mitochondria and sensitize cells to apoptotic signals.70 This process represents an amplifying loop for inducing massive apoptotic cell death. Apoptosis induced by the proapoptotic protein Bax reduced autophagy by enhancing caspase-mediated cleavage of Beclin 1 at D149 (Luo and Rubinsztein71). Non-cleavable Beclin 1 as well as Bcl-XL restores Bax-reduced autophagy.71 Moreover, TRAIL, a death receptor ligand, triggers the caspase-mediated cleavage of Beclin 1 in HeLa cells. However, recent studies indicate that active caspase-8, a death receptor effector, can be degraded by autophagy,72 suggesting that a feedback mechanism that cross-regulates autophagy and apoptosis exists. Indeed, autophagy and apoptosis share common stimuli and signaling pathways; therefore cell life or death, depends on the cell response and which process is dominant. Although apoptosis-associated cleavage of Beclin 1 and Atg5 inactivates autophagy, the cleavage of Atg4D by caspase-3 generates a fragment with increased autophagic activity. Further investigation of these cleavage events will be important for gaining greater knowledge of the interrelationship between autophagy and apoptosis.
Figure 3.
Crosstalk between apoptosis and autophagy. Autophagy and apoptosis share common stimuli and signaling pathways, and exhibit some degree of mutual inhibition. During sustained exposure to apoptotic stimuli, caspase-mediated cleavage of Beclin 1 generates fragments (‘N' and ‘C') that lose their ability to induce autophagy. The C-terminal fragment translocates to the mitochondria and sensitizes cells to apoptotic signals. Although apoptosis-associated cleavage of Beclin 1 and Atg5 inactivates autophagy, the cleavage of Atg4D by caspase-3 generates a fragment with increased autophagy activity. Moreover, autophagy inhibits apoptosis partly by degrading active caspase-8 or preventing activation of Bid by Beclin 1
Conclusion
Through the analysis of Beclin 1 and its guests at the table of self-eating, great advances have been made in our understanding of the mechanism and roles of autophagy in cell homeostasis. Intriguingly, Beclin 1 can intervene at every major step in autophagic pathways, from autophagosome formation, to autophagosome/endosome maturation. Many of these effects are mediated through the activation of specific Beclin 1-binding proteins, including autophagic inducers and autophagic inhibitors. However, the assembly of the Beclin 1 complexes appears in a cell- or tissue-dependent fashion. One possible explanation is that their interaction with Beclin 1 may be relatively unstable, transient or occur only under specific conditions. Beclin 1 likely has additional functions other than autophagy. Further studies will reveal the precise role of each of the identified Beclin 1 complexes in autophagy and specific disease contexts, and determine whether stimulation or inhibition of autophagy is more beneficial. Another important task in the coming years will be matching differences between autophagic and apoptotic variants with the individual conditions that result in tumor development.
Acknowledgments
We thank the numerous colleagues in the field of autophagy, who through their animated discussions have helped shape this review. We would particularly like to thank Dr. Beth Levine who generously critiqued the article and improved it based on her insights and pioneering work in the area of Beclin 1 biology. We also thank Christine Heiner for careful editing of the paper. The development of a metabolism and mitochondria in disease working group with Dr. Bennett Van Houten, who has provided reflection and encouragement for our studies is appreciated. Work in our laboratory is generously supported by the University of Pittsburgh Department of Surgery (Dr. Timothy Billiar), the University of Pittsburgh Cancer Institute (Dr. Nancy Davidson) and the National Institutes of Health via a grant from the National Cancer Institute (P01 CA 101944-04 to Michael T Lotze, MD).
Glossary
- Ambra1
activating molecule in Beclin 1-regulated autophagy
- BH
Bcl-2-homology
- Bif-1
endophilin B1/Bax-interacting factor 1
- CCD
coiled-coil domain
- Cdk1
cyclin-dependent kinase 1
- DAPK
death-associated protein kinase
- ER
endoplasmic reticulum
- ECD
evolutionarily conserved domain
- ERK
extracellular signal-regulated kinase
- GluRδ2
glutamate receptor δ2
- HMGB1
high mobility group box 1
- IP3
inositol-1,4,5 trisphosphate
- JNK
c-Jun N-terminal kinase
- LC3
microtubule-associated protein light chain 3
- mTOR
mammalian target of rapamycin
- NES
nuclear export signal
- NAF-1
nutrient-deprivation autophagy factor-1
- nPIST
neuronal isoform of protein-interaction, specifically with TC10
- NF-κB
nuclear factor-κB
- PI3KC3
class III type phosphoinositide 3-kinase
- PAS
pre-autophagosomal structure
- PINK1
PTEN-induced putative kinase 1
- ROS
reactive oxygen species
- RAGE
receptor for advanced glycation end products
- Rubicon
RUN domain protein as Beclin 1 interacting and cysteine-rich containing
- TRAF6
tumor necrosis factor receptor-associated factor 6
- TP53INP2
tumor protein 53-induced nuclear protein 2
- TLR
Toll-like receptor
- UVRAG
UV radiation resistance-associated gene
- Vps
vacuolar protein sorting
- VMP1
vacuole membrane protein 1
The authors declare no conflict of interest.
Footnotes
Edited by H-U Simon
References
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, et al. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009;5:303–306. doi: 10.4161/auto.5.3.7491. [DOI] [PubMed] [Google Scholar]
- Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- Adi-Harel S, Erlich S, Schmukler E, Cohen-Kedar S, Segev O, Mizrachy L, et al. Beclin 1 self-association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. J Cell Biochem. 2010;110:1262–1271. doi: 10.1002/jcb.22642. [DOI] [PubMed] [Google Scholar]
- Noble CG, Dong JM, Manser E, Song H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin 1. J Biol Chem. 2008;283:26274–26282. doi: 10.1074/jbc.M804723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, et al. The IKK complex contributes to the induction of autophagy. Embo J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- Kusama Y, Sato K, Kimura N, Mitamura J, Ohdaira H, Yoshida K. Comprehensive analysis of expression pattern and promoter regulation of human autophagy-related genes. Apoptosis. 2009;14:1165–1175. doi: 10.1007/s10495-009-0390-2. [DOI] [PubMed] [Google Scholar]
- Wang B, Ling S, Lin WC. 14-3-3Tau regulates Beclin 1 and is required for autophagy. PLoS ONE. 2010;5:e10409. doi: 10.1371/journal.pone.0010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ, III, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28:2128–2141. doi: 10.1038/onc.2009.60. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. Embo J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27 (Suppl 1:S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Wang WJ, Kuo JC, Tsai HC, Lin JR, Chang ZF, et al. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. Embo J. 2005;24:294–304. doi: 10.1038/sj.emboj.7600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Kimchi A. DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ. 2007;14:1908–1915. doi: 10.1038/sj.cdd.4402212. [DOI] [PubMed] [Google Scholar]
- Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. Embo J. 2010;29:606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin 1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, et al. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6:863–870. doi: 10.4161/auto.6.7.13033. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, et al. Beclin 1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Zeh HJ, Lotze MT.HMGB1, oxidative stress, and disease Antioxid Redox Signal 2010. E-pub ahead of print 24 October 2010, doi: 10.1089/ars.2010.3356 [DOI]
- Kang R, Livesey KM, Zeh HJ, Loze MT, Tang D. HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6:1209–1211. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, et al. DAMP-mediated autophagy contributes to drug resistance. Autophagy. 2010;7:112–114. doi: 10.4161/auto.7.1.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 2010;17:666–676. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Lotze MT, Zeh HJ. Apoptosis to autophagy switch triggered by the MHC class III-encoded receptor for advanced glycation endproducts (RAGE) Autophagy. 2011;7:91–93. doi: 10.1038/cdd.2009.149. [DOI] [PubMed] [Google Scholar]
- Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35:921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem. 2007;282:37124–37133. doi: 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- Vaccaro MI, Ropolo A, Grasso D, Iovanna JL. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy. 2008;4:388–390. doi: 10.4161/auto.5656. [DOI] [PubMed] [Google Scholar]
- Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pebusque MJ, Vaccaro MI, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–881. doi: 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SB, Romero X, Ma C, Wang G, Faubion WA, Liao G, et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat Immunol. 2010;11:920–927. doi: 10.1038/ni.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- Khan MT, Joseph SK. Role of inositol trisphosphate receptors in autophagy in DT40 cells. J Biol Chem. 2010;285:16912–16920. doi: 10.1074/jbc.M110.114207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, et al. The Parkinson-associated protein PINK1 interacts with Beclin 1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- Niu TK, Cheng Y, Ren X, Yang JM. Interaction of Beclin 1 with survivin regulates sensitivity of human glioma cells to TRAIL-induced apoptosis. FEBS Lett. 2010;584:3519–3524. doi: 10.1016/j.febslet.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29:1717–1719. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–277. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Futter M, Jahreiss L, Korolchuk VI, Lichtenberg M, Luo S, et al. Mammalian macroautophagy at a glance. J Cell Sci. 2009;122 (Part 11:1707–1711. doi: 10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Morselli E, Kepp O, Marino G, Michaud M, Vitale I, et al. Oncosuppressive functions of autophagy Antioxid Redox Signal 2010. E-pub ahead of print 4 December 2010, doi: 10.1089/ars.2010.3478 [DOI] [PubMed]