Abstract
Although much has been learned in recent years about the apoptotic machinery, the mechanisms underlying survival and death choices during development of metazoans remain less clearly understood. During early oogenesis in Drosophila, a small excess in the number of specialized somatic cells, called polar cells (PCs), produced at follicle extremities is reduced to exactly two cells through apoptosis by mid-oogenesis. We have found that PCs destined to die first lose their apical contacts and then round up and shrink progressively until they disappear. Caspases are activated only once the cells have begun to shrink, suggesting that they are implicated in this part of the process, but not in the initial loss of cell polarity. Loss-of-function analyses based on mutant, clonal and RNAi approaches show that among the RHG family of pro-apoptotic factors, Hid is specifically necessary for PC apoptosis, as well as the initiator caspase Dronc and its adaptor Dark/Apaf-1, and likely several effector caspases, in particular Drice. In addition, we show that Hid protein and transcripts accumulate specifically in PCs destined to die, while the anti-apoptotic factor Diap1 is downregulated in these cells in a hid-dependent manner. Therefore, our results implicate the Hid–Diap1 module as an important regulatory point in a developmental case of apoptosis.
Keywords: apoptosis, Drosophila, Hid, Diap1, polar cells, oogenesis
In metazoans, apoptosis is widely used during development to reach proper cell number by elimination of targeted individual cells.1 In Drosophila, apoptosis has been implicated in several specific cell number reduction processes, such as the selection of exactly three glial cells from nine precursors at the ventral midline of each embryonic segment2 and the survival of exactly six secondary pigment cells per omatidium allowing formation of the highly ordered hexagonal lattice of the fly compound eye.3 The components of the apoptosis machinery have been shown to be highly conserved during evolution,4 but the mechanisms underlying these types of exquisitely precise survival versus death choices in the context of the whole organism are not as yet fully understood.
Apoptosis is caused by proteolytic cleavage of vital proteins by a group of aspartate-specific cysteine proteases, called caspases.4 The mammalian genomes encode 14 distinct caspases, while the Drosophila genome bears 7 caspase-encoding genes. In Drosophila, caspases are maintained in an inactive state in virtually all cells by a specific class of proteins called inhibitors of apoptosis (IAPs) that are bound to caspases and inhibit their activity.5, 6 In the presence of apoptotic stimuli, IAP antagonists are activated and competitively bind to IAPs through an IAP-binding motif (IBM) leading to IAP ubiquitination and consequent degradation of both IAPs and IBM-domain proteins.7, 8 In mammals, both IAPs and IBM-domain proteins of the Smac/Diablo and Omi/HtrA2 families are also present, but they are ubiquitously expressed. In contrast, the IBM-domain proteins Reaper, Hid and Grim (RHG) in Drosophila are transcriptionally activated in response to many different hormonal, stress and developmental signals.7 Post-transcriptional regulation has been also reported for Hid.2 Most of these studies are based on in vitro analysis or in vivo models in which apoptosis is induced. Thus, how the cell death machinery is activated in vivo during development is still poorly understood.
In the Drosophila ovary, polar cells (PCs) form clusters of specialized somatic cells embedded in the follicular epithelium at anterior and posterior follicle poles and in contact with the germline (Figure 1a). PCs have an important organizer function at several stages during oogenesis (Figure 1a).9, 10 In particular, PCs are the only ovarian cells to secrete Unpaired1 (Upd1), a specific ligand of the JAK/STAT signaling pathway. By activating this signaling pathway in adjacent follicle cells during mid-oogenesis, anterior PCs induce their differentiation into border cells and, subsequently, PCs and border cells migrate together through the egg chamber to attain the oocyte and subsequently form the micropyle, the sperm entry point into the oocyte (Figure 1a).11, 12 During early stages of oogenesis, PCs undergo a highly regulated cell death program, whereby a small excess of PCs that is produced (1–4 supernumerary cells) is eliminated by apoptosis such that by mid-oogenesis 100% of the follicles contain exactly two PCs at each anterior and posterior extremity.13 This restriction fulfills a physiological role as inducing prolonged survival of supernumerary PCs by blocking apoptosis leads to follicular cell patterning defects including recruitment of excess border cells and perturbed migration of these cells.13 PCs thus represent an interesting model to study how cell survival/death choices are made, in particular, in the context of a whole organ. We have undertaken a genetic and cellular analysis of PC apoptosis to identify which of the known components of the core cell death machinery are specifically involved in PC apoptosis and which represents regulatory points.
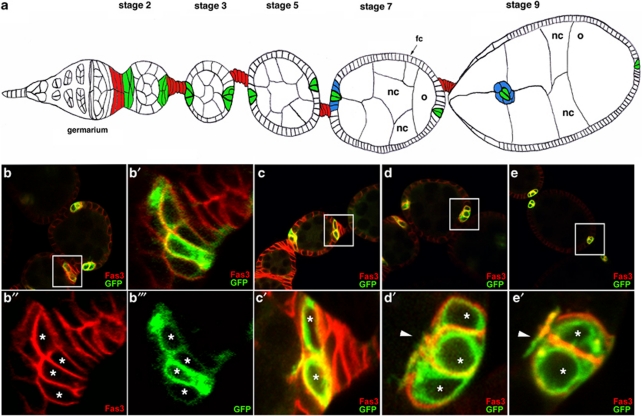
Figure 1.
Polar cells are produced in excess and restrict to a pair of cells during oogenesis. (a) Schematic drawing of an ovariole showing the anteriorly positioned germarium from which follicles (or egg chambers) are formed, subsequently maturing progressively as they move toward the posterior (stages 2 through 9 follicles shown of 14 total stages). Each follicle is composed of 15 nurse cells (nc) and one posteriorly positioned oocyte (o) all of germline origin, surrounded by a monolayered epithelium of somatic follicular cells (fc). Specialized somatic cells are indicated by different colors, polar cells in green, interfolicular cells in red and border cells in blue. Groups of supernumerary polar cells restrict to only a pair of cells between stages 2 and 5. As of stage 7, anterior polar cells induce differentiation of adjacent border cells and during stage 9, the border/polar cell group delaminates from the follicular epithelium and migrates between the nurse cells to reach the oocyte. (b–e) Confocal images of upd-Gal4; UAS-mCD8:GFP follicles in which polar cells are identified by membrane mCD8:GFP whose expression is driven only in these cells (green) and immunodetection of endogenous Fas3 (red). (b′–e′) Panels are magnified views of boxed areas in the corresponding (b–e) panels. Polar cells are marked with asterisks. The anterior of all follicles is to the left and the apical side of polar cells in (c′–e′) also to the left. (b and c) At early stage 2, the membrane mCD8:GFP marker allows specific identification of polar cells, while the Fas3 marker is expressed in both polar cells and surrounding follicular cells. At this stage, polar cells are flattened with their long axis perpendicular to the antero-posterior axis of the follicle. Some follicle extremities contain more than two polar cells (four in b) or two polar cells (c). (d) At stage 3, Fas3 and mCD8:GFP both show specific polar cell accumulation. At this stage, polar cells in clusters of three cells acquire a more rounded shape, are equivalent in size and organized around an axis of radial symmetry, all maintaining apical contact with the germline (arrowhead), as well as basal contacts with the basement membrane. (e) At stage 6, follicles present two equivalent polar cells at each extremity, each contacting the germline apically (arrowhead)
Results
Ovarian PC death involves apical constriction, rounding up and exclusion of dying cells during which caspase activation occurs
The number of PCs per follicle extremity between stages 3 and 10 of oogenesis was assayed by immunostaining for Fasciclin III (Fas3) in the presence and absence of the caspase inhibitor p35 (Figure 1d and e, red; 2F–J, red) (Table 1). In parallel, the morphology of the individual PCs comprising a cluster was determined by immunodetection of either PC-expressed membrane-addressed mCD8:GFP (upd-GAL4 driver) or endogenous membrane Fas3. During early stage 2, the PC clusters (3–5 cells) or PC pairs, at both the anterior and posterior poles, are comprised of flattened cells with their long axis perpendicular to the anterior–posterior axis of the ovariole (Figure 1b and c, green and data not shown). As of stage 3 (Figure 1d and e, red and green), PCs change shape, becoming polarized along the follicular epithelial apical/basal axis with points of convergence between the cells at the apical poles (Figure 1d′ and e′, red, arrowhead). In some groups of three or more PCs, all cells in the group are equivalent in size and maintain both apical–basal contacts (Figures 1d′, 2A and F). In other groups of more than two PCs, the supernumerary cell or cells exhibit constriction at the apical contact, progressive lengthening of the apical constriction (Figure 2B–D and G–I, white arrows), rounding up and reduction in size (Figure 2B–D and G–I, yellow arrows) and basal displacement (Figure 2B, D, I and J). Importantly, the use of specific antibodies demonstrated that the shrinking PCs specifically accumulate an activated form of caspases (Figure 2B–D, green) and are, therefore, the PCs destined to die. Interestingly, the intermediate steps in the morphological changes in PCs destined to die were even more apparent upon expression of p35 in these cells, possibly because the process of cell elimination is blocked at a downstream step (Figure 2F–J). Finally, even when blocking caspase function by p35 leads to prolonged survival of supernumerary PCs (Table 1), the PCs destined to die nonetheless undergo morphological and polarity changes (Figure 2D and E, rightmost yellow arrow and F–J).
Table 1. An excess in polar cell number is produced in egg chambers upon overexpression of the baculovirus caspase inhibitor p35 in these cells.
|
Stage 3 |
Stage 4 |
Stages 5–6 |
Stages 7–8 |
Stages 9–10 |
Stages 7–10 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | A | P | A | P | A | P | A | P | A | P | (A+P) |
| n=51 | n=48 | n=71 | n=50 | n=60 | n=220 | ||||||
| upd-gal4/+ UAS-mCD8:GFP/+ | |||||||||||
| Chambers with two PCs (%) | 78 | 67 | 100 | 98 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Chambers with three PCs (%) | 20 | 33 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chambers with four PCs (%) | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Average nb of PCs | 2.2 | 2.4 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 | 2.0 |
| |
|
|
|
|
|
|
|
|
|
|
|
| n=49 | n=21 | n=45 | n=40 | n=23 | n=126 | ||||||
| upd-gal4/+UAS-p35/UAS-mCD8:GFP | |||||||||||
| Chambers with two PCs (%) | 21 | 57 | 29 | 24 | 38 | 36 | 30 | 30 | 27 | 45 | 32 |
| Chambers with three PCs (%) | 73 | 37 | 62 | 62 | 53 | 55 | 55 | 55 | 73 | 55 | 58 |
| Chambers with four PCs (%) | 6 | 6 | 9 | 14 | 7 | 7 | 15 | 13 | 0 | 0 | 8 |
| Chambers with five PCs (%) | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 1 |
| Chambers with six PCs (%) | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 |
| Average nb of PCs | 2.9 | 2.5 | 2.8 | 2.9 | 2.7 | 2.8 | 2.8 | 2.8 | 2.7 | 2.5 | 2.7 |
In females ectopically expressing the baculovirus caspase inhibitor p35 specifically in polar cells, the number of polar cells (PCs), as indicated by Fas3 immunostaining, both at the anterior (A) and posterior (P) ends of each chamber, was determined from stages 3 to 10 of oogenesis. Results are expressed as the percentage of total chambers (n) of a given genotype with 2, 3, 4, 5 or 6 polar cells. The average number of polar cells at each pole for each stage is indicated at the bottom of each column
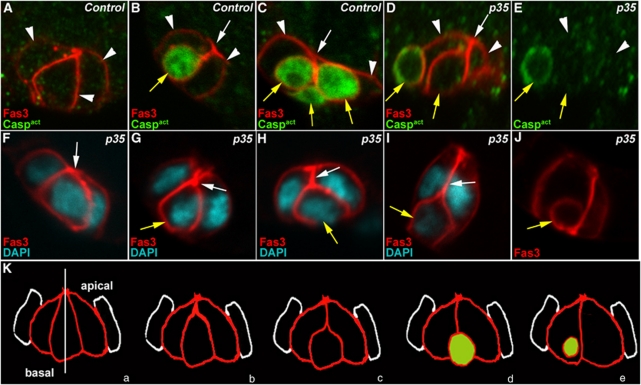
Figure 2.
Supernumerary PCs die by apoptosis in a stereotyped manner. Confocal images of control upd-Gal4/+ follicles (A–C) and upd-Gal4/+ UAS-p35/+ follicles in which PCs express the baculovirus caspase inhibitor, p35 (D–J). The apical side of PCs is always to the top. In all images, except (E), the morphology of PCs is revealed by immunostaining specifically for membrane Fas3 (red). Immunodetection of activated caspases (green), which confirms the identity of dying polar cells, was carried out in (A–E). (D) and (E) are views of the same group of PCs. DAPI staining was used to reveal nuclei in (F–I). (F–J) represent a series of different groups of three PCs, which illustrate what may be the temporal progression of morphological changes occurring during PC elimination, in particular, initial apical constriction (F and G, white arrows), followed by apical detachment (H and I, white arrows) as the supernumerary PC rounds up, reduces in size and takes a basal position (H–J, yellow arrows). (K) Model for supernumerary PC elimination. PCs are in red and adjacent follicle cells in white. (a–e) Represent successive steps during elimination of one cell in a group of three PCs. (a) Group of equivalent cells. The white vertical line indicates the axis of radial symmetry around which the group of equivalent cells is organized. The shrinking supernumerary PC progressively loses its apical contact (b–d) and becomes spherical (c–e). (c–e) The green color indicates the timing of caspase activation detection in the dying cell
Among effector caspases, Drice is specifically implicated in PC apoptosis
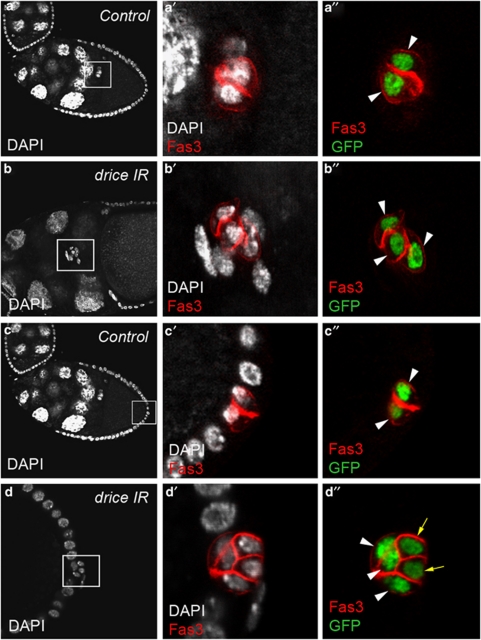
We used UAS-RNAi transgenic lines targeting the caspase-encoding genes drice, dcp1, decay, damm and strica that had been previously validated upon hid-induced apoptosis in the Drosophila eye.14 We induced expression of the UAS-RNAi transgenic constructs specifically as of stage 2 of oogenesis using either the neur-Gal4 or the upd-Gal4 drivers (see Figure 1b–e for the upd-Gal4 expression profile and data not shown) and determined the number of PCs using the Fas3 marker and expression of a UAS-H2B:YFP transgene. Only RNAi directed against the drice gene resulted in prolonged survival of supernumerary PCs (up to five cells) at both anterior and posterior follicle poles after stage 6 of oogenesis (Figure 3b and d compare with a and c) (Table 2). At the anterior pole, the presence of an excess number of PCs was associated with a delay in PC/border cell migration at stage 10 (Figure 3b) compared with wild type (Figure 3a) (see Supplementary Table 1). A second drice RNAi transgenic line also gave high levels of supernumerary PCs with the same driver and a second PC driver, neur-Gal4, as well (data not shown).
Figure 3.
Drice is implicated in PC apoptosis. Confocal images of control upd-Gal4/+ UAS-H2B:YFP/+ (a and c) and upd-Gal4/+ UAS-driceIR/+ UAS-H2B:YFP/+ (b and d) stage 10 ovarian follicles stained with DAPI (white), Fas3 antibodies (red) and GFP antibodies (green) that, respectively, label all nuclei, PC membranes and PC nuclei. Anterior is to the left, as well as the apical side of PCs in (a, c and d). The PCs and surrounding border cells are migrating between nurse cells in (b). (a′–d′) and (a″–d″) are magnified views of the boxed areas in the corresponding a–d panels. Note that in the control, pairs of PCs are present at a given extremity of stage 10 follicle (a″ and c″, white arrowheads), while expression of a drice RNAi transgene leads to the presence of more than two PCs at both anterior and posterior poles at the same stage (b″ and d″, white arrowheads and yellow arrows to indicate the supernumerary PCs that have begun to shrink and shift basally). Also note that upon expression of a drice RNAi transgene, excess anterior PCs surrounded by border cells are delayed in their migration toward the oocyte at stage 10 (b and b′) compared with control PC pairs that have already reached the oocyte at the same stage (a and a′)
Table 2. An excess in polar cell number is produced in egg chambers upon expression of RNAi constructs targeting either a member of the RHG family (hid) or the effector caspase (drice).
|
Stage 3 |
Stage 4 |
Stages 5–6 |
Stages 7–8 |
Stages 9–10 |
Stages 7–10 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | A | P | A | P | A | P | A | P | A | P | A+P |
| n=31 | n=31 | n=37 | n=37 | n=41 | n=41 | n=38 | n=38 | n=37 | n=37 | n=150 | |
| upd-gal4/+ ; UAS-H2B:YFP/+ | |||||||||||
| Chambers with two PCs (%) | 58 | 55 | 86 | 86 | 97 | 100 | 100 | 100 | 97 | 100 | 99 |
| Chambers with three PCs (%) | 42 | 45 | 14 | 14 | 3 | 0 | 0 | 0 | 3 | 0 | <1 |
| Average nb of PCs |
2.42 |
2.45 |
2.13 |
2.13 |
2.02 |
2.0 |
2.0 |
2.0 |
2.02 |
2.0 |
2.01 |
| n=18 | n=21 | n=29 | n=29 | n=38 | n=38 | n=42 | n=42 | n=40 | n=47 | n=171 | |
| upd-gal4/+UAS-driceIRa/+ UAS-H2B:YFP/+ | |||||||||||
| Chambers with two PCs (%) | 17 | 57 | 14 | 21 | 37 | 26 | 47 | 32 | 34 | 34 | 37 |
| Chambers with three PCs (%) | 61 | 37 | 62 | 55 | 60 | 55 | 47 | 52 | 55 | 47 | 50 |
| Chambers with four PCs (%) | 22 | 6 | 17 | 24 | 3 | 19 | 5 | 14 | 11 | 19 | 12 |
| Chambers with five PCs (%) | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 |
| Average nb of PCs |
3.05 |
3.0 |
3.17 |
3.17 |
2.66 |
3.02 |
2.67 |
2.86 |
3.4 |
2.85 |
2.76 |
| n=21 | n=21 | n=25 | n=25 | n=29 | n=29 | n=29 | n=29 | n=32 | n=32 | n=122 | |
| upd-gal4/+ ; UAS-hidIRa/UAS-H2B:YFP | |||||||||||
| Chambers with two PCs (%) | 10 | 62 | 52 | 32 | 66 | 52 | 52 | 76 | 69 | 63 | 65 |
| Chambers with three PCs (%) | 76 | 33 | 40 | 64 | 31 | 45 | 45 | 24 | 25 | 37 | 33 |
| Chambers with four PCs (%) | 14 | 5 | 8 | 4 | 3 | 3 | 3 | 0 | 6 | 0 | 2 |
| Average nb of PCs | 3.04 | 2.43 | 2.56 | 3.17 | 2.38 | 2.52 | 2.52 | 2.24 | 2.38 | 1.95 | 2.37 |
The strains carrying RNAi constructs used were obtained from NIG-FLY
(UAS-hidIR: 5123R2 and
UAS-driceIR: 7788R1).
In females ectopically expressing the RNAi transgenes UAS-driceIR and UAS-hidIR specifically in polar cells, the number of polar cells (PC), as indicated by Fas3 and YFP immunostaining, both at the anterior (A) and posterior (P) ends of chambers, was determined from stages 3 to 10 of oogenesis. The results are expressed as the percentage of total chambers (n) of a given genotype with 2, 3, 4 or 5 polar cells. The average number of polar cells at each pole for each stage is indicated at the bottom of each column
However, we next recovered females homozygous for the amorphic sublethal driceΔ1 allele and found that dissected ovaries immunostained for Fas3 did not exhibit prolonged survival of more than two PCs after stage 6 of oogenesis (data not shown). As the drice RNAi transgenic construct used also targets dcp1 sequences (strong off target), we tested the strong hypomorphic dcp1prev allele, which is homozygous viable; however, homozygous females did not display supernumerary PC after stage 6 of oogenesis (data not shown). We were not able to recover driceΔ1; dcp1prev double mutants due to fully penetrant lethality. We, therefore, recombined the driceΔ1 allele with an FRT insertion on the same chromosome and generated homozygous driceΔ1 mitotic clones (FLP–FRT system) in PCs heterozygous for dcp1prev. In this genetic context, we did not observe an effect on PC number during late stages of oogenesis (data not shown).
The initiator caspase Dronc and its adaptor Dark are required for PC apoptosis
We expressed transgenic RNAi lines to inactivate the function of the Drosophila caspase-encoding genes dronc15, 16 and dredd17 in PCs and only RNAi directed against the dronc gene resulted in prolonged survival of excess PCs at both anterior and posterior poles (data not shown).
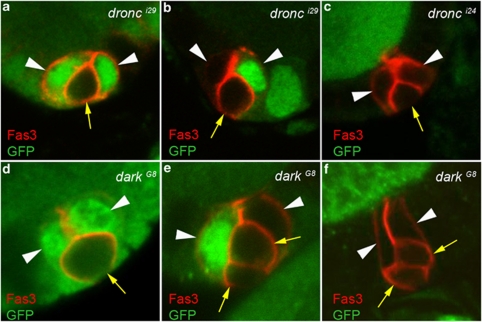
As amorphic alleles for the dronc gene are available (dronci29 and dronci24), as well as for dark, encoding its specific adaptor,18 (darkG8 and darkH16), we tested the effect of these mutations on PC apoptosis. These mutations are lethal during development, so we induced homozygous mutant mitotic clones in the ovaries using the FLP–FRT system and loss of ubi-GFP expression as a clone marker. Under these conditions, we found groups of Fas3+ PCs with more than two cells (either three or four) after stage 6 of oogenesis at both anterior and posterior poles and in each case at least one PC in the group exhibited no GFP expression and, therefore, was homozygous mutant for either dronc or dark (Figure 4a–f). When only one PC in a group was homozygous mutant for dronc or dark, this cell often exhibited the morphological characteristics of a PC destined to die, that is diminished apical contacts with the other cells, rounding up and reduced size (Figure 4a and d, arrows). In addition, in all mosaic groups composed of both GFP+ and GFP− PCs, the cell or cells in the process of being extruded were always GFP− (Figure 4a, b, d and e, yellow arrows). These results indicate cell autonomous function of dronc and the adaptor-encoding dark within the PCs destined die to allow this process to occur.
Figure 4.
The initiator caspase Dronc and its adaptor Dark are required for PC apoptosis. Confocal images of stage 7–10 hsp-flp/+; +/+; FRT42D dronci24/i29/FRT42D P[ubi-GFP] (a–c) and hsp-flp/+; +/+; FRT42D darkG8/FRT42D P[ubi-GFP] (d–f) ovarian follicles stained with Fas3 antibodies (red) to label PC membranes and GFP antibodies (green) to identify homozygous dronc and dark mutant clones (absence of green staining). The apical side of the follicular epithelium in which the PCs are embedded is toward the top. White arrowheads and yellow arrows mark PCs destined to survive or die, respectively, as indicated by the round shape, smaller size and more basal position, indicating progressive exclusion of the latter from the group. The presence of PC groups with more than two cells (three or four) is always associated with the presence of at least one or two dronc or dark homozygous mutant PCs
Among the RHG family members, Hid is specifically required for PC apoptosis
We next tested the possible function of three RHG family members, reaper, hid and grim by generating FLP–FRT mitotic clones, marked by the absence of GFP expression, of the H99 deletion that covers all three genes. We found groups of PCs with more than two PCs (three to five cells) marked by Fas3 during late stages of oogenesis at both anterior and posterior poles and in each case at least one PC was homozygous for the H99 deletion (Figure 5a and b).
Figure 5.
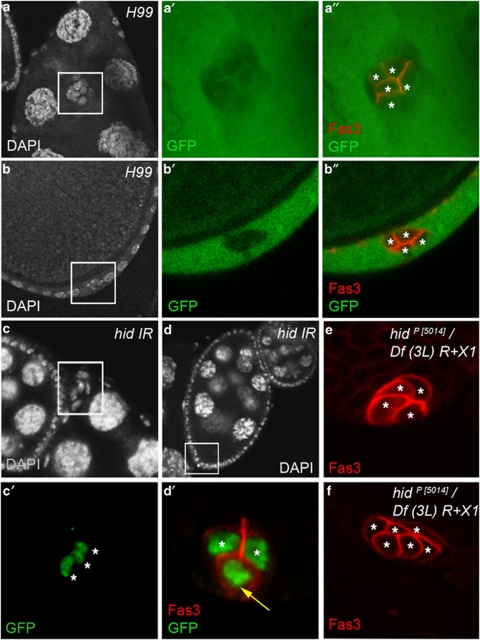
Among the members of the RHG family, Hid is specifically necessary for PC apoptosis. (a and b) Confocal images of hsp-flp/+; FRT79D H99/FRT79D P[ubi-GFP] stage 10 ovarian follicles stained with DAPI (white), Fas3 antibodies (red) and GFP antibodies (green) that, respectively, label nuclei, PC membranes and homozygous H99 mutant clones (absence of green), respectively. (a′ and b′) and (a″ and b″) are magnified views of the boxed areas in the corresponding a and b panels. A GFP− clone of five anterior Fas3+ PCs is visualized in (a′ and a″) (asterisks) and a GFP− clone of four posterior Fas3+ PCs is visualized in (b′ and b″) (asterisks). (c and d) upd-Gal4/+; UAS-hid-IR/UAS-H2B:YFP stage 10 ovarian follicles stained with DAPI (white), Fas3 antibodies (red, only in d′) and GFP antibodies (green) that, respectively, label nuclei, PC membranes and PC nuclei. (c′) and (d′) are magnified views of the boxed areas in the corresponding c and d panels. Note that in (c), a cluster of three PCs within the boxed area (each indicated by an asterisk) has not delaminated from the anterior end of stages 9–10 follicle. In (d), three PCs are present at the posterior end of stage 9 follicle. The two PCs that have maintained apical contact with the germline are indicated with asterisks, while the third PC (yellow arrow) that has lost apical contact and is positioned basally is presumably the supernumerary cell destined to die. (e and f) hidP(5014)/Df(3L)R+X1 ovarian follicles after stage 6 stained with Fas3 antibodies, which indicate the presence of four (e) and six (f) PCs each marked with an asterisk
We next expressed UAS-RNAi transgenic lines in PCs to test, individually, the functions of hid, grim and sickle, a fourth RHG not covered in the H99 deletion, in PC apoptosis. Only RNAi directed against the hid gene, using two different RNAi transgenic constructions (NIG and VDRC collections) and two different PC drivers, upd-Gal4 and neur-Gal4, led to prolonged survival of more than two PCs at late stages of oogenesis at both anterior and posterior poles (Figure 5c, d and data not shown) (Table 2). At the anterior pole, the presence of excess PCs was associated with a significant delay in PC/border cell migration (Figure 5c compare with wild type in Figure 3a; see Supplementary Table 1).
We also analyzed both reaper and hid null escapers for PC number after stage 6 of oogenesis. The reaper XR38/Df(3L)H99 female escapers showed no PC number phenotype (data not shown), while hidP[5014]/Df(3L)R+X1 and hidP[5014]/Df(3L)H99 female escapers displayed more than two PCs at 100% of follicle extremities, anterior and posterior, after stage 6 (Figures 5e, f and 7g, h). The hid mutant phenotype was, therefore, highly penetrant, and as the majority of follicle poles had more than three PCs (four, five or six PCs; Figure 5e, f and data not shown), highly expressive as well. These results, therefore, indicate that, among RHG family members, Hid is specifically required for PC apoptosis. In some clusters of three or four PCs, the supernumerary PCs nonetheless exhibited loss of apical of apical contact, rounding up and shrinking (Figure 7g and h, yellow arrows).
hid transcription is specifically activated in PCs destined to die
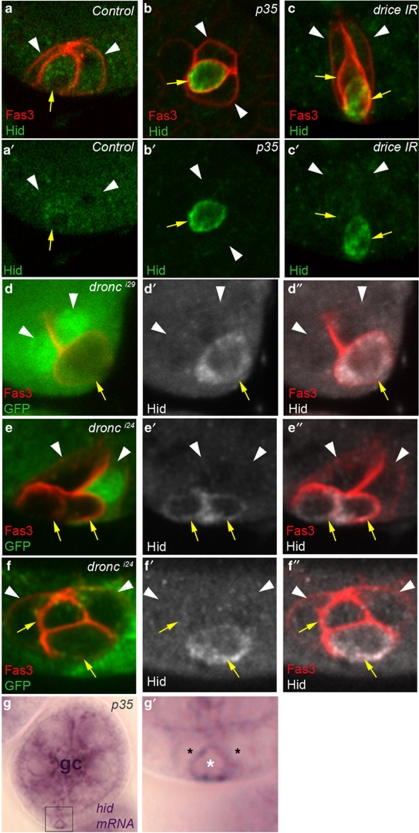
We determined the Hid expression profile in the ovary using specific anti-Hid antibodies and marked PCs specifically by Fas3 accumulation. In wild-type ovaries, no Hid protein accumulation was detected in any cell type (Figure 6a, a′ and data not shown). However, blocking PC apoptosis by overexpression of the p35 caspase inhibitor specifically in PCs led to detectible accumulation of Hid in supernumerary PCs (Figure 6b and b′, yellow arrow). It was also possible to detect Hid in supernumerary PCs by removing caspase function using RNAi or amorphic mutations directed against dronc or drice (Figure 6c–f, yellow arrows). In all cases, Hid was detected in PCs shrinking and undergoing exclusion from the group, therefore, indicating that Hid is specifically expressed in PCs destined to die. Interestingly, when two supernumerary PCs were present, both cells did not necessarily accumulate Hid, indicating that the process of PC elimination is not synchronized within the PC group (Figure 6c, e and f, yellow arrows). Taken together with the results of the analysis of hid mutants indicating that hid is necessary for PC apoptosis, these results strongly suggest that PC apoptosis is induced by specific upregulation of Hid in PCs destined to die.
Figure 6.
Hid is specifically expressed in supernumerary PCs. Confocal images of ovarian follicles stained with Fas3 antibodies to identify PCs (a–f), Hid antibodies (a–f) and GFP antibodies to identify homozygous mutant dronc clones (absence of GFP) (d–f). The apical side of the follicular epithelium in which the PCs are embedded is toward the top. White arrowheads and yellow arrows mark PCs destined to survive or die, respectively, as indicated by the round shape, smaller size and more basal position, indicating progressive exclusion of the latter from the group. (g) Image using light micrcoscopy of stages 4–5 follicle stained with an antisense riboprobe complementary to hid mRNA. (g′) is a magnified view of the boxed area in the corresponding g panel in which the position of three PCs is indicated by asterisks. The white asterisk indicates the PC destined to die that has already lost apical contact, is smaller in size than the other two PCs and accumulates hid signal. The germline cells of this follicle are indicated by gc. (a) upd-Gal4/+. (b and g) upd-Gal4/+; UAS-p35/+. (c) upd-Gal4/+; UAS-drice-IR/+. (d) hsp-flp/+; FRT 82B dronci29/FRT 82B P[ubi-GFP]. (e and f) hsp-flp/+; FRT82B dronci24/FRT 82B P[ubi-GFP]. In the wild-type control, Hid protein is never detected (a), while inducing prolonged survival of supernumerary PCs by inhibiting caspase function leads to accumulation of Hid protein (b–f) and transcripts (g) in PCs destined to die
In order to determine whether specific accumulation of Hid in PCs destined to die is regulated at the transcript level, we performed RNA in situ hybridization on ovaries using a hid riboprobe. The specificity of the hid probe was verified in the embryo as the hid expression profile has been characterized in this tissue (http://www.fruitfly.org/cgi-bin/ex/insitu.pl and data not shown). In the wild-type ovary, relatively strong hid germline signal (equivalent to what is observed in Figure 6g – gc) was observed, as has been previously reported,19 but no expression was evident in any somatic cells including the PC groups at any stage (data not shown). However, when caspase activity was blocked by overexpression of p35 in PCs, then significant accumulation of hid transcripts was detected specifically in one somatic cell at poles of follicles from stage 4 onwards (Figure 6g and g′, white asterisk). The position of these cells at follicle extremities and the fact that their shape differs from that of other somatic cells in the follicular epithelium in that they display apical constriction and rounding up strongly suggest that these are PCs destined to die. These results thus allow us to propose that Hid protein is upregulated in PCs destined to die because of specific hid transcript accumulation in these cells.
Hid controls Diap1 post-translational downregulation in PCs destined to die
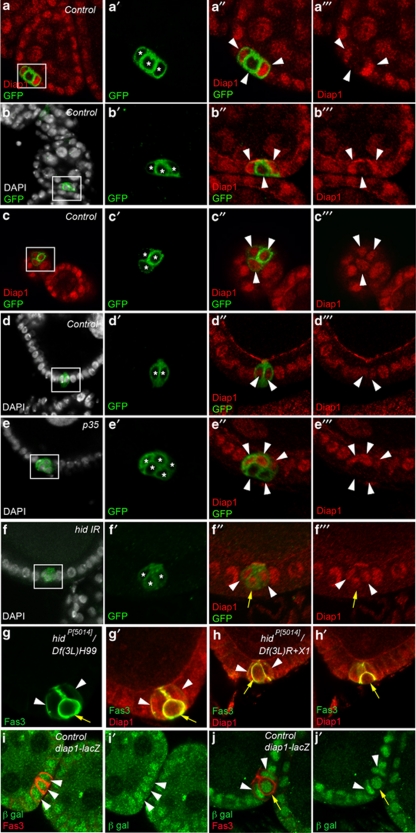
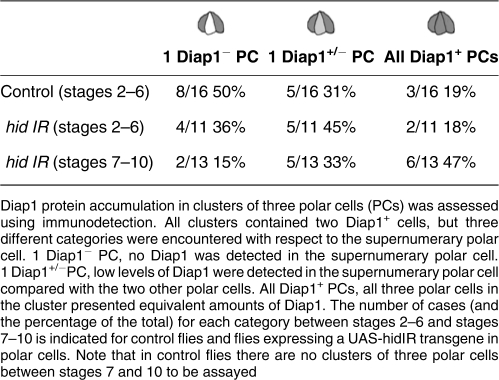
We determined the expression pattern of Diap1 protein in the ovary and found that Diap1 accumulates in all follicle cells (Figure 7a–c), in mature PC pairs (Figure 7d, white arrowheads) and in germline nurse cells (Figure 7a). The specificity of the Diap1 antibodies was confirmed by testing them on ovaries expressing an RNAi construct targeting diap1 sequences specifically in PCs (see Supplementary Figure 1a). In clusters of three PCs during early oogenesis two PCs displayed high Diap1 levels, but three different situations were encountered for the supernumerary PC. In 50% of the clusters of three PCs, one out of the three PCs presented no detectable Diap1 protein (Figure 7a″ and a′″, lower white arrowhead), while in 31% of the clusters, Diap1 was present at low levels in one out of the three PCs (Figure 7b″ and b′″, lower white arrowhead) and in only a minority of cases (19%) all three PCs of a cluster exhibited similar high levels of Diap1 protein (Figure 7c″ and c′″, white arrowheads) (Table 3). Upon ectopic expression of the anti-apoptotic factor p35, a high proportion of follicle poles present PC clusters with three or four PCs (Table 1). Under these conditions, 100% of PC clusters with more than two PCs after stage 6 (n=85) exhibited at least two Diap1+ PCs (Figure 7e″ and e′″, top arrowheads) and one or two PCs with no detectable or extremely low Diap1 accumulation (Figure 7e″ and e′″, two bottom arrowheads). In these cases as well, the Diap1− supernumerary PCs were often round and basally positioned, indicating that they are the PCs that were destined to die (Figure 7e″ and e′″, two bottom arrowheads). Taken together, these results strongly suggest that Diap1 is differentially regulated in the PC cluster such that two PCs accumulate Diap1 affording them protection from apoptosis, while supernumerary PCs undergo downregulation of Diap1, thereby allowing caspase activation and death to occur.
Figure 7.
Hid controls Diap1 downregulation in PCs destined to die. Confocal images of ovarian follicles stained with DAPI to label nuclei (white), GFP antibodies to label PC membranes (green) and Diap1 antibodies (red) (a–f) or with Fas3 antibodies to label PC membranes (green) and Diap1 antibodies (red) (g and h) or with Fas3 antibodies to label PC membranes (red) and β-galactosidase antibodies (green) to detect diap1-lacZ enhancer trap expression (i and j). The apical side of the follicular epithelium in which the PCs are embedded is toward the top. (a′–f′), (a″–f″) and (a′″–f′″) are magnified views of the boxed areas in the corresponding a–f panels. In (a′–f′), each PC is indicated by an asterisk. In (a″–e″), (a′″–e′″) and (i and i′), it is not possible to distinguish between PC destined to survive or die, so all PCs are indicated by white arrowheads. In (f″, f′″, g, g′, h, h′, j and j′), white arrowheads and yellow arrows mark PCs destined to survive or die, respectively, as indicated by the round shape, smaller size and more basal position, indicating progressive exclusion of the latter from the group. (a–d) Control: upd-Gal4/+ UAS-mCD8:GFP/+, (e) upd-Gal4/+ UAS-mCD8:GFP/UAS-p35, (f) upd-Gal4/+ UAS-mCD8:GFP/+ UAS-hid-IR/+, (g and h) hidP(5014)/ Df(3L)H99 (g) and hidP(5014)/Df(3L)R+X1 (h) escapers and (i and j) yw; thj5C8/TM3Sb ovaries
Table 3. Diap1 protein accumulation in polar cells during oogenesis in control and hid RNAi contexts.
We next assayed Diap1 accumulation in PC clusters in which hid function is knocked down by transgenic RNAi. Clusters of three (or four) PCs survived after stage 6 of oogenesis (Table 2), and this was associated with a greater proportion of clusters exhibiting Diap1 accumulation in all cells of the cluster compared with wild type (Table 3). Expression of hid RNAi leads to a majority of clusters from stages 7 to 10 with three Diap1+ PCs (47% compared with only 19% in the wild-type control; Figure 7f), and only a minority exhibited complete Diap1 downregulation in the supernumerary PC(s) (15% compared with 50% in the wild-type control). Diap1 accumulation was also assayed in ovaries from hid mutant escapers of two different genotypes, hidP[5014]/Df(3L)H99, which removes one copy of reaper as well as affecting both copies of hid and hidP[5014]/Df(3L)R+X1, which only affects hid and similar results were obtained in both cases. Almost all follicle extremities after stage 6 exhibited groups of more than two PCs as detected by membrane accumulation of Fas3 (Figure 7g, three PCs and Figure 7h and h′, four PCs observed in two different focal planes) and Diap1 protein was never absent in the supernumerary PCs (Figure 7g′, h and h′, yellow arrows), in contrast to the situation in controls in which Diap1 was often low or absent in these cells (Figure 7a, b and e). These results, therefore, indicate that hid function is necessary for downregulation of Diap1 in supernumerary PCs destined to die. In addition, as supernumerary PCs devoid of hid function and presenting Diap1 accumulation nevertheless exhibited loss of apical contact, rounding up and shrinkage (Figure 7g and h, yellow arrows), it would seem that these processes do not require hid function or Diap1 downregulation.
A diap1-lacZ reporter construct, shown to be correctly regulated in other tissue contexts in Drosophila,20 was expressed in all cells of PC clusters of three or more cells, even when one PC was already showing signs of apical detachment and shrinking (Figure 7i and j). Therefore, downregulation of Diap1 in PCs destined to die likely occurs post-transcriptionally.
Discussion
We show that PC apoptosis involves a canonical Hid–Diap1–Dronc/Dark–Drice apoptotic pathway like the one described by Leulier et al.14 in the Drosophila eye upon ectopic expression of Hid and consequent apoptosis. Our study, therefore, provides the confirmation that this pathway is functional in a physiological case of developmental apoptosis. Leulier et al.14 also found evidence for a second pathway involving Hid-induced activation of the caspases Strica and Decay, independent of Diap1. Using the same genetic tools they used (transgenic RNAi lines), however, we found no involvement of Strica or Decay functions in PC apoptosis. This difference might reflect a different level of Hid expression in the two systems. The high level of Hid induced in the eye by transgene expression may allow activation of Strica and Decay, while the physiological level of Hid in supernumerary PCs may not be sufficient to do so.
A role for effector caspases in PC apoptosis is strongly suggested by p35-induced survival of supernumerary PCs and specific caspase activation in PCs destined to die. In addition, among the transgenic RNAi lines targeting four effector caspase-encoding genes, drice, dcp1, decay and damm (previously validated by Leulier et al.14), only RNAi directed against drice led to prolonged survival of excess PCs. However, females homozygous for the null driceΔ1 or the strong hypomorphic dcp1prev1 allele, as well as driceΔ1/Δ1; dcp1prev1/+ females and driceΔ1/Δ1 PC mitotic clones in a dcp1prev1/+ background undergo PC apoptosis normally. As redundant function between these two genes has been reported in several cell types,21 the best interpretation for our results is that drice and dcp1 (and possible decay and/or damm as well) exhibit redundant function for PC apoptosis.
Among Dredd, Dronc and Strica, which resemble inititator caspases,22 Dronc is specifically required for many of the known developmental cell death programs, with specific exceptions, as well as in radiation-induced apopotosis15, 16, 23 along with the adaptor protein, Apaf-1/Dark.18, 24, 25 Our analysis of dronc and dark null mutations indicates that both are necessary for PC apoptosis cell autonomously in PCs destined to die. The partnership between Dronc and Dark thus seems maintained in the case of PC apoptosis, further supporting the idea that Dronc and Dark are universal partners for induction of apoptosis.
Control of caspase activation in PCs likely involves Diap1 function for several reasons. Indeed, Diap1 protein, present in all other germline and somatic cells of the ovary, is specifically downregulated in PCs destined to die. This regulation is likely to be post-translational as we show that expression of a diap1-lacZ enhancer trap is ubiquitous even in clusters in which supernumerary PCs are undergoing elimination. In addition, inactivation of diap1 by RNAi specifically in PCs leads to progressive degradation and demise of PC pairs, indicating that Diap1 is necessary for survival of mature PCs (Supplementary Figure 1b–e). In view of the known function of Diap1 in inhibition of caspase activity,5 our results allow us to propose that specific downregulation of Diap1 in PCs destined to die leads to caspase activation in these cells and consequent apoptosis.
We have also implicated a specific member of the RHG family of known Diap1 antagonists in PC apoptosis.26 Among several genes tested, we show that the function of hid is specifically necessary for PC apoptosis. The functions of reaper, hid and grim have been shown to be largely redundant in developmental cell death, though some specific phenotypes are associated with single gene mutations.27, 28, 29, 30 Our results clearly indicate a specific, essential function for hid in PC apoptosis and thus distinguish individual developmental roles for members of the RHG family. However, a role for some of the other RHGs, in particular Reaper, though possibly redundant or secondary, is not to be excluded. Indeed, we have previously shown that a lacZ transcriptional reporter for the reaper gene is activated in PCs destined to die when PC apoptosis is blocked by expression of p35.13 It is thus possible that hid function is necessary for primary induction of PC apoptosis and that reaper function participates to a secondary amplification loop that insures completion of PC elimination.
In addition, we have been able to establish a link between hid function and Diap1 regulation in PC apoptosis. Significantly, upon RNAi knockdown of hid function specifically in the PC group, or in hid mutant escapers, the supernumerary PCs that survive until late stages of oogenesis show maintained Diap1 expression likely explaining their prolonged survival. Although RHG family regulation of Diap1 levels by promoting Diap1 autoubiquitination and degradation has been shown through biochemical studies in vitro and upon ectopic overexpression of RHG proteins,25, 27, 31, 32, 33 this is the first demonstration, combining both genetic evidence and analysis of protein accumulation in situ, of Hid-mediated Diap1 downregulation in a case of developmental apoptosis involving physiological levels of both of these proteins. Some studies report a significantly higher capacity for Reaper than for Hid in blocking Diap1 activity,32 but our results show a specific role for Hid in Diap1 regulation for PC apoptosis.
Finally, we identify hid as an important regulatory point in the apoptotic cascade responsible for PC death. The accumulation of Hid has been shown to be regulated at both the transcriptional and post-transcriptional level depending on the tissue.34 In PCs, both Hid protein and hid transcripts accumulate specifically in supernumerary PCs destined to die, indicating upregulation of hid transcription and/or stabilization of hid mRNA. It is important to note, however, that detection of hid expression in PCs was not possible in a wild-type context. It was necessary to block PC apoptosis by knocking-down caspase function (ectopic p35 expression or mutations in caspase-encoding genes) in order to reveal the presence of Hid protein or hid transcripts in supernumerary PCs normally destined to die. Therefore, in the wild type, both hid transcripts and Hid protein are likely short-lived and/or transient, and induction of prolonged survival of supernumerary PCs by blocking caspase activity may allow increased accumulation, and thus detection, of these gene products. The accumulated Hid protein in supernumerary PCs induced in these genetic contexts appears to be functional as these cells exhibit highly efficient downregulation of Diap1.
Polar and border cell migration is also delayed to the same extent by mutations affecting different apoptotic players (hid, drice, caspases in general) (Supplementary Table 1). In addition, in all three genetic contexts tested, clusters with four PCs were more perturbed in their migration than clusters with three PCs (Supplementary Table 1). These results suggest that these migration defects are likely a consequence of the elevated number of PCs surviving into late stages of oogenesis, rather than reflecting a non-apoptotic role for these molecules in migration per se.
During the PC elimination process, PCs destined to die exhibit apical constriction followed by apical detachment from the germline and reduction in the contact with the other PCs leading to progressive shrinking of these cells. These morphological and polarity events may not depend on the Hid-mediated caspase activation, as they seem to occur before caspase activation and in the loss-of-function contexts we tested including hid escapers, which exhibited almost 100% of follicle poles with more than two PCs. It will be interesting to develop an in vivo real-time approach, which may reveal the sequence of events during PC apoptosis.
Materials and Methods
Fly strains
The following strains were generated for PC-specific expression of GFP or YFP: the enhancer trap upd-Gal4 X (provided by S Noselli) combined with UAS-mCD8:GFP III or with UAS-H2B:YFP III and neup72-Gal4; UAS–H2B:YFP (provided by M Gho). The following chromosomes were used in this study: UAS-p35,35 FRT80B dronci29 and FRT80B dronci24,16 FRT42D darkH16 and FRT42D darkG8,25 FRT79D Df(3L)H99,36 P (PZ) hid0514,27 hidWR+X1,37 XR38,28 yw; thj5C8/TM3Sb (lacZ enhancer trap in thread/diap1),20 UAS-diap1IR, tub-Gal 80ts (provided by F Leulier), driceΔ1,38 drice17,21 dcp1prev 39 and strains carrying RNAi constructs from NIG-FLY and VDRC collections: UAS-driceIR (NIG 7788R1, R2), UAS-hidIR (NIG 5123 R2, R3 and VDRC 7912, 8269), UAS-dammIR (NIG 118188 R1), UAS-decay-IR (NIG 14902 R2), UAS-dcp1IR (NIG 5370R and VDRC 34328, 34330), UAS-stricaIR (NIG 7863R) and UAS-dronc (VDRC 23033, 23035). FRT82B, driceΔ1 recombinants were generated between the P(neo FRT)82B (Bloomington stock number 2004) and driceΔ1 chromosomes. Flies carrying the recombinant chromosomes were selected for the presence of the neomycin-resistance gene by using fly medium supplemented with 0.1 mg/ml G418 (Gibco BRL Life Technologies, Inc., Carlsbad, CA, USA) and tested for the presence of the deletion corresponding to the driceΔ1 sequence by PCR (primers: 5′-CATGGACGCCACTAACAATG-3′ and 5′-GAGAATCAAAGAGGCGCAAG-3′).
Egg chamber staining procedure
Ovaries were dissected in PBS1X, then fixed for 20 min in 4% formaldehyde and rinsed three times in PBT (0.3% Triton X-100). The following primary antibodies, diluted in PBT, were used in this study: mouse monoclonal anti-Fas3 (1:20 DSHB), rabbit polyclonal anti-β-galactosidase (1:200, MP Biomedicals, Santa Ana, CA, USA), rabbit anti-GFP (1 : 500-Interchim), rabbit anti-human cleaved caspase-3 (1 : 20-Ozyme), guinea pig polyclonal anti-Hid (1 : 100 – provided by HD Ryoo), mouse monclonal anti-Diap1 (1 : 200 – provided by B Hay), mouse monoclonal anti-Singed (1 : 20 – DSHB). Fluorescence conjugated secondary antibodies were purchased from Invitrogen. All were used at a 1 : 200 dilution. In experiments in which anti-Fas3 and anti-Diap1 mouse monoclonal antibodies were used on the same tissue sample, first anti-Fas3 immunodetection was carried out with anti-mouse Alexa-488 secondary antibodies for fluorescent detection in green, samples were then thoroughly rinsed and anti-Diap1 immunodetection was next carried out with anti-mouse Cy3 secondary antibodies for fluorescent detection in red. All samples were mounted in citifluor (Kent, London, UK).
The hid riboprobe was generated using the 5A1 cDNA clone (provided by J Abrams).27 In situ hybridization with hid riboprobe was performed as described in Suter and Steward,40 with the following changes: Ovaries were dissected and fixed with 4% paraformaldehyde in PBST; temperature of hybridization was 65°C and sheep serum was used instead of bovine serum albumin. Detection was performed using the Dig Nucleic Acid Detection kit (Roche, Basel, Switzerland).
RNAi analysis
Crosses between RNAi lines and the upd-Gal4 driver were carried out at 25°C for 2 days, then progeny was transferred to 29°C for the rest of development as well as for adulthood. Females were dissected 4–5 days after eclosion.
upd-Gal4/+ UAS-diap1IR, tub-Gal 80ts/females were maintained at 18°C until eclosion, at which point they were shifted to 29°C and ovaries were dissected 5 days later.
Clonal analysis
For clonal analysis, females of the following genotypes were analyzed: hsp-flp/+ FRT80B dronci24,29/FRT80B ubi-GFP, hsp-flp/+ FRT42D darkG8,H16/FRT42D ubi-GFP and hsp-flp/+ FRT79D H99/FRT79D ubi-GFP. Females raised at 25°C were heat shocked at mid-pupae, upon eclosion and at 2 days of age for 1 h at 37°C each time, and ovaries were dissected 4 days later.
Microscopy and image processing
Immunolabeled ovary Z-stack images were taken on a Leica Sp2 confocal microscope housed by the Imagif Platform (https://www.imagif.cnrs.fr) and Z-projections were made using ImageJ.
Acknowledgments
We are grateful to J Abrams, A Bergmann, B Hay, P Kerner, F Leulier, K McCall, HD Ryoo, L Théodore and K White for providing us with precious reagents and fly stocks without which this work could not have been carried out. We appreciate the NIG-Fly, VDRC, DSHB (http://dshb.biology.uiowa.edu) and the Bloomington Stock Center for providing reagents and fly stocks. Imaging was carried out in part on the Imagif platform of the CNRS (Centre National de Recherche Scientifique) at Gif-sur-Yvette (https://www.imagif.cnrs.fr). AK and PG were funded by doctoral fellowships from the Ministère de l′enseignement supérieur et de la recherche (MESR). The work was financed by CNRS ATIP, Association pour la Recherche contre le Cancer (ARC) and Fondation pour la Recherche Médicale (FRM) grants to AMP.
Glossary
- CARD
caspase activation and recruitment domain
- DArk
Drosophila Apaf-1-related killer
- Dcp-1
death caspase-1
- Df(3L)
deficiency on the left arm of chromosome 3
- Diap1
Drosophila inhibitor of apoptosis protein 1
- DrICE
Drosophila ICE
- Dronc
Drosophila Nedd-2-like caspase
- DSHB
Developmental Studies Hybridoma Bank
- Fas3
Fasciclin III
- FLP
Flippase
- FRT
Flippase recombination target
- GFP
green fluorescent protein
- hid
head involution defective
- H2B
histone 2B
- hsp
heat shock protein 70 promoter
- IAP
inhibitor of apoptosis proteins
- IR
inverted repeat
- JAK/STAT
Janus kinase/signal transducer and activator of transcription
- mCD8
mouse CD8
- neur
neuralized
- NIG
National Institute of Genetics (Japan)
- PCs
polar cells
- RHG
Reaper, Hid and Grim
- RNAi
RNA interference
- TM3SB
third chromosome multiply inverted Stubble
- UAS
upstream activating sequence
- ubi
ubiquitin promoter
- upd
unpaired
- VDRC
Vienna Drosophila Research Center
- YFP
yellow fluorescent protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Baehrecke EH. How death shapes life during development. Nat Rev Mol Cell Biol. 2002;3:779–787. doi: 10.1038/nrm931. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Tugentman M, Shilo BZ, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2:159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- Monserrate JP, Brachmann CB. Identification of the death zone: a spatially restricted region for programmed cell death that sculpts the fly eye. Cell Death Differ. 2007;14:209–217. doi: 10.1038/sj.cdd.4401947. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Abrams JM. Caspase activation – stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- Orme M, Meier P. Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis. 2009;14:950–960. doi: 10.1007/s10495-009-0358-2. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, et al. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Yang AY, Srivastava M. Regulators of IAP function: coming to grips with the grim reaper. Curr Opin Cell Biol. 2003;15:717–724. doi: 10.1016/j.ceb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Eckelman BP, Drag M, Snipas SJ, Salvesen GS. The mechanism of peptide-binding specificity of IAP BIR domains. Cell Death Differ. 2008;15:920–928. doi: 10.1038/cdd.2008.6. [DOI] [PubMed] [Google Scholar]
- Grammont M, Irvine KD. Organizer activity of the polar cells during Drosophila oogenesis. Development. 2002;129:5131–5140. doi: 10.1242/dev.129.22.5131. [DOI] [PubMed] [Google Scholar]
- Torres IL, Lopez-Schier H, St Johnston D. A Notch/Delta-dependent relay mechanism establishes anterior-posterior polarity in Drosophila. Dev Cell. 2003;5:547–558. doi: 10.1016/s1534-5807(03)00272-7. [DOI] [PubMed] [Google Scholar]
- Beccari S, Teixeira L, Rorth P. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech Dev. 2002;111:115–123. doi: 10.1016/s0925-4773(01)00615-3. [DOI] [PubMed] [Google Scholar]
- Xi R, McGregor JR, Harrison DA. A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev Cell. 2003;4:167–177. doi: 10.1016/s1534-5807(02)00412-4. [DOI] [PubMed] [Google Scholar]
- Besse F, Pret A-M. Apoptosis-mediated cell death within the ovarian polar cell lineage of Drosophila melanogaster. Development. 2003;130:1017–1027. doi: 10.1242/dev.00313. [DOI] [PubMed] [Google Scholar]
- Leulier F, Ribeiro PS, Palmer E, Tenev T, Takahashi K, Robertson D, et al. Systematic in vivo RNAi analysis of putative components of the Drosophila cell death machinery. Cell Death Differ. 2006;13:1663–1674. doi: 10.1038/sj.cdd.4401868. [DOI] [PubMed] [Google Scholar]
- Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Yamamoto-Goto Y, Tokushige N, Kanda H, Miura M. Down-regulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and DRONC. J Biol Chem. 2002;277:23103–23106. doi: 10.1074/jbc.C200222200. [DOI] [PubMed] [Google Scholar]
- Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A. The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ. 2006;13:1697–1706. doi: 10.1038/sj.cdd.4401920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum JS, Arama E, Steller H, McCall K. The Drosophila caspases Strica and Dronc function redundantly in programmed cell death during oogenesis. Cell Death Differ. 2007;14:1508–1517. doi: 10.1038/sj.cdd.4402155. [DOI] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu W-J, Mills K, Daish T, et al. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, et al. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K. Reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thummel CS. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. PNAS. 2004;101:8022–8027. doi: 10.1073/pnas.0402647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. PNAS. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colón-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Yan N, Wu J-W, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Bilak A, Su TT. Regulation of Drosophila melanogaster pro-apoptotic gene hid. Apoptosis. 2009;14:943–949. doi: 10.1007/s10495-009-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Abbott MK, Lengyel JA. Embryonic head involution and rotation of male terminalia require the Drosophila locus head involution defective. Genetics. 1991;129:783–789. doi: 10.1093/genetics/129.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, et al. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development. 2006;133:3305–3315. doi: 10.1242/dev.02495. [DOI] [PubMed] [Google Scholar]
- Laundrie B, Peterson JS, Baum JS, Chang JC, Fileppo D, Thompson SR, et al. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 2003;165:1881–1888. doi: 10.1093/genetics/165.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Steward R. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell. 1991;67:917–926. doi: 10.1016/0092-8674(91)90365-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.