Abstract
Loss of parkin function is responsible for the majority of autosomal recessive parkinsonism. Here, we show that parkin is not only a stress-protective, but also a stress-inducible protein. Both mitochondrial and endoplasmic reticulum (ER) stress induce an increase in parkin-specific mRNA and protein levels. The stress-induced upregulation of parkin is mediated by ATF4, a transcription factor of the unfolded protein response (UPR) that binds to a specific CREB/ATF site within the parkin promoter. Interestingly, c-Jun can bind to the same site, but acts as a transcriptional repressor of parkin gene expression. We also present evidence that mitochondrial damage can induce ER stress, leading to the activation of the UPR, and thereby to an upregulation of parkin expression. Vice versa, ER stress results in mitochondrial damage, which can be prevented by parkin. Notably, the activity of parkin to protect cells from stress-induced cell death is independent of the proteasome, indicating that proteasomal degradation of parkin substrates cannot explain the cytoprotective activity of parkin. Our study supports the notion that parkin has a role in the interorganellar crosstalk between the ER and mitochondria to promote cell survival under stress, suggesting that both ER and mitochondrial stress can contribute to the pathogenesis of Parkinson's disease.
Keywords: ATF4, c-Jun, ER stress, parkin, Parkinson's disease, UPR
Mitochondrial dysfunction has long been implicated in the pathogenesis of Parkinson's disease (PD). Mitochondrial toxins targeting complex I of the electron transport chain can induce acute parkinsonism in humans and are being used to model PD in animals. More recently, several PD-associated genes have been shown to influence mitochondrial function, morphology, dynamics and turnover (reviewed in Winklhofer and Haass1 and Schapira2). In addition to mitochondrial dysfunction, several lines of evidence indicate that endoplasmic reticulum (ER) stress may contribute to the pathogenesis of PD (reviewed in Wang and Takahashi3 and Lindholm et al.4). First, toxins such as MPTP, 6-OHDA or rotenone, used to induce parkinsonism in animal models, have been shown to cause ER stress.5, 6, 7 Second, ER stress accounts for at least some aspects of α-synuclein toxicity. α-Synuclein has been shown to block ER to Golgi vesicular trafficking in different model systems,8 and overexpression of α-synuclein can induce ER stress.8, 9 Finally, ER stress has been documented in dopaminergic neurons of the parkinsonian brain, exemplified by increased levels of phospho-PERK, phospho-eIF2α and caspase-4.10, 11 The link between PD and ER dysfunction was recently reinforced by the observation that the E3 ubiquitin ligase parkin can protect cells from ER stress-induced cell death induced by the overexpression of Pael-R, a putative parkin substrate prone to misfolding within the secretory pathway.12, 13 Mutations in the parkin gene were identified as a cause of early onset PD in Japanese families.14 Since then, more than 100 mutations have been described in patients of diverse ethnic backgrounds, accounting for the majority of autosomal recessive parkinsonism.
ER stress originates from the accumulation of unfolded secretory proteins, perturbations in calcium homeostasis or redox status, alterations in glycosylation or energy deprivation. The ER has evolved sophisticated stress response signaling pathways collectively called the unfolded protein response (UPR), destined to increase the ER folding capacity, to reduce the folding load and to restore ER homeostasis (reviewed in Ron and Walter15). Conversely, when ER stress conditions are severe or persistent, apoptotic cell death is induced (reviewed in Kim et al.16).
Recent research revealed that the ER physically and functionally interacts with mitochondria to influence key aspects of cellular physiology and viability (reviewed in Pizzo and Pozzan17). Interactions between these organelles allow the exchange of metabolites and are implicated in the regulation of calcium signaling and cell death pathways. Our study shows that parkin expression is increased on ER stress and mitochondrial stress through the PERK/ATF4 branch of the UPR. Increased expression of parkin prevents ER stress-induced mitochondrial damage and cell death, providing evidence for a functional link between parkin, ER stress and mitochondrial integrity.
Results
Mitochondrial membrane dissipation causes ER stress and leads to transcriptional upregulation of parkin
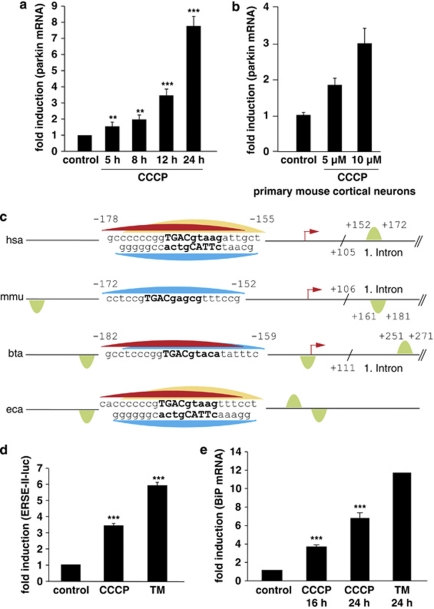
The protonophore CCCP is being used to induce mitochondrial damage in various model systems. CCCP renders the mitochondrial inner membrane permeable for protons and causes dissipation of the proton gradient. We previously observed that the complex I inhibitor rotenone induces an upregulation of parkin mRNA and protein levels.18 CCCP has recently been shown to cause the translocation of parkin to damaged mitochondria, which are then removed by autophagy.19 We, therefore addressed the question whether CCCP might also have an impact on the transcriptional regulation of parkin. Parkin transcripts were significantly increased both in CCCP-treated SH-SY5Y cells and primary mouse cortical neurons (Figures 1a and b). To get insight into the underlying mechanism, we screened the parkin promoter for possible binding sites (BSs) of stress-regulated transcription factors by using TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html).20 We discovered a putative CREB/ATF site in the parkin promoter located from −169 to −161 bp relative to the transcriptional start site (TSS). A further in-depth analysis identified this site as a possible binding sequence for ATF4 (Figures 1c and 3a). The CREB/ATF site within the parkin promoter is conserved among species (Figure 1c), supporting the notion that this site might be functionally relevant. An additional ATF4-binding site is located downstream of the TSS within the first intron of human, bovine and rodent parkin (Figure 1c).
Figure 1.
Mitochondrial stress induced by CCCP activates the UPR and leads to an upregulation of parkin. (a) Parkin mRNA levels are increased in response to mitochondrial membrane dissipation, induced by CCCP. SH-SY5Y cells were incubated with 10 μM CCCP for the indicated time. Cells were collected and total cellular RNA was isolated and subjected to quantitative RT-PCR using parkin-specific primers. The amount of RNA of each sample was normalized with respect to the endogenous housekeeping gene β-actin. Shown is the fold increase of parkin-specific mRNA compared with untreated control cells. (b) Parkin mRNA is upregulated upon CCCP treatment in primary mouse cortical neurons. Primary cortical neurons derived from embryonic mouse brain were incubated with CCCP (10 μM) for 12 h and analyzed as described in (a). (c) Human, mouse, bovine and equine promoter sequences of parkin, which are elongated downstream of the transcription start site (TSS) by 150 bp. Red arrow indicates the TSS and positions are denoted with relative to the TSS. The CREB/ATF-binding sites are indicated by semicircles. Red, yellow and blue semicircles are predicted by three different binding motifs, which correspond to a Genomatix-defined family of 14 matrices describing the CREB/ATF-binding site. The red and yellow colored binding sites are conserved between Homo sapiens, Bos taurus and Equus caballus, and H. sapiens and E. caballus, respectively, whereas the blue binding site is conserved across all four species. The green semicircles (not conserved) are additional binding sites. Downstream of the TSS, in the first intron of the parkin gene, an additional CREB/ATF-binding site is located in H. sapiens, Mus musculus and B. taurus. The consensus ATF4-binding site is written in bold letters. hsa, Homo sapiens; mmu, Mus musculus; bta, Bos taurus; eca, Equus caballus. (d and e) CCCP activates the UPR and causes ER stress. (d) The ER stress luciferase reporter construct ER stress-response element II (ERSE-II-luc) is activated by CCCP. HEK293T cells were transfected with the ERSE-II-luc reporter. At 24 h after transfection, the cells were treated with 10 μM CCCP for 24 h. As a positive control, the cells were treated with the ER stressor tunicamycin (2 μg/ml, 24 h). Shown is the fold induction of luciferase activity in CCCP-treated cells in comparison with non-treated control cells. Quantification is based on triplicates of at least three independent experiments. (e) BiP expression is increased in response to CCCP treatment. As an indicator of ER stress, BiP mRNA levels were analyzed in SH-SY5Y cells treated with CCCP (10 μM) for the indicated time by quantitative RT-PCR as described in Figure 1a. Tunicamycin (2 μg/ml) was used as a positive control to induce ER stress. ***P<0.001, **P<0.01
ATF4 is a transcription factor, which is activated under ER stress. Some toxins used to model PD have been associated with ER stress, therefore we analyzed whether mitochondrial membrane dissipation induced by CCCP can cause ER stress. Regulation of gene expression in response to ER stress is mediated by the UPR, a stress-response program that re-establishes cellular homeostasis by the combinatorial action of specific transcription factors, binding to ER stress-responsive elements in the regulatory regions of UPR target genes (reviewed in Ron and Walter15). To test whether CCCP induces ER stress, we cloned a luciferase reporter construct containing the ER stress-responsive element ERSE-II (ERSE-II-luc, Supplementary Figure 5). Treatment of cells transiently expressing the luciferase reporter construct with tunicamycin (TM) induced an increase in luciferase expression, demonstrating the functionality of this ER stress reporter construct (Figure 1d). Moreover, CCCP was also able to significantly increase luciferase expression from ERSE-II-luc (Figure 1d). In addition, we quantified mRNA levels of the ER chaperone BiP, which is a major target of the UPR. CCCP treatment caused an upregulation of BiP mRNA, indicating that the mitochondrial toxin CCCP can induce ER stress (Figure 1e).
Parkin gene expression is upregulated in response to ER stress
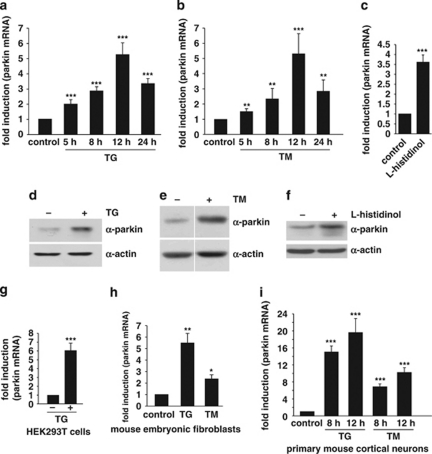
In a next step, we tested whether expression of parkin is influenced by ER stress. SH-SY5Y cells were incubated with either the ER Ca2+-ATPase inhibitor thapsigargin (TG) or the N-glycosylation inhibitor TM. Both ER stressors significantly increased the levels of parkin mRNA with a maximum at 12 h after treatment (Figures 2a and b). Another classical inducer of ER stress is amino acid starvation. We therefore made use of -histidinol, a histidine analog, which inhibits activation of histidine by histidyl-tRNA synthetase. Also in this ER stress paradigm, parkin mRNA levels increased (Figure 2c). Notably, the upregulation of parkin under ER stress, induced by TG, TM or -histidinol, was also observed on the protein level (Figures 2d–f), and was not restricted to SH-SY5Y cells, as a significant upregulation of parkin in response to TG or TM treatment was also observed in HEK293T cells, mouse embryonic fibroblasts (MEF) and primary mouse cortical neurons (Figures 2g–i). Interestingly, the upregulation of parkin upon ER stress in not dependent on PINK1 expression (data not shown).
Figure 2.
Parkin gene expression is upregulated in response to ER stress. (a and b) Parkin mRNA levels are increased under ER stress induced by thapsigargin or tunicamycin. SH-SY5Y cells were incubated with 1 μM thapsigargin (TG) (a) or 2 μg/ml tunicamycin (TM) (b) for the indicated time. Cells were collected and total cellular RNA was isolated and subjected to quantitative RT-PCR using parkin-specific primers. The amount of RNA of each sample was normalized with respect to the endogenous housekeeping gene β-actin. The same results were obtained when 18sRNA was used as a control gene (data not shown). Shown is the fold increase of parkin-specific mRNA compared with untreated control cells. (c) Amino acid starvation leads to an upregulation of parkin mRNA. SH-SY5Y cells were treated with 2 mM -histidinol in cell culture medium, containing 10% dialysed FCS, for 14 h. The cells were then collected and total cellular RNA was isolated and subjected to quantitative RT-PCR using parkin-specific primers as described under Figure 1a. (d–f) Parkin protein expression is increased after ER stress induced by TG, TM, or amino acid starvation. Expression of endogenous parkin after treatment of SH-SY5Y cells with TG (d), TM (e) or -histidinol (f) for 14 h was analyzed by western blotting using the anti-parkin mAb PRK8. Loading was controlled by re-probing the blots for β-actin. The western blot image (e) was re-arranged by excluding one line, as indicated by a white line; all samples originate from one gel. (g–i) Parkin mRNA is upregulated on ER stress in HEK293T cells, mouse embryonic fibroblasts and primary mouse cortical neurons. HEK293 T cells (g), mouse embryonic fibroblasts (h), or primary cortical neurons derived from embryonic mouse brain (i) were incubated with TG (1 μM) or TM (2 μg/ml; primary cortical neurons: 3 μg/ml) for 12 or 8 h and 12 h (primary cortical neurons) and analyzed as described in (a). ***P<0.001, **P<0.01, *P<0.05
Parkin is a target of the UPR through the PERK/ATF4 pathway
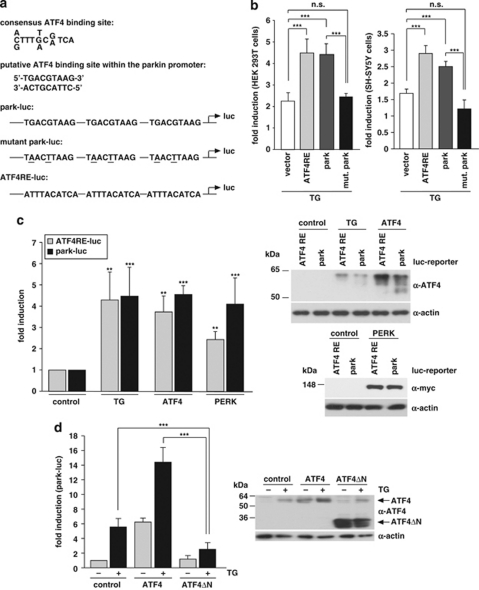
To test whether the putative ATF4-binding site within the parkin promoter mediates upregulation of parkin in response to ER stress, we created a luciferase reporter construct using the pGL3-luc promoter vector containing the putative ATF4-binding site (in triplicate) in front of a sequence coding for luciferase (park-luc, Figure 3a). As a positive control, we cloned the confirmed ATF4-binding site of the IGFBP1 promoter21 analogously to the park-luc construct. This control construct was termed ATF4-responsive element (ATF4RE)-luc (Figure 3a). As a negative control, we cloned a mutant park-luc construct, harboring two point mutations in the putative ATF4-binding motif (mut. park-luc, Figure 3a). We tested park-luc in comparison with ATF4RE-luc under ER stress conditions and observed that luciferase expression from both reporter constructs was increased to a similar extent, whereas mutant park-luc behaved like the pGL3-luc vector control, both in HEK293T and SH-SY5Y cells (Figure 3b). Of note, CCCP treatment also induced increased transcription from both ATF4RE-luc and park-luc (Supplementary Figures 1A and B). Moreover, forced expression of ATF4 or upstream PERK also activated transcription from park-luc (Figure 3C). In line with this observation, dominant-negative ATF4ΔN, which lacks the N-terminal transcriptional activation domain,22 significantly interfered with the ER stress-induced activation of park-luc (Figure 3d).
Figure 3.
Transcriptional upregulation of parkin under ER stress is mediated by ATF4. (a) Schematic representation of the consensus ATF4-binding site, the putative ATF4-binding site within the parkin promoter and the luciferase reporter constructs cloned for the analysis described in the following: park-luc contains the putative ATF4-binding site of the parkin promoter in triplicate, mutant park-luc habors two point mutations within the putative ATF4-binding site, and ATF4RE-luc contains the confirmed ATF4-binding site of the insulin growth factor binding protein 1 (IGFBP1) promoter in triplicate. Of note, the putative binding site for ATF4 within the parkin promoter is located on the complementary strand in 5′ → 3′ direction. (b) The park-luc reporter construct is induced after ER stress. HEK293 T cells or SH-SY5Y cells were transfected with either the control luciferase reporter construct pGL3-luc (vector), the ATF4RE-luc construct containing the confirmed ATF4-binding site, the park-luc construct or the park-luc construct with a mutated ATF4 binding site (mut. park). At 8 h after transfection, cells were incubated with 1 μM thapsigargin (TG) and collected after 14 h of treatment. Shown is the fold induction of luciferase activity in stressed cells compared with the non-stressed control based on triplicates of at least three independent experiments. (c) Increased expression of ATF4 or upstream PERK induces transcription from the park-luc reporter construct. HEK293T cells were co-transfected with the ATF4RE-luc reporter plasmid or the park-luc reporter plasmid and ATF4, PERK or GFP (as a control). As a positive control, one set of cells was treated with TG as described under (b) Shown is the fold induction of luciferase activity compared with GFP-expressing control cells based on triplicates of at least three independent experiments (left panel). Expression levels of ATF4 and PERK were analyzed by immunoblotting using the anti-ATF4 pAb C-20 or the anti-myc mAb 9E10 (right panels). Notably, TG treatment (1 μM, 14 h) induced the increased expression of endogenous ATF4. Loading was controlled by re-probing the blots for β-actin. (d) A dominant-negative mutant of ATF4 (ATF4ΔN) interferes with the activation of the park-luc reporter construct in response to ER stress. HEK293T cells were co-transfected with the park-luc reporter plasmid and ATF4, ATF4ΔN, or GFP (as a control). At 8 h after transfection, cells were incubated with 1 μM TG for 14 h. Shown is the fold induction of luciferase activity in comparison with GFP-expressing control cells based on triplicates of at least three independent experiments (left panel). Expression levels of ATF4 and ATF4ΔN were analyzed by immunoblotting using the anti-ATF4 pAb C-20 (right panel). Loading was controlled by re-probing the blots for β-actin. ***P<0.001, **P<0.01, n.s.=not significant
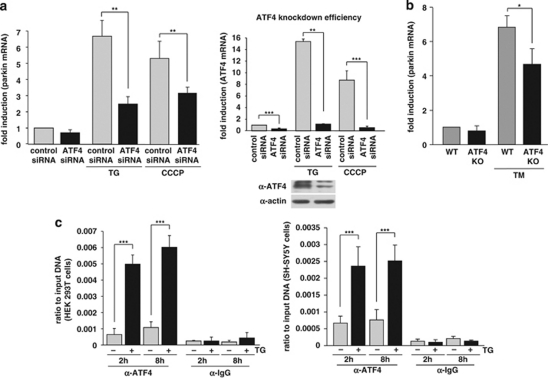
To increase experimental evidence for a role of endogenous ATF4 in mediating the transcriptional upregulation of parkin under ER stress, we knocked down ATF4 expression by RNAi. SH-SY5Y cells were transfected with ATF4 small-interfering RNA (siRNA) or control siRNA, and the efficiency of the ATF4 knockdown was verified at the mRNA and protein level (Figure 4a, right panel). Of note, ATF4 is specifically induced after ER stress, whereas under non-stress conditions expression levels are low. Remarkably, in ATF4-deficient cells, the upregulation of parkin induced by ER stress (TG) or mitochondrial stress (CCCP) was significantly reduced (Figure 4a, left panel). The same results were obtained employing two different ATF4-specific siRNA duplexes (data not shown). Moreover, we analyzed the transcriptional regulation of parkin in primary cortical neurons derived from ATF4-knockout mice.23 The upregulation of parkin on ER stress was significantly reduced in ATF4-deficient neurons (Figure 4b), indicating that ATF4 indeed has an important role in the stress-induced regulation of parkin expression. Interestingly, in our experimental approach, a transient downregulation of ATF4 had a more severe effect on the stress-induced regulation of parkin than a stable knockout of ATF4. It is conceivable that compensatory effects in ATF4-knockout mice account for this observation.
Figure 4.
ATF4 binds to the parkin promoter and mediates parkin upregulation in response to ER and mitochondrial stress. (a) ER and mitochondrial stress-induced upregulation of parkin is impaired in ATF4-deficient cells. SH-SY5Y cells were transfected with ATF4-specific or control siRNA duplexes. Two days later, cells were re-transfected with siRNA duplexes and then incubated with 1 μM TG or 10 μM CCCP for 14 h. The cells were collected and analyzed as described in Figure 1a by quantitative RT-PCR using parkin-specific or ATF4-specific primers. The amount of RNA of each sample was normalized with respect to β-actin. Shown is the fold increase of parkin mRNA in response to TG or CCCP treatment (left panel). The efficiency of ATF4 downregulation was determined by quantitative RT-PCR (right panel) and western blotting (lower right panel) using the anti-ATF4 pAb C-20. (b) ER stress-induced upregulation of parkin is impaired in primary cortical neurons from ATF4-knockout mice. Primary cortical neurons from ATF4-deficient or wild-type mice were treated with tunicamycin (3 μg/ml) for 8 h. Total RNA was isolated and analyzed using parkin-specific primers as described in Figure 1a. (c) ATF4 binds to the parkin promoter in vivo. HEK293T cells or SH-SY5Y cells incubated with or without 300 nM TG for 2 and 8 h were used to perform a ChIP analysis using a pAb specific for ATF4 in comparison with a nonspecific rabbit IgG. For the final real-time PCR step, primers specific for the parkin promoter region were used. ***P<0.001, **P<0.01, *P<0.05
Finally, binding of ATF4 to the parkin promoter could be demonstrated by chromatin immunoprecipitation (ChIP) assays using a polyclonal anti-ATF4 antibody. A rabbit polyclonal antibody (pAb) against chicken IgG was used as a nonspecific control. After isolation of crosslinked chromatin from cells incubated with or without TG, immunoprecipitated DNA was analyzed by real-time PCR. The ChIP analysis revealed specific binding of ATF4 to the parkin promoter after 2 and 8 h of TG treatment in both HEK293T and SH-SY5Y cells (Figure 4c).
c-Jun acts as a transcriptional repressor of parkin and has a dominant effect on ATF4
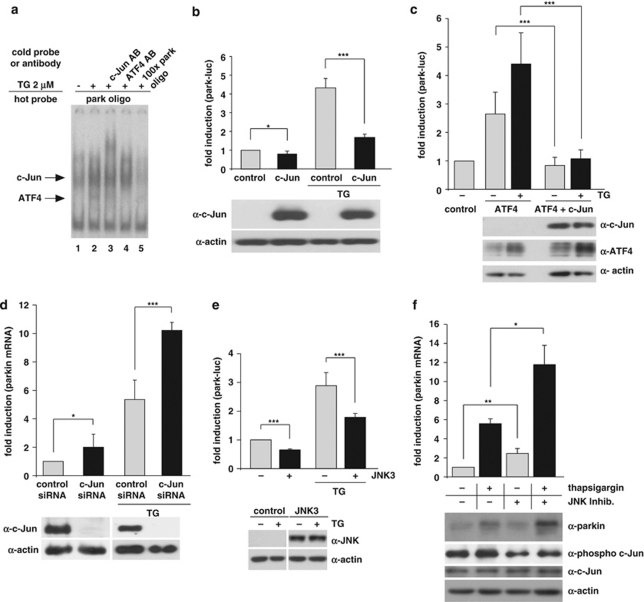
When we performed electrophoretic mobility shift assays using the putative ATF4-binding site of the parkin promoter as a radiolabeled probe (park oligo), we observed a second complex in addition to the ATF4–DNA complex. This complex showed a reduced mobility in comparison with the ATF4–park oligo complex, and its relative intensity was increased after ER stress (Figure 5a, lanes 1, 2). By testing various antibodies against transcription factors that could bind to the CREB/ATF site for their potential to supershift the upper band, we found that c-Jun is also able to bind to the park oligo. The respective band was supershifted by an anti-c-Jun antibody (Figure 5a, lane 3) and was competed by an excess of unlabeled park oligo (Figure 5a, lane 5). To test whether c-Jun might have an impact on the transcriptional regulation of parkin, we first analyzed the effect of c-Jun on the park-luc reporter construct. Interestingly, increased levels of c-Jun significantly reduced transcription from park-luc under ER stress (Figure 5b). Moreover, c-Jun suppressed the ATF4-induced activation of park-luc, both under basal and ER stress conditions, indicating a dominant-negative effect (Figure 5c). Next we performed a c-Jun knockdown approach by RNAi. c-Jun-specific siRNA duplexes were transfected into SH–SY5Y cells, resulting in a reduction of c-Jun mRNA by ∼87% under normal conditions and by ∼69% under ER stress conditions (Supplementary Figure 2). The knockdown efficiency was also verified at the protein level (Figure 5d, lower panel). Notably, in c-Jun-deficient cells, parkin upregulation in response to ER stress was significantly increased and also in non-stressed cells more parkin was expressed when c-Jun was silenced (Figure 5d, upper panel). These results indicate that c-Jun can bind to the ATF4-binding site within the parkin promoter to mediate repression of parkin expression. c-Jun is a major target of JNKs, a subfamily of the MAPK superfamily. JNK1 and JNK2 are ubiquitously expressed, whereas JNK3 is primarily found in brain, heart and testes. As JNK3 has been linked to cell death in several models of neurodegeneration (reviewed in Waetzig and Herdegen24), we tested whether it has an impact on parkin expression. Increased expression of JNK3 indeed significantly suppressed transcription from the park-luc construct (Figure 5e). Moreover, treatment of cells with the JNK inhibitor SP600125 increased parkin mRNA and protein levels both under stress and non-stress conditions (Figure 5f).
Figure 5.
c-Jun suppresses the upregulation of parkin after ER stress. (a) c-Jun binds to the parkin oligonucleotide harboring the CREB/ATF-binding site. HEK293 T cells were incubated with 2 μM thapsigargin (TG) and collected after 3 h. Nuclear extracts were prepared and tested for binding to the 32P-labeled oligonucleotide comprising the putative ATF4-binding site within the parkin promoter (park oligo; lanes 1–5) by an electrophoretic mobility shift assay (EMSA). The labeled oligonucleotides were incubated with nuclear extracts in the absence or presence of a 100-fold excess of unlabeled park oligo (lane 5) to compete with the binding reaction. To test for supershift activity, the anti-c-Jun pAb (N) sc-45X (lane 3) or the anti-ATF4 pAb C-20 (lane 4) was added to the binding reaction. (b) c-Jun decreases transcription from the park-luc reporter after ER stress. HEK293 T cells were co-transfected with the park-luc reporter construct and c-Jun or GFP (as a control). Eight hours after transfection, the cells were treated with 1 μM TG for 14 h. Shown is the fold induction of luciferase activity in c-Jun-expressing cells in comparison with GFP-expressing control cells based on triplicates of at least three independent experiments. Expression levels of c-Jun were analyzed by immunoblotting using the anti-c-Jun pAb (N) sc-45 (lower panel). Protein (3 μg) of total cell lysates was loaded. Loading was controlled by re-probing the blots for β-actin. (c) c-Jun suppresses the ATF4-mediated activation of the park-luc construct. HEK293 T cells were co-transfected with the park-luc reporter plasmid and either GFP (as a control), ATF4 plus GFP or ATF4 plus c-Jun. Eight hours after transfection, the cells were treated with 1 μM TG for 14 h. Shown is the fold induction of luciferase activity in ATF4-expressing cells in comparison with ATF4- and c-Jun-expressing cells based on triplicates of at least three independent experiments. Expression levels of ATF4 and c-Jun were analyzed by immunoblotting using the anti-ATF4 pAb C-20 or the anti-c-Jun pAb (N) sc-45. Protein (3 μg) of total cell lysates was loaded. Loading was controlled by re-probing the blots for β-actin (lower panel). (d) ER stress-induced upregulation of parkin is increased in c-Jun-deficient cells. SH-SY5Y cells were transfected with c-Jun-specific or control siRNA duplexes. One day later, cells were re-transfected with siRNA duplexes and incubated with 1 μM TG for 14 h. The cells were collected and analyzed as described in Figure 1a by quantitative RT-PCR using parkin-specific primers. The amount of RNA of each sample was normalized with respect to β-actin. Shown is the fold increase of parkin mRNA in response to TG treatment (upper panel). The efficiency of c-Jun downregulation was determined by western blotting using the anti-c-Jun anti-c-Jun pAb (N) sc-45 (lower panel). Protein (30 μg) of total cell lysates was loaded. (e) JNK3 decreases transcription from the park-luc reporter. HEK293 T cells were co-transfected with the park-luc reporter plasmid and JNK3 or GFP (as a control). Twenty four hours after transfection, the cells were treated with 1 μM TG for 8 h. Shown is the fold induction of luciferase activity in JNK3-expressing cells in comparison with GFP-expressing control cells based on triplicates of at least three independent experiments. Expression levels of JNK3 were analyzed by immunoblotting using an anti-JNK pAB (lower panel). Loading was controlled by re-probing the blots for β-actin. (f) The JNK inhibitor SP600125 increases parkin upregulation in response to ER stress. SH-SY5Y cells were treated with or without the JNK inhibitor SP600125 (10 μM) for 24 h. Thapsigargin was added after 10 h for additional 14 h. To quantify parkin-specific mRNA, cells were collected and analyzed as described in Figure 1a for quantitative RT-PCR using parkin-specific primers. The amount of RNA of each sample was normalized with respect to β-actin. Shown is the fold increase of parkin mRNA. Parkin protein levels were analyzed by immunoblotting using an anti-parkin PRK8 mAb (lower panel). The efficiency of SP600125 was controlled by blotting against phosphorylated c-Jun using the phospho-specific anti-c-Jun antibody X (Ser63) II pAb. Protein (15 μg) of total cell lysates was loaded. Loading was controlled by re-probing the blots for c-Jun and β-actin. ***P<0.001, **P<0.01, *P<0.05
Parkin protects cells from ER stress-induced cell death
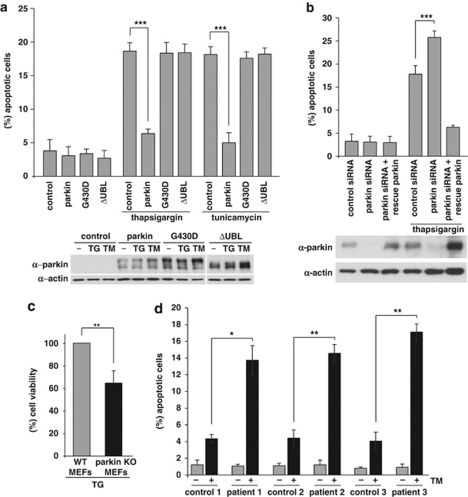
Conceptually, upregulation of parkin in response to ER stress might help to preserve cellular function and survival within the adaptive phase of the UPR. To test the capacity of parkin to protect cells from ER stress-induced toxicity, we treated SH-SY5Y cells transiently expressing wildtype (wt) or mutant parkin with TG or TM. Cells undergoing apoptosis were analyzed by indirect fluorescence using an antibody specific for activated caspase-3. In contrast to control cells and cells expressing pathogenic parkin mutants, cells overexpressing wt parkin were protected against apoptosis induced by ER stress (Figure 6a). To increase evidence for a role of endogenous parkin in coping with ER stress, we analyzed the consequences of a parkin knockdown induced by RNAi. Parkin knockdown cells showed a significant increase in apoptotic cells after ER stress in comparison with control siRNA-transfected cells (Figure 6b). A decrease in the viability of parkin-deficient SH-SY5Y and HEK293T cells after ER stress was also observed by employing the MTT assay (Supplementary Figures 3A and B). Notably, the increased vulnerability of parkin-deficient cells to ER stress-induced cell death could be rescued by the expression of siRNA-resistant parkin, confirming a parkin-specific effect (Figure 6b). In addition, we analyzed the viability of MEFs derived from parkin-knockout mice25 under ER stress conditions. In comparison with MEFs derived from wt mice, the parkin-knockout MEFs showed a decreased viability under ER stress induced by TG (Figure 6c). Moreover, primary skin fibroblasts from three patients with mutations in the parkin gene displayed a significant increase in cell death in response to TM treatment compared with age- and gender-matched control fibroblasts from healthy individuals (Figure 6d). Of note, levels of CHOP, phospho-c-Jun and phospho-JNK, which have been associated with the proapoptotic branch of ER stress pathways, were increased in patient fibroblasts under ER stress (Supplementary Figure 4).
Figure 6.
Parkin protects cells from ER stress-induced cell death. (a) Increased expression of wild-type (wt) parkin protects cells from ER stress-induced cell death. SH-SY5Y cells were co-transfected with EYFP (as a control) and wt parkin or the pathogenic parkin mutants G430D or ΔUBL. Twenty four hours after transfection, cells were incubated with 10 μM thapsigargin (TG) or 5 μg/ml tunicamycin (TM) at 37 °C for 8 h, fixed, permeabilized, and then the activation of caspase-3 was analyzed by indirect immunofluorescence using an anti-active caspase-3 pAb. Shown is the percentage of apoptotic cells among transfected cells. Parkin expression levels were determined by immunoblotting using the anti-parkin PRK8 mAb. Loading was controlled by re-probing the blots for β-actin (lower panel). (b) Parkin-deficient cells are more vulnerable to ER stress-induced cell death. SH-SY5Y cells were transfected with parkin-specific or control siRNA duplexes and co-transfected with EYFP (as a control) or siRNA-resistant wt parkin (rescue parkin). Three days later, the cells were stressed with TG (10 μM) for 8 h fixed, permeabilized, and then the activation of caspase-3 was analyzed by indirect immunofluorescence as described in A. Parkin expression levels were determined by immunoblotting using the anti-parkin PRK8 mAb. Loading was controlled by re-probing the blots for β-actin (lower panel). (c) Mouse embryonic fibroblasts (MEFs) derived from parkin-knockout mice are more vulnerable to ER stress than wt MEFs. MEFs from wt or parkin-knockout (ko) mice were stressed with TG (10 μM) for 16 h and then cellular viability was determined by the MTT assay. Shown is the relative viability of ko MEFs in comparison with wt MEFs after TG treatment. Quantification is based on five independent experiments. (d) Skin fibroblasts of patients carrying pathogenic mutations in the parkin gene are more vulnerable to ER stress. Skin fibroblasts from patients and control indivduals were stressed with tunicamycin (TM, 10 μM) for 24 h, fixed, permeabilized, and then the activation of caspase-3 was analyzed by indirect immunofluorescence as described in (a). ***P< 0.001, **P<0.01, *P<0.05
Parkin does not decrease the level of ER stress and functions independently from the proteasome
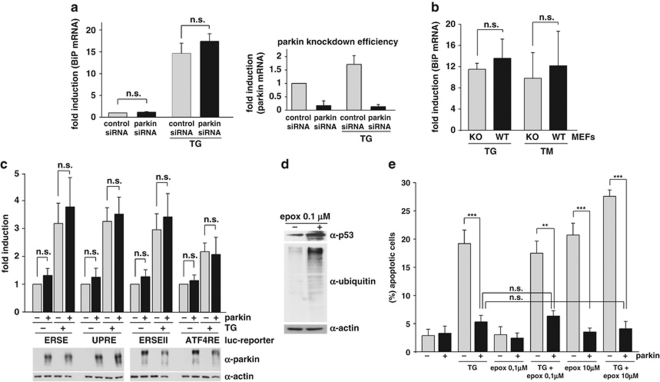
The experiments described above established a protective role of parkin in response to ER stress. To address the question whether parkin may have an effect on the severity of ER stress, we quantified the mRNA levels of the ER chaperone BiP in parkin-knockdown cells in response to ER stress. After ER stress induced by TG, BiP, mRNA was highly upregulated (∼15-fold compared with untreated cells). Interestingly, downregulation of parkin had no significant impact on BiP mRNA levels, both under basal conditions and ER stress (Figure 7a). In addition, parkin-knockout MEFs did not show increased levels of BiP mRNA after TG or TM treatment when compared with control MEFs (Figure 7b). These results indicated that the transient or stable loss of parkin does not cause ER stress. In line with these results, increased expression of parkin did not significantly influence the level of ER stress as monitored by luciferase reporter assays using four different ER stress-responsive elements (ESRE, ERSE-II, UPR and ATF4RE), which cover all branches of the UPR (Figure 7c and Supplementary Figure 5). In a next step, we tested whether the protective activity of parkin under ER stress is dependent on the proteasome. SH-SY5Y cells were exposed to TG in the presence of the proteasomal inhibitor epoxomycin. First, we used a non-toxic concentration of epoxomycin (0.1 μM), which efficiently inhibited the proteasome, as shown by an increase in endogenous p53 levels and an accumulation of ubiquitylated proteins (Figure 7d). Notably, the efficiency of parkin to protect against ER stress-induced cell death was not impaired when the proteasome was inhibited. Furthermore, increasing parkin expression also prevented cell death induced by a toxic concentration of epoxomycin (10 μM), indicating that protein degradation through the proteasome is not required for the acute prosurvival effect of parkin (Figure 7e).
Figure 7.
Parkin has no direct effect on ER stress. (a) Parkin deficiency does not increase the level of ER stress. SH-SY5Y cells were transfected with parkin-specific or control siRNA duplexes. Three days later, the cells were stressed with 1 μM thapsigargin (TG) for 5 h. As an indicator of ER stress BiP mRNA levels were analyzed by quantitative RT-PCR as described in Figure 1a (left panel). To test for the efficiency of parkin knockdown, parkin mRNA levels were quantified in parallel (right panel). (b) The level of ER stress is not increased in mouse embryonic fibroblasts (MEFs) derived from parkin-knockout mice. MEFs from wildtype (wt) or parkin-knockout (ko) mice were stressed with 1 μM TG or 2 μg/ml tunicamycin (TM) for 5 h. The levels of BiP mRNA were analyzed by RT-PCR as described in Figure 1a. (c) Overexpression of parkin has no influence on the ER stress level determined by ER stress reporter constructs. HEK293 T cells were co-transfected with the ER stress reporter plasmids indicated (Supplementary Figure 5) and either parkin or GFP (as a control). Twenty four hours after transfection, the cells were treated with 1 μM TG for 8 h. Shown is the fold induction of luciferase activity in parkin-expressing cells in comparison with GFP-expressing control cells. Quantification is based on triplicates of at least three independent experiments. Expression levels of parkin were analyzed by immunoblotting using the anti-parkin pAb 2132 (lower panel). Loading was controlled by re-probing the blots for β-actin. (d and e) The protective activity of parkin after ER stress is independent of the proteasome. (d) The efficiency of proteasomal inhibition by epoxomycin was demonstrated by an accumulation of endogenous p53 and ubiquitylated proteins. For immunoblotting, an anti-p53 and anti-ubiquitin mAb was used. Loading was controlled by re-probing the blots for β-actin. (e) Proteasomal inhibition does not impair the protective activity of parkin. SH-SY5Y cells were cotransfected with EYFP (as a control) or wild-type parkin. Twenty four hours after transfection, cells were incubated with 10 μM thapsigargin (TG) and/or epoxomycin (epox, 0.1 or 10 μM ) for 8 h, fixed, permeabilized, and then activation of caspase-3 was analyzed as described in Figure 6a. ***P<0.001, **P<0.01, n.s.=not significant
Parkin interferes with ER stress-induced mitochondrial damage
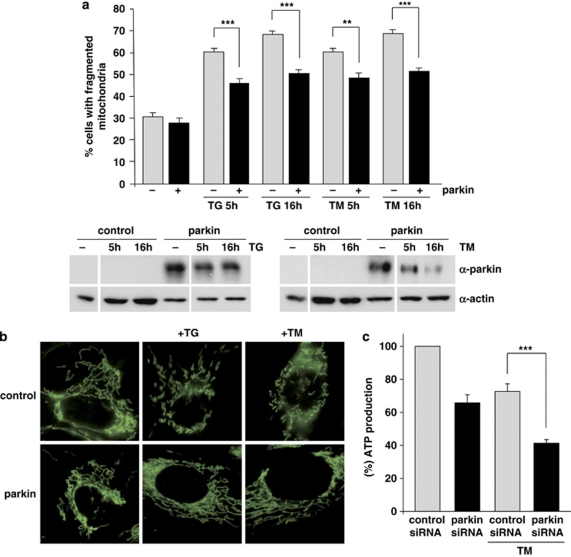
Obviously, parkin does not reduce ER stress per se, but it can protect cells from ER stress-induced cell death. Based on the fact that parkin has an impact on mitochondrial integrity,26 we analyzed the effect of ER stress on mitochondrial morphology and determined whether parkin might have a role in this pathway. SH-SY5Y cells were incubated with the fluorescent dye DiOC6(3) to visualize mitochondria in living cells by fluorescence microscopy. Under normal conditions, when mitochondrial fusion and fission activities are balanced, cells show a tubular mitochondrial network. When fission is increased with relative to fusion, small rod-like or spherical mitochondria can be observed, which are classified as fragmented. Typically, about 70% of SH-SY5Y cells show a tubular mitochondrial network under normal conditions, the remaining 30% are characterized by fragmented mitochondria.26, 27 ER stress, induced by either TM or TG increased the percentage of cells with fragmented mitochondria up to 70% (Figure 8a). Remarkably, enhanced expression of parkin significantly reduced ER stress-induced mitochondrial fragmentation (Figures 8a and b). Moreover, we observed that parkin loss of function increases the vulnerability of cells to ER stress-induced mitochondrial dysfunction, as cellular ATP production in response to ER stress was significantly reduced in parkin-deficient SH-SY5Y cells (Figure 8c). Thus, parkin maintains mitochondrial integrity under ER stress, and prevents alterations in mitochondrial morphology and bioenergetics.
Figure 8.
Parkin prevents mitochondrial damage and dysfunction induced by ER stress. (a and b) Increased parkin expression suppresses ER stress-induced mitochondrial fragmentation. SH-SY5Y cells were transfected with parkin or mCherry (as a control). One day after transfection, the cells were treated with thapsigargin (TG, 1 μM for 5 or 16 h) or tunicamycin (TM, 2 μg/ml for 5 or 16 h). Mitochondria were visualized by life cell microscopy after incubating cells with the fluorescent dye DiOC6(3). (a) The mitochondrial morphology was classified as tubular or fragmented (small rod-like or spherical mitochondria). Shown is the percentage of cells with fragmented mitochondria. Quantifications were based on triplicates of three independent experiments. For each experiment ⩾300 cells per coverslip of triplicate samples were assessed. Expression levels of parkin were analyzed by immunoblotting using the anti-parkin pAb 2132 (lower panel). Loading was controlled by re-probing the blots for β-actin. (b) Examples of mitochondrial morphologies of the experiment described in (a). Treatment of cells with TG or TM cause a disruption of the tubular mitochondrial network, which can be suppressed by increased parkin expression. (c) Parkin deficiency increases ATP depletion in response to ER stress. SH-SY5Y cells were transfected with parkin or control siRNA duplexes. On day 2 after transfection, the cells were shifted to low-glucose medium containing 3 mM glucose instead of 25 mM. On day 3, the cells were treated with 2 μg/ml tunicamycin (TM) for 5 h and the steady-state cellular ATP levels were measured by a bioluminescence assay. Cultured cells derived from tumors derive almost all of their energy from aerobic glycolysis rather than mitochondrial oxidative phosphorylation; in addition, stimulation of glycolysis in the presence of glucose inhibits mitochondrial respiration. Therefore, low glucose concentrations in the medium forces the cells to relay on oxidative phosphorylation to generate sufficient ATP. ***P<0.001, **P<0.01
Discussion
In this study, we show that parkin is transcriptionally upregulated by mitochondrial and ER stress via the UPR to promote viability under cellular stress. We identified the transcription factor ATF4 as a key factor in the stress-induced regulation of parkin expression, which binds to a CREB/ATF site within the parkin promoter. An essential role for ATF4 in the regulation of parkin expression was substantiated by the following observations: First, a dominant-negative ATF4 mutant prevented ER stress-induced upregulation of parkin. Second, in ATF4-knockdown cells and primary neurons from ATF4-knockout mice, parkin upregulation in response to ER stress was significantly impaired. Finally, we could demonstrate by a ChIP assay that ATF4 indeed binds to the parkin promoter. Our analysis also revealed that c-Jun acts as a transcriptional repressor of parkin. Intriguingly, c-Jun can bind to the same site within the parkin promoter as ATF4 to induce downregulation of parkin expression. What might be the physiological relevance of this observation? Conceptually, c-Jun may terminate the ATF4-mediated upregulation of parkin expression by competing with ATF4 on the parkin promoter. In line with this scenario, we observed that c-Jun has a dominant effect on ATF4; it can suppress ATF4-mediated upregulation of parkin expression. It is also conceivable that severe or prolonged ER stress leads to a preferential binding of c-Jun to the parkin promoter to suppress cytoprotective pathways and to favor the elimination of irreversibly damaged cells by proapoptotic pathways. In support of this view, CHOP, a proapoptotic transcription factor activated upon severe or prolonged ER stress, suppressed parkin expression (Supplementary Figure 6). Parkin transcription was also repressed by JNK3, a kinase upstream of c-Jun, which has been implicated in neuronal cell death pathways in dopaminergic neurons.28, 29 Indeed, our data show that the use of a JNK inhibitor is able to increase parkin expression, particularly under stress conditions. Interestingly, parkin has been reported to suppress JNK activity in cellular models and Drosophila melanogaster.30, 31 In line with these studies, we observed that phospho-JNK and phospho-c-Jun levels are increased in parkin-deficient patient fibroblasts under ER stress (Supplementary Figure 4). Our finding that the JNK pathway can negatively regulate parkin gene expression suggests a reciprocal interaction between parkin and JNK3, which might be instrumental in the dichotomy of cell survival and death. Depending on the cellular context and the severity of stress conditions, parkin can shift the balance towards cell survival by attenuating JNK3 signaling, whereas JNK3 gaining the upper hand, suppresses prosurvival pathways, either directly or indirectly, for example by inhibiting the expression of parkin.
The increased expression of parkin in response to ER stress clearly serves as a cytoprotective function. Cells overexpressing wt but not mutant parkin are protected against ER stress-induced cell death, whereas parkin-deficient cells show an increased vulnerability to ER stress. It will now be interesting to determine which cell types are affected in their response to stress by the loss of parkin function. On the basis of the emerging concept of non-cell autonomous cell death in neurodegeneration, it is conceivable that an impaired stress response in glial cells in the neighborhood of dopaminergic cells can contribute to nigrostriatal degeneration.
Neither an acute nor a permanent loss of parkin function per se causes ER stress. Furthermore, increased parkin expression does not decrease the severity of ER stress, arguing against a direct role of parkin in the ERAD pathway. In support of this notion, the antiapoptotic activity of parkin is independent of the proteasome. We observed that proteasomal inhibition does not impair the ability of parkin to prevent ER stress-induced cell death. Which activity of parkin might then be responsible for its protective effect under ER stress? In this context, it is important to note that organellar stress within cells cannot be regarded in an isolated manner. In fact, our study adds evidence for an interaction between the ER and mitochondria in response to stress.32 ER stress can induce mitochondrial stress, resulting in a bioenergetic deficit and mitochondrial fragmentation. Our working model would suggest that, at this interplay, parkin seems to enter the stage, preventing the pathophysiological consequences of ER stress on mitochondrial integrity (Figure 9). The next important step will be to figure out what the precise role of parkin might be in the communication between the ER and mitochondria.
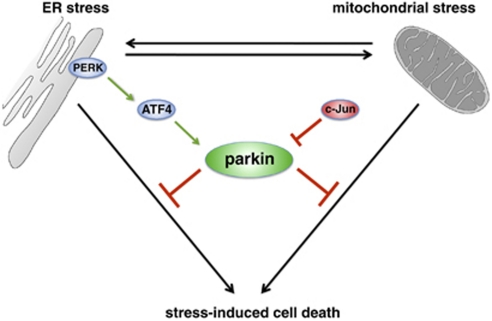
Figure 9.
Interplay between ER stress, mitochondrial stress and parkin. ER stress can induce mitochondrial damage, such as alterations in mitochondrial morphology and bioenergetics. Conversely, mitochondrial stress can induce ER stress, reflected by the induction of the unfolded protein response (UPR). Parkin is transcriptionally upregulated in response to both mitochondrial and ER stress by ATF4, a transcription factor of the UPR. The stress-induced transcriptional upregulation of parkin is antagonized by c-Jun, which is activated by the JNK pathway. Increased expression of parkin under stress conditions protects cells from stress-induced cell death, explaining the high vulnerability of parkin-deficient cells to cellular stress
Materials and Methods
DNA constructs
The following constructs were described previously: wt human parkin, G430D, ΔUBL mutant parkin,18 ATF4,33 ATF4ΔN,22 PERK,34 c-Jun,35 JNK3,36 Bcl-2-FLAG37 and mCherry.26 To generate siRNA-resistant parkin, four silent mutations were introduced into human wt parkin, to prevent binding of parkin-specific siRNA1 (Invitrogen, Karlsruhe, Germany). The following mutations were introduced by standard PCR cloning techniques: C 1038 to T, G 1044 to A, C 1053 to A and A 1059 to G. The luciferase reporter constructs were cloned by subcloning the UPR element (UPRE), the ER stress response element (ERSE), ERSE-II, the ATF4 binding site of the IGFBP-1 promoter (ATF4RE) or the ATF4 binding element of the parkin promoter (PARK) in triplicate flanked by NheI and BglII restriction sites into the pGL3 promoter vector (Promega, Mannheim, Germany). To generate the renilla luciferase construct, the SV40 promoter from the pGL3 vector was subcloned into the phRL-TK vector (Promega), thereby replacing the HSV-TK promoter. The plasmids encoding enhanced yellow fluorescent protein (EYFP) and enhanced green fluorescent protein (EGFP) were purchased from Clontech (Mountain View, CA, USA).
Cell culture and transfections
SH-SY5Y (DSMZ number ACC 209) and HEK293T (ATCCC number CRL-1573) cells were transfected with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. For RNA interference, SH-SY5Y or HEK293T cells were reversely transfected with Stealth siRNA (Invitrogen) using Lipofectamine RNAiMAX (Invitrogen) for SH-SY5Y cells and Lipofectamine 2000 (Invitrogen) for the HEK293T cells.
Antibodies and reagents
The following antibodies were used: anti-parkin mouse monoclonal antibody (mAb) PRK8 (Millipore, Schwalbach, Germany); anti-parkin pAb 2132, anti-ubiquitin P4D1 pAb (Cell Signaling Technology, Danvers, MA, USA); anti-SAPK/JNK pAb, anti-phospho-SAPK/JNK (Thr183/Tyr185) pAb and anti-phospho-c-Jun (Ser63) II rabbit pAb (Cell Signaling Technology); anti-CHOP mAb (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-β-actin mAb, anti-Flag M2 mAb and anti-myc 9E10 mAb (Sigma, Taufkirchen, Germany); anti-c-Jun (N) sc45 rabbit pAb, anti-c-Jun (N) sc45X rabbit pAb, anti-CREB-2 C-20 rabbit pAb and anti-TRAF6 (H-274) rabbit pAb (Santa Cruz Biotechnology); anti-ATF4 pAB (Cocalico Biologicals, Reamstown, PA, USA); anti-p53 mAb (Calbiochem/Merck, Darmstadt, Germany); anti-active caspase-3 rabbit pAb (Promega); Alexa 555-conjugated goat anti-rabbit pAb (Sigma); horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit IgG antibody (Promega). Thapsigargin was purchased from Sigma, TM from Fluka/Sigma (Taufkirchen, Germany), epoxomycin from Calbiochem, -histidinol from Sigma, the JNK inhibitor SP600125 from Enzo Life Sciences (Loerrach, Germany), and dialysed FCS and 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3)) from Invitrogen. The mounting medium Mowiol (Calbiochem) was supplemented with 4′ 6-diamidino-2-phenylindole (Sigma).
Western blot analysis
SDS-PAGE and western blotting was described earlier.18 Antigens were detected with the enhanced chemiluminescence detection system (Amersham Biosciences, Freiburg, Germany) or the Immobilon Western chemoluminiscent HRP substrate (Millipore).
Mouse embryonic fibroblast cultures
Primary mouse fibroblasts were isolated from parkin-knockout and wt mice with the same genetic background.25 E12.5 mouse embryos were extracted, and the head and inner organs were removed. The remaining tissue was dissociated by trypsination and trituration.
Primary cortical neuronal culture
Cortical neurons were cultured from wt or ATF4 transgenic mouse embryos at E14.5–E15.5 days of gestation and individually dissected. Briefly, neurons from each embryo were plated individually, into six-well dishes (∼3 million cells/well) coated with poly--lysine (100 μg/ml) in serum-free medium (MEM/F12 (1:1) supplemented with 6 mg/ml -glucose, 100 μg/ml transferrin, 25 μg/ml insulin, 20 nM progesterone, 60 μM putrescine and 30 nM selenium) as described previously.38 At 2 days in vitro, cortical neurons were treated with TM (3 μg/ml) for 8 h. Stock solutions of TM were prepared in DMSO (Sigma Aldrich, Oakville, ON, Canada) and diluted in culture media immediately before addition. Total RNA was isolated from cells at indicated time points using TRIzol reagent as per manufacturer's instructions (Invitrogen). Concentrations of RNA were measured on a spectrophotometer at λ260 nm.
Human primary fibroblast cultures
Punch-skin biopsy samples were taken from PD patients with compound heterozygous mutations (c101delAG, het. del ex3/4; patients 1 and 2) or a heterozygous mutation (het. dupl. ex7; patient 3) in the parkin gene. Genotyping was performed using direct DNA sequencing and the Multiplex ligation-dependent probe amplification parkin gene dosage kits (P051; MRC-Holland, Amsterdam, Holland), covering all exons of the parkin gene, as well as other known Mendelian PD genes. Sex and age-matched control fibroblasts from healthy individuals were provided by the Department of Orthopedics (Universitätsklinik für Orthopädie, Eberhard Karls University, Tübingen, Germany). Chopped tissue pieces were placed into a tissue culture flask and carefully covered with maintenance medium (RPMI medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, 1 mM sodium pyruvate). Cultures were kept at 37 °C in a humidified atmosphere containing 5% CO2. Collected fibroblasts were aliquoted and frozen for storage between passages 2 and 10.
Real-time RT-PCR
Real-time RT-PCR was performed, as described before.18 Briefly, SH-SY5Y cells were incubated with 1 μM TG, 2 μg/ml TM, 10 μM CCCP or 10 μM SP600125 for the indicated time. Total cellular RNA was isolated according to manufacturer's instructions (RNaesy mini kit, QIAGEN, Hilden, Germany). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad, München, Germany). For the quantification of human parkin mRNA, the TaqMan Gene Expression Assay (parkin: Hs01038827-m1; beta actin: P/N 4326315E; 18sRNA: P/N 4319413E) (Applied Biosystems, Foster City, CA, USA) was used. For all other mRNA quantifications, PCR reactions were performed with 2 × Power SYBR Green PCR Master Mix (Applied Biosystems) and 1 μM of each primer pair; mouse (m) parkin forward primer (F): 5′-AAACCGGATGAGTGGTGAGT-3′ m-parkin reverse primer (R): 5′-AGCTACCGACGTGTCCTTGT-3′ m-actin-β F: 5′-AGCCTTCCTTCTTGGGTATG-3′ m-actin-β R: 5′-GGTCTTTACGGATGTCAACG-3′ m-BiP F: 5′-GCCTCATCGGACGCACTT-3′ m-BiP R: 5′-GGGGCAAATGTCTTGGTT-3′ human (h) ATF4: F: 5′-CCCTTCACCTTCTTACAACCTC-3′ h-ATF4 R: 5′-GTCTGGCTTCCTATCTCCTTCA-3′ h-c-Jun F 5′-CGCCTGATAATCCAGTCCA-3′ h-c-Jun R: 5′-CCTGCTCATCTGTCACGTTC-3′ ; h-BIP F: 5′-GCTCGACTCGAATTCCAAAG-3′ h-BIP R: 5′-GATCACCAGAGAGCACACCA-3′ h-actin-β F: 5′-TGGACTTCGAGCAAGAGATG-3′ h-actin-β R: 5′-AGGAAGGAAGGCTGGAAGAG-3′. Quantification was performed with 7500 Fast Real Time System (Applied Biosystems) based on triplicates per primer set. Gene expression was normalized with respect to endogenous housekeeping control genes, β-actin and 18sRNA, which were not influenced by ER stress. Relative expression was calculated for each gene using the ΔΔCT method. Statistical analysis of RT-PCR data is based on at least three independent experiments with tripicates samples.
Luciferase assays
HEK293T or SH-SY5Y cells transiently expressing renilla luciferase and firefly luciferase reporter plasmids were subjected to the stress treatment indicated. Luciferase activity of cell lysates was determined luminometrically using an LB96V luminometer (Berthold Technologies, Bad Wildbad, Germany) by the dual luciferase assay system (Promega) as specified by the manufacturer. The measured values were analyzed with WinGlow Software (Berthold Technologies). Quantification was based on at least three independent experiments. For each experiment, each transfection was performed at least in triplicate.
Apoptosis and cell viability assays
Activation of caspase-3 was determined as described previously.37 Briefly, SH-SY5Y cells or skin fibroblasts were grown on glass coverslips. Twenty four hours after transfection (for parkin knockdown 3 days later), cells were incubated with TG, TM and/or epoxomycin as indicated. The cells were then fixed, and activated caspase-3 detected by indirect immunofluorescence using an anti-active caspase-3 antibody. To detect cells undergoing apoptosis, the number of activated caspase-3-positive cells out of at least 300 transfected cells was determined using a Zeiss Axioscope 2 plus microscope (Carl Zeiss, Göttingen, Germany). Quantifications were based on triplicates of at least three (SH-SY5Y cells) or two (human skin fibroblasts) independent experiments. For each experiment ⩾300 cells per coverslip of triplicate samples were assessed. For the cell viability assays, SH-SY5Y cells, HEK293T cells or MEFs were plated into 12-well plates. SH-SY5Y cells and HEK293T cells were reversely transfected with parkin or control siRNA. Two days later, the cells were stressed with TM or TG as indicated and the Vybrant MTT Cell Proliferation Assay was performed according to manufacturer's instructions (Invitrogen).
Electrophoretic mobility shift assay
Nuclear extracts were prepared as described earlier.18 For the binding reaction, 10 μg of extracts were incubated with 10 mM HEPES (pH 7.9), 50 mM NaCl, 5 mM MgCl2, 2 mM DTT, 0.1 mM EDTA, 5% glycerol, 10 μg BSA, 2 μg poly(dI-dC) and 0.2 ng (20 000 cpm) of 32P-labeled, double-stranded park oligonucleotide (5′-CCCCGGTGACGTAAGATTGC-3′) in a final volume of 20 μl. For supershift assays, 0.2 μg of the ATF4 antibody or 2 μg of the c-Jun (N) sc45X antibody, for competition experiments 50 ng (100 × ) of cold-park oligonucleotide was added to the binding reaction. After binding on ice for 30 min, mixtures were loaded onto non-denaturating 4% polyacrylamide gels in 0.5 × TBE (45 mM Tris borate and 1 mM EDTA). Gels were electrophoresed at 4°C for 4 h at 160 V, dried, and exposed for autoradiography at −80°C.
Chromatin immunoprecipitation
HEK293T or SH-SY5Y cells were replenished with fresh medium 12 h before initiating all treatments to ensure that the cells were in the basal state. To trigger the UPR, fresh medium containing 300 nM TG was added. To monitor ATF4 binding to the parkin gene, a ChIP assay was performed as previously described.39 The ATF4 antibody was a rabbit polycolonal antibody. Enrichment of DNA at the parkin promoter region that contains the potential ATF4 binding site was analyzed with quantitative real-time PCR. A 5 μl aliquot of DNA was mixed with 62.5 pmol of each PCR primer and 12.5 μl of SYBR Green PCR master mix (Applied Biosystems) in a 25 μl total volume. The real-time PCR was performed with a DNA Engine Opticon 3 system (Bio-Rad). The reaction mixtures were incubated at 95°C for 15 min, followed by amplification at 95°C for 15 s and 60°C for 60 s for 35 cycles. All experiments were performed in triplicate and each sample was subjected to PCR in duplicate. The primers used were: forward: 5′-GTTGCTAAGCGACTGGTCAA-3′ and reverse: 5′-CAGCCCCCCACCGCCGCC-3′.
Fluorescent staining of mitochondria
SH-SY5Y cells were grown on 15 mm glass coverslips, and were fluorescently labeled with 0.1 μM DiOC6(3) in cell culture medium for 15 min. After washing the coverslips with medium, living cells were analyzed for mitochondrial morphology by fluorescence microscopy as described previously26 using a Leica DMRB microscope (Leica, Wetzlar, Germany). Cells were categorized in two classes according to their mitochondrial morphology: tubular or fragmented. Quantifications were based on three independent experiments. For each experiment, the mitochondrial morphology of ⩾300 transfected cells per coverslip of triplicate samples was assessed.
Measurement of cellular ATP levels
A quantitative determination of ATP in SH-SY5Y cells was performed using the ATP Bioluminescence Assay Kit HS II (Roche, Mannheim, Germany) according to the manufacturer's instructions. Briefly, SH-SY5Y cells were reversely transfected with the indicated siRNA duplexes. Twenty four hours before collecting, the culture medium was replaced by low-glucose medium containing 3 mM instead of 25 mM glucose. Five hours, before the measurement, cells were treated with 2 μg/ml TM. Cells were washed twice with PBS, scraped off the plate and then lysed according to the provided protocol. The ATP content of the samples was determined using an LB96 V luminometer (Berthold Technologies), analyzed with WinGlow Software (Berthold Technologies) and normalized to total protein levels. Quantification was based on at least three independent experiments. For each experiment, each transfection was performed at least in triplicate.
Bioinformatics
Transcription factor (TF) BSs were identified by the TFSEARCH20 and the MatInspector40 program. All sequences were derived from the promoter sequence retrieval database ElDorado 02-2010 (Genomatix, Munich, Germany). Promoter sequences of parkin from four different mammalian species were aligned with the DiAlign TF program40 in the Genomatix software suite GEMS Launcher to evaluate the overall promoter similarity and to identify conserved CREB/ATF BSs. The promoter sequences were defined as in ElDorado and elongated at the 3′ end of the promoter (downstream) by 150 bp. Position weight and matrices were used according to Matrix Family Library Version 8.2 (Genomatix) (January 2010) for promoter analyses. The BSs were considered as ‘conserved BSs' if the promoter sequences of human and the orthologs can be aligned in the region of CREB/ATF BSs with the help of the DiAlign TF program (using default settings).
Statistical analysis
Data were expressed as means±S.E. Statistical analysis was carried out using ANOVA. *P<0.05, **P<0.01, ***P<0.001.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 596 ‘Molecular Mechanisms of Neurodegeneration' to KFW, JT and WW), the German Ministry for Education and Research (NGFN plus ‘Functional Genomics of Parkinson's Disease' to KFW, TG and WW), the Helmholtz Alliance ‘Mental Health in an Ageing Society' (to KFW, TG and WW), the Hans and Ilse Breuer Foundation (to LB), the Heart and Stroke Foundation of Canada, Heart and Stroke Foundation of Ontario and the Canadian Institutes of Health Research (to DSP). Funding for MSK was from the National Institutes of Health (DK-52064). II was supported by a postdoctoral fellowship from Canadian Institutes of Health Research (CIHR). We are grateful to Christian Haass for continuous support and stimulating discussions. We thank Dr Tim Townes for providing the ATF4-knockout mice, Alexis Brice and Olga Corti for the parkin-knockout mice, Torsten Kluba for human skin fibroblasts from control individuals, Carsten Culmsee for primary mouse cortical neurons, Daniel Krappmann for the c-Jun plasmid, Kimitoshi Kohno and Hiroto Izumi for the ATF4 plasmid, Gerald Thiel for the ATF4ΔN and CHOP plasmids, David Ron for the PERK plasmid and Vicky Wätzig and Thomas Herdegen for the JNK3 plasmid. We thank Kerstin Lämmermann and Christian Naumann for technical assistance and Jonathan Lin for advice on ATF4 western blotting.
Glossary
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assays
- ERAD
ER-associated degradation
- IGFBP1
insulin growth factor binding protein 1
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MEFs
mouse embryonic fibroblasts
- RNAi
RNA interference
- TG
thapsigargin
- TM
tunicamycin
- TTS
transcriptional start site
- UPR
unfolded protein response
- wt
wildtype
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Piacentini
Supplementary Material
References
- Winklhofer KF, Haass C. Mitochondrial dysfunction in Parkinson′s disease. Biochimica Et Biophysica Acta. 2010;1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- Wang HQ, Takahashi R. Expanding insights on the involvement of endoplasmic reticulum stress in Parkinson's disease. Antioxid Redox Signal. 2007;9:553–561. doi: 10.1089/ars.2006.1524. [DOI] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Ghribi O, Herman MM, Pramoonjago P, Savory J. MPP+ induces the endoplasmic reticulum stress response in rabbit brain involving activation of the ATF-6 and NF-kappaB signaling pathways. J Neuropathol Exp Neurol. 2003;62:1144–1153. doi: 10.1093/jnen/62.11.1144. [DOI] [PubMed] [Google Scholar]
- Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D, Greene LA. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson's disease. J Neurosci. 2002;22:10690–10698. doi: 10.1523/JNEUROSCI.22-24-10690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz WA, O'Malley KL. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem. 2003;278:19367–19377. doi: 10.1074/jbc.M211821200. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, et al. Alpha-synuclein blocks ER-golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Jiang H, Pei Z, Tanaka Y, Morita H, Sawa A, et al. Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Hum Mol Genet. 2005;14:3801–3811. doi: 10.1093/hmg/ddi396. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM, Scheper W. Activation of the unfolded protein response in Parkinson's disease. Biochem Biophys Res Commun. 2007;354:707–711. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Moran LB, Croisier E, Duke DC, Kalaitzakis ME, Roncaroli F, Deprez M, et al. Analysis of alpha-synuclein, dopamine and parkin pathways in neuropathologically confirmed parkinsonian nigra. Acta Neuropathol (Berl) 2007;113:253–263. doi: 10.1007/s00401-006-0181-6. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- Pizzo P, Pozzan T. Mitochondria-endoplasmic reticulum choreography: structure and signaling dynamics. Trends Cell Biol. 2007;17:511–517. doi: 10.1016/j.tcb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, et al. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand A, Tomkiewicz C, Magne L, Barouki R, Garlatti M. Endoplasmic reticulum stress induction of insulin-like growth factor-binding protein-1 involves ATF4. J Biol Chem. 2006;281:19124–19133. doi: 10.1074/jbc.M602157200. [DOI] [PubMed] [Google Scholar]
- Steinmuller L, Thiel G. Regulation of gene transcription by a constitutively active mutant of activating transcription factor 2 (ATF2) Biol Chem. 2003;384:667–672. doi: 10.1515/BC.2003.074. [DOI] [PubMed] [Google Scholar]
- Masuoka HC, Townes TM. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood. 2002;99:736–745. doi: 10.1182/blood.v99.3.736. [DOI] [PubMed] [Google Scholar]
- Waetzig V, Herdegen T. Context-specific inhibition of JNKs: overcoming the dilemma of protection and damage. Trends Pharmacol Sci. 2005;26:455–461. doi: 10.1016/j.tips.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lammermann K, et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandebring A, Thomas KJ, Beilina A, van der Brug M, Cleland MM, Ahmad R, et al. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS one. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunot S, Vila M, Teismann P, Davis RJ, Hirsch EC, Przedborski S, et al. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson′s disease. Proc Natl Acad Sci USA. 2004;101:665–670. doi: 10.1073/pnas.0307453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht S, Kirchhof R, Chromik A, Willesen M, Nicolaus T, Raivich G, et al. Specific pathophysiological functions of JNK isoforms in the brain. Eur J Neurosci. 2005;21:363–377. doi: 10.1111/j.1460-9568.2005.03857.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Treis A, Patenge N, Fiesel FC, Springer W, Kahle PJ. Parkin protects against tyrosinase-mediated dopamine neurotoxicity by suppressing stress-activated protein kinase pathways. J Neurochem. 2008;105:1700–1715. doi: 10.1111/j.1471-4159.2008.05277.x. [DOI] [PubMed] [Google Scholar]
- Cha GH, Kim S, Park J, Lee E, Kim M, Lee SB, et al. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc Natl Acad Sci USA. 2005;102:10345–10350. doi: 10.1073/pnas.0500346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom JR, Gewandter JS, Michael L, Sheu SS, Yoon Y. Thapsigargin induces biphasic fragmentation of mitochondria through calcium-mediated mitochondrial fission and apoptosis. J Cell Physiol. 2007;212:498–508. doi: 10.1002/jcp.21051. [DOI] [PubMed] [Google Scholar]
- Tanabe M, Izumi H, Ise T, Higuchi S, Yamori T, Yasumoto K, et al. Activating transcription factor 4 increases the cisplatin resistance of human cancer cell lines. Cancer Res. 2003;63:8592–8595. [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Krappmann D, Wulczyn FG, Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- Waetzig V, Herdegen T. A single c-Jun N-terminal kinase isoform (JNK3-p54) is an effector in both neuronal differentiation and cell death. J Biol Chem. 2003;278:567–572. doi: 10.1074/jbc.M207391200. [DOI] [PubMed] [Google Scholar]
- Rambold AS, Miesbauer M, Rapaport D, Bartke T, Baier M, Winklhofer KF, et al. Association of Bcl-2 with misfolded prion protein is linked to the toxic potential of cytosolic PrP. Mol Biol Cell. 2006;17:3356–3368. doi: 10.1091/mbc.E06-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleyasin H, Cregan SP, Iyirhiaro G, O'Hare MJ, Callaghan SM, Slack RS, et al. Nuclear factor-(kappa)B modulates the p53 response in neurons exposed to DNA damage. J Neurosci. 2004;24:2963–2973. doi: 10.1523/JNEUROSCI.0155-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J Biol Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.