Abstract
Ultraviolet (UV) radiation-induced DNA damage evokes a complex network of molecular responses, which culminate in DNA repair, cell cycle arrest and apoptosis. Here, we provide an in-depth characterization of the molecular pathway that mediates UV-C-induced apoptosis of meiotic germ cells in the nematode Caenorhabditis elegans. We show that UV-C-induced DNA lesions are not directly pro-apoptotic. Rather, they must first be recognized and processed by the nucleotide excision repair (NER) pathway. Our data suggest that NER pathway activity transforms some of these lesions into other types of DNA damage, which in turn are recognized and acted upon by the homologous recombination (HR) pathway. HR pathway activity is in turn required for the recruitment of the C. elegans homolog of the yeast Rad9-Hus1-Rad1 (9-1-1) complex and activation of downstream checkpoint kinases. Blocking either the NER or HR pathway abrogates checkpoint pathway activation and UV-C-induced apoptosis. Our results show that, following UV-C, multiple DNA repair pathways can cooperate to signal to the apoptotic machinery to eliminate potentially hazardous cells.
Keywords: apoptosis, C. elegans, HR, NER, UV-C
Eukaryotic cells possess several surveillance mechanisms that, upon sensing DNA damage, initiate signaling cascades that lead to response programs such as cell cycle arrest, DNA repair and apoptosis, to protect the organism against the introduction of new mutations. Disruption of such pathways results in increased genomic instability, a hallmark of most types of cancers.1, 2
Genetic and biochemical studies have provided a thorough mechanistic understanding of the various repair processes initiated upon recognition of specific types of lesions. For example, the nucleotide excision repair (NER) pathway removes cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts generated upon exposure to ultraviolet (UV-C) light, whereas the homologous recombination (HR) machinery repairs double-strand DNA breaks (DSBs) induced by treatments such as ionizing radiation (IR).3
Mutations in NER components underlie the syndromes xeroderma pigmentosum (XP), trichothiodystrophy or Cockayne syndrome. Patients are hypersensitive to sunlight and exhibit a variety of clinical features, including developmental defects, predisposition to skin cancer or internal tumors, neurological disorders and highly accelerated aging.4 Studies in cell culture and mouse models,5, 6 as well as in yeast have led to the detailed molecular characterization of the NER factors.3 Besides repair, many of these factors participate in the signaling network that ultimately balances cellular DNA damage responses between genome maintenance, senescence and death. Accordingly, loss of their function has consequences for DNA repair, cellular proliferation and survival.6, 7, 8
Simple model organisms are very useful to decipher complex DNA damage responses. In Caenorhabditis elegans, the effects of IR have been studied extensively.9, 10 We previously reported a genetic pathway that induces both apoptotic cell death of meiotic cells and cell cycle arrest of proliferating mitotic cells following UV-C treatment.11 We identified several new genes required for these responses and genetically ordered them into a signaling pathway that overlaps with, but is distinct from the pathway(s) activated upon IR.
In this study, we investigate the molecular mechanism by which UV-C triggers apoptosis in the C. elegans adult hermaphrodite germ line. We show that lesions caused by UV-C are not pro-apoptotic per se; rather, they first require processing by the NER machinery before they can activate apoptosis. A fraction of UV-C lesions is likely transformed by NER into DNA intermediates that are substrates for the HR machinery. Activation of the latter, in turn, leads to recruitment of the C. elegans homolog of the yeast Rad9-Hus1-Rad1 (9-1-1) complex, activation of downstream checkpoint kinases and initiation of p53-dependent apoptosis. Our results indicate that UV-C-induced apoptosis in C. elegans requires the sequential activation of at least two distinct DNA damage response pathways.
Results
Components of the HR pathway are required for UV-C-induced apoptosis
We previously showed that in C. elegans UV-C radiation induces cell cycle arrest in mitotic germ cells and apoptosis of meiotic cells at the pachytene stage.11 We also reported that XPA-1 and XPC-1, two components of the NER pathway, are required for the activation of these responses, with XPA-1 acting upstream of the 9-1-1 complex protein HUS-1 and the p53 homolog CEP-1 in UV-induced apoptosis.11
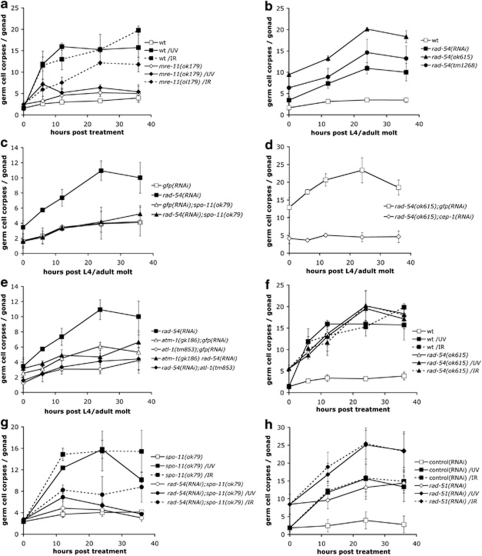
How does XPA-1 function promote p53 activation following UV-C exposure? One possibility is that NER processes a fraction of UV-C-induced damage into other types of DNA structures, which might in turn activate apoptosis. Given our previous observation that many genes involved in response to DSBs are also activated following UV-C, we focused our initial attention on the HR pathway. mre-11(ok179) mutants, which lack the exonuclease responsible for the initial processing of DSBs during meiosis, showed a compromised apoptotic response to UV-C, suggesting a requirement for MRE-11 in the initiation of UV-C-induced apoptosis (Figure 1a). By contrast, IR-induced apoptosis was only slightly reduced (Figure 1a), in agreement with previous reports,12 possibly owing to alternative processing pathways.
Figure 1.
Several components of the HR repair pathway are required for UV-C-induced germ cell apoptosis. (a) mre-11 mutants fail to induce apoptosis in response to UV-C. Staged young adult mre-11(ok179) animals were treated with either 100 J/m2 UV-C or 120 Gy X-rays and germ cell corpses were scored at the indicated time points. (b–e) Genetic characterization of rad-54 mutants. (b) rad-54(lf) results in increased levels of germ cell apoptosis. Germ cell corpses were scored every 12 h until 36 post the L4/adult molt in staged rad-54(ok615) or rad-54(tm1268) mutants, or rad-54(RNAi)-treated animals. (c–e) rad-54(lf)-induced germ cell death depends on spo-11 (c), cep-1 (d) and ATM/ATR function (e). (f) rad-54 mutants fail to induce apoptosis in response to UV-C and IR. Staged young adult rad-54(ok615) animals were treated with either 100 J/m2 UV-C or 120 Gy X-rays and germ cell corpses were scored at the indicated time points. (g) Loss of spo-11 function does not restore UV-C- and IR-induced apoptosis in rad-54(RNAi) animals. Staged spo-11(ok79) L1 larvae were raised on bacteria expressing rad-54 or gfp dsRNA, and were treated with either 100 J/m2 UV-C or 120 Gy X-rays as young adults. Germ cell corpses were scored at the indicated time points. (h) rad-51(lf) animals normally induce apoptosis in response to UV-C. Staged wild-type L1 larvae were raised on bacteria expressing rad-51 or control dsRNA, and were treated with either 100 J/m2 UV-C or 120 Gy X-rays as young adults. Germ cell corpses were scored at the indicated time points. Data shown in all cases represent the average±S.D. of two or three independent experiments (n>20 animals for each experiment)
The homologs of RAD-54 and RAD-51 functionally interact during recombinational repair in yeasts and mammals.13, 14 We analyzed two previously uncharacterized, likely null rad-54 mutants, tm1268 and ok615 (Supplementary Figure 1a). As homozygous rad-54 progeny from heterozygous mothers show fully penetrant maternal-effect lethality, we analyzed first-generation homozygotes for germline apoptosis. Under normal growth conditions, both rad-54(ok615) and rad-54(tm1268) mutants exhibit a strong increase in germline apoptosis, an effect that could be phenocopied by RNAi (Figure 1b). This increase was dependent on the endonuclease SPO-11, which specifically generates the DSBs that initiate meiotic recombination (Figure 1c),12 suggesting that the increased germ cell death is due to the accumulation of unresolved recombination intermediates. rad-54(rf)-induced apoptosis was abrogated in atl-1(tm853) or cep-1(RNAi) animals, and reduced in atm-1(gk186) mutants (Figures 1d and e). These results suggest that rad-54(lf) mutants accumulate DSBs, which activate apoptosis in an ATL-1/ATM-1- and CEP-1-dependent manner.
Interestingly, exogenous damage by X-rays or UV failed to further increase the levels of germ cell apoptosis in rad-54(ok615) mutants (Figure 1f), suggesting either that the DNA damage response pathway (or the apoptotic machinery to which it signals) is already ‘saturated' by the strong signal generated by the unresolved meiotic intermediates in rad-54 mutants or that the HR pathway is required for IR- and UV-induced apoptosis. To address this issue, we blocked SPO-11 endonuclease function. We found that rad-54(RNAi);spo-11(ok79) animals showed a greatly reduced response to IR and, surprisingly, also to UV (Figure 1g). Importantly, spo-11(ok79) mutants respond normally to both IR and UV, showing that the DNA damage response pathways are functional (Figure 1g). The HR pathway therefore plays an important role in promoting apoptosis in response to UV-C.
Finally, we tested animals lacking the recombinase RAD-51. In the human and yeast homologs, RAD-51 acts downstream of MRE-11 and RAD-54 to promote strand exchange during HR. rad-51(RNAi) animals also fail to complete meiotic recombination, resulting in increased basal levels of apoptosis (Figure 1h). However, unlike in rad-54 mutants, this increase can be suppressed only by atl-1(tm853), but not atm-1(gk186) (Supplementary Figure 2). In contrast to mre-11 and rad-54 mutants, rad-51(lf) animals did not show any defect in either UV- or IR-induced apoptosis (Figure 1h).15 Moreover, UV-C treatment led to RAD-51 foci formation (Supplementary Figure 3a), independent of both xpa-1 and rad-54 (Supplementary Figures 3b and c). These observations suggest that UV-C-induced RAD-51 foci formation and apoptosis are independent events, and that RAD-51 is not essential for UV-C-induced apoptosis.
Taken together, our results support the idea that the proteins involved in early stages of HR also act to promote apoptosis in response to UV-C damage.
The HR pathway acts downstream of the NER pathway to promote UV-C-induced apoptosis
The data above suggest that both NER and HR pathways are required for UV-C-induced apoptosis. These two pathways might act in parallel and be simultaneously required for the induction of apoptosis. Alternatively, they might act in succession, with NER recognizing and processing the initial UV-C lesions and HR relaying the signal to the apoptotic machinery. To distinguish between these two possibilities, we determined the effect of mutations in the NER or HR pathways on the subcellular localization of selected DNA damage response proteins.
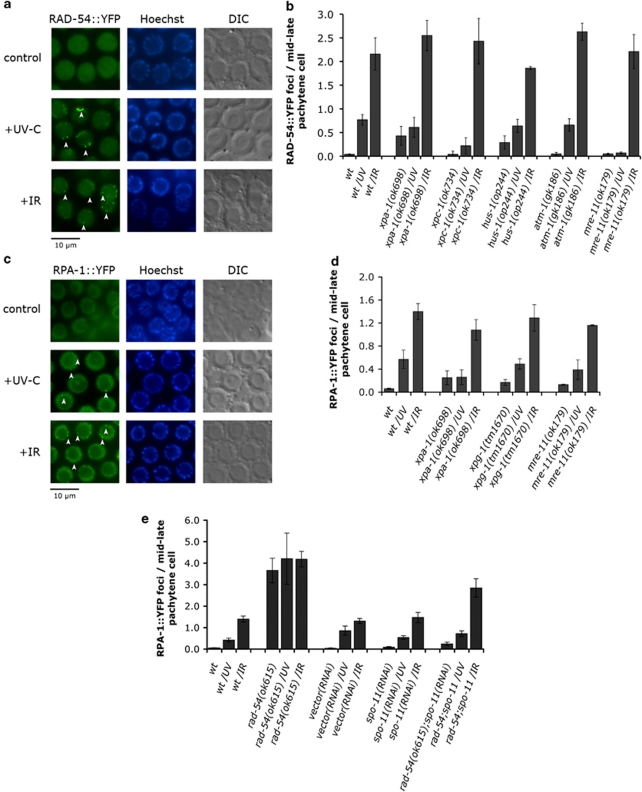
We started by examining the distribution of a functional RAD-54::YFP fusion protein (Supplementary Figure 1b). RAD-54::YFP formed foci following exposure to both IR or UV-C, consistent with our observation that the HR pathway participates in UV-C-induced responses (Figure 2a). In xpa-1 and xpc-1 mutants, foci accumulation following UV treatment, but not IR, was dramatically reduced (Figure 2b), suggesting that the NER component XPA-1 acts upstream of RAD-54 recruitment in the UV-C response pathway.
Figure 2.
The HR pathway acts downstream of the NER pathway to induce UV-C-induced germ cell apoptosis. (a and c) Fluorescent microscopy of mid–late pachytene germ cells expressing RAD-54::YFP (a) or RPA-1::YFP (c). Germ cell nuclei from staged young adult wild-type animals were scored for the presence of YFP, 3 h after exposure to 100 J/m2 of UV-C or 120 Gy of X-rays. RAD-54 or RPA-1 show a diffuse nuclear staining in control animals, but relocalize into distinct foci following treatment (arrowheads). (b) XPA-1, XPC-1 and MRE-11 are required for RAD-54 foci formation in response to UV-C, but not IR. (d and e) UV-C-induced RPA-1 foci require XPA-1 (d), but not MRE-11 (d) or RAD-54 (e). RAD-54::YFP and RPA-1::YFP foci were quantified as described in Materials and Methods. Data shown represent the average±S.D. of at least two experiments (n≥15 worms for each experiment)
We previously showed that xpa-1 or xpc-1 mutants exhibit increased basal levels of apoptosis in the germ line,11 probably because accumulation of unrepaired endogenous damage leads to apoptosis through the activation of (an)other DNA damage signaling pathway(s). Interestingly, the basal levels of RAD-54::YFP foci were also elevated in the absence of xpa-1 (Figure 2b), suggesting that the HR pathway mediates the increased apoptosis observed in xpa-1 mutants. Consistent with this hypothesis, mutations in genes that act downstream of DNA DSBs, such as hus-1 and atl-1 (Supplementary Table S1), as well as atm-1 and cep-111 abrogated the apoptosis phenotype of xpa-1 mutants.
We also examined the localization of RAD-54::YFP in atm-1, hus-1 and mre-11 mutants. Foci formation was essentially wild type in atm-1 mutants and only mildly affected in hus-1 mutants, indicating that ATM-1 and HUS-1 are either not required for or act downstream of RAD-54 recruitment to sites of DNA damage. By contrast, UV-C-treated mre-11 animals failed to show any focal accumulation of RAD-54::YFP in the mid–late pachytene cells (Figure 2b), whereas IR-treated animals exhibited wild-type foci numbers throughout the gonad (Figure 2b). Therefore, two (or more) distinct mechanisms for RAD-54 recruitment to sites of damage might exist, which differ in their requirement for MRE-11. Interestingly, Hayashi and co-workers16 have shown a similar difference in the requirement of RAD-50 for RAD-51 loading onto sites of DSBs.
Next, we examined the distribution of the replication protein A subunit homolog in C. elegans (RPA-1). RPA is a heterotrimeric single-stranded DNA-binding protein highly conserved in eukaryotes that plays an important role in DNA replication and repair.17, 18, 19, 20 Transgenic animals expressing YFP-tagged RPA-1, the homolog of the largest human RPA subunit p70, showed foci as early as 30 min after UV-C or IR treatment, reaching a plateau 3.5 h later (Figures 2c and d; Supplementary Figures 4a and b). Loss of XPA-1 completely blocked the increase of RPA-1 foci following UV, but not IR, suggesting that the NER machinery is required for UV-C-, but not for IR-induced accumulation of RPA-1 (Figure 2d).
In rad-54 mutants, RPA-1::YFP foci numbers were increased even in the absence of treatment (Figure 2e), likely marking sites of unresolved meiotic recombination intermediates.21 Surprisingly, UV-C and IR treatment did not increase much further the number of RPA-1::YFP foci in rad-54 mutants (Figure 2e). To distinguish between saturation in DNA damage signaling and a requirement for RAD-54 in damage-induced foci formation, we used a spo-11-deficient background. Both the excess in apoptosis and the high levels of RPA-1::YFP foci were abrogated in rad-54(ok615);spo-11(RNAi) animals (Figures 1g and 2e), and exposure to UV-C now led to a robust increase in foci number, suggesting that RPA-1 foci formation occurs upstream of, or in parallel to, RAD-54 in the UV response. RPA-1::YFP foci similarly increased following UV-C in mre-11(ok179) mutants (Figure 2d). Thus, most RPA-1::YFP foci following UV-C require the NER machinery and occur upstream of the HR components tested here.
In summary, our results so far show that components of the HR pathway act downstream of the NER DNA damage recognition step to trigger UV-C-induced apoptosis. NER pathway activity leads to focal recruitment of RPA-1 and RAD-54 and signaling to the apoptotic machinery.
The NER components XPG, XPB and XPD are required to promote UV-C-induced apoptosis by recruitment of HR proteins
During NER, initial recognition of the lesion, carried out by XPA and XPC, is followed by local unwinding of the DNA, performed by two DNA helicases with opposite polarities, XPB and XPD.22 This enables the excision step carried out by two structure-specific endonucleases, XPF and XPG.23
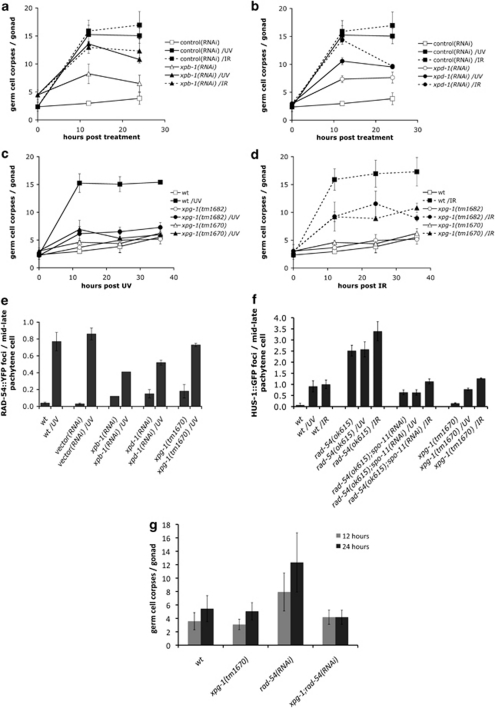
To determine whether NER steps downstream of DNA lesion recognition also play a role in UV-C-induced apoptosis, we analyzed germ cell apoptosis in animals with compromised or abolished XPB, XPD or XPG function. As with xpa-1 and xpc-1, RNAi knockdown of xpb-1 (Y66D12A.15) and xpd-1 (Y50D7A.2) gave rise to increased basal levels of apoptosis (Figures 3a and b). These findings strengthen the notion that in the absence of NER, unrepaired endogenous damage can activate (an)other DNA damage signaling pathway(s) that lead to apoptosis. xpb-1(RNAi)- and xpd-1(RNAi)-treated animals showed reduced apoptosis levels upon both UV-C and IR (Figures 3a and b), suggesting that these two helicases are also involved in promoting UV-C-induced apoptosis.
Figure 3.
C. elegans NER component XPG-1 is required for UV-C-induced germ cell apoptosis, but not for recruitment of RAD-54. Staged wild-type L1 larvae were raised on bacteria expressing xpb-1 (a), xpd-1 (b) or control dsRNA, and were treated with either 100 J/m2 UV-C or 120 Gy X-rays as young adults. Germ cell corpses were scored at the indicated time points. We could not score the 36-h time point in these animals, as the germ lines started to degenerate. This is possibly owing to the fact that both XPB-1 and XPD-1 are components of TFIIH, and thus might also affect mRNA transcription. (c and d) Apoptosis was scored in xpg-1(tm1670) or xpg-1(tm1682) mutant animals after treatment with 100 J/m2 UV-C (c) or 120 Gy X-rays (d). (e) XPG-1 is not required for UV-C-induced RAD-54 foci formation. RAD-54::YFP foci were quantified 3 h after treatment with 100 J/m2 UV-C as described in Materials and Methods. (f) RAD-54, but not XPG-1, is required for UV-C-induced HUS-1 foci formation. HUS-1::GFP foci were quantified in mid–late pachytene germ cell nuclei from staged young adult animals expressing HUS-1::GFP (opIs34) 3 h after exposure to 100 J/m2 of UV-C or 120 Gy of X-rays. Data shown in all cases represent the average±S.D. of three independent experiments (n>15 animals for each experiment). (g) Increased levels of apoptosis in rad-54 mutants are suppressed by xpg-1. Germ cell apoptosis was scored 12 and 24 h post the L4/adult molt in staged wild-type (wt), xpg-1(tm1670), rad-54(RNAi) and xpg-1(tm1670);rad-54(RNAi) animals. Data shown represent the average±S.D. of two independent experiments (n>20 animals for each experiment)
We also analyzed the effect on apoptosis of two likely null alleles of the endonuclease gene xpg-1 (F576B10.6) (Supplementary Figure 1c). UV-C-induced apoptosis was abolished in xpg-1 mutants (Figure 3c). The observed defect in xpg-1 (and xpa-1) mutants is not due to a general defect in germline physiology, which could reduce the number of cells that can undergo apoptosis: loss of xpg-1 (or xpa-1) did not reduce apoptosis in animals defective in the antiapoptotic Bcl-2 homolog CED-9, nor did it affect the egg laying rate (Supplementary Figures 5a and b). IR-induced apoptosis still occurred in xpg-1 mutants, although with reduced levels (Figure 3d). As IR-induced apoptosis is normal in other NER mutants (e.g., xpa-1, xpc-1), XPG-1 likely participates in other repair pathways following IR. Indeed, human XPG has been suggested to also function in base excision repair, as a cofactor for hNth1 DNA-glycosylase-AP-lyase during oxidative DNA damage repair.24 Remarkably, the increased apoptosis in rad-54(RNAi) animals (Figure 1b) was suppressed by the loss of xpg-1 function (Figure 3g), suggesting that XPG-1 acts downstream of, or in parallel to, RAD-54 to promote apoptosis in response to unresolved recombination intermediates. This effect was specific to rad-54, as xpg-1(tm1670) did not suppress the increased apoptosis in rad-51(RNAi) animals (Supplementary Figure 3d).
We showed above that XPA-1 is required for UV-C-induced RAD-54::YFP foci formation. Similarly, foci formation was reduced by twofold or more in xpb-1(RNAi) or xpd-1(RNAi) animals (Figure 3e). By contrast, loss of XPG-1 function did not have any strong effect (Figure 3e). Notably, untreated animals had elevated foci numbers in all three cases, likely reflecting accumulation of unrepaired lesions. Similarly, loss of xpg-1 did not inhibit recruitment of RPA-1::YFP (Figure 2d), which we showed to be recruited upstream of RAD-54 upon UV-C (Figure 2e).
Taken together, our findings suggest that recruitment of RAD-54 onto sites of UV-C damage does not require the whole complement of NER genes we analyzed: the DNA-binding protein XPA-1, and the XPB-1 and XPD-1 helicases appear sufficient for this step.
The HUS-1 complex functions downstream of the HR machinery to promote UV-C-induced apoptosis
We previously showed that recruitment of the HUS-1-containing 9-1-1 complex in response to UV-C requires XPA-1 function.11 Similarly, elimination of rad-54 blocked accumulation of HUS-1::GFP foci following UV treatment (Figure 3f). By contrast, we observed a modest increase in foci numbers following IR (Figure 3f). Proteins involved in early stages of HR, therefore, act upstream of and are important for 9-1-1 recruitment in UV response. By contrast, lack of xpg-1 did not affect HUS-1::GFP foci formation following either UV-C or IR treatment (Figure 3f). Thus, XPG-1 must impinge on the apoptotic signaling pathway at a step downstream of, or in parallel to, 9-1-1 recruitment.
The HR repair factors are involved in a signaling process to the apoptotic machinery following UV-C
Our finding that HR components are involved in UV-C-induced apoptosis led us to speculate on their exact role: are these proteins merely involved in signaling to the apoptotic apparatus, or do they also participate in the repair of the initial UV-C lesions?
To address this, we assessed the DNA repair activity in mutants defective in either NER or HR. We performed in vivo immunofluorescence to measure the presence of CPDs – one of the two major types of lesions caused by UV exposure – at defined time intervals in wild-type animals. Pachytene germ cells showed a maximum CPD signal immediately following UV-C (Figures 4a and b), which gradually disappeared with a half-life of approximately 10 h. These repair kinetics fall within the range reported for other systems.25, 26 Interestingly, CPDs were cleared at the same rate in rad-54(ok615) mutants as in wild type (Figure 4b), consistent with the notion that RAD-54 is not involved in CPD lesion repair. By contrast, xpa-1 mutants showed a greatly reduced ability to remove CPDs (Figure 4b), confirming the role of C. elegans xpa-1 in NER.
Figure 4.
CPD repair kinetics in NER and HR pathway mutants. (a) Immunofluorescence images of germ cell nuclei from UV-C-irradiated wild-type, rad-54(ok615) and xpa-1(ok698) worms. Gonads were extruded and fixed immediately following UV-C radiation or after 14 h. Images in the upper panel represent the CPD staining (green), the nuclei DAPI staining (magenta) and an overlay of the CPD staining and DAPI in wild-type animals, resulting in a white ring where UV lesions are detected on germ cell chromatin. Although in the absence of NER in xpa-1(ok698) mutants nuclei remain white (lower panel), CPDs are removed in rad-54(ok615) mutants similar to wild type mutants (chromatin turns magenta). (b) CPD signal intensity after irradiation with 100 J/m2 UV-C was determined by fluorescence image analysis of isolated gonads. Intensity before irradiation was defined as 0 and the value immediately after UV-C as 1 for each strain, and later time points were expressed as a fraction of the initial UV-C-induced intensity. At least 10 animals were scored per data point. Error bars represent the S.E.M. Repair kinetics for CPD lesions in transition cell nuclei of wild-type hermaphrodites (t1/2 approx. 10 h) are in good agreement with repair kinetics reported for other systems. xpa-1(ok698) mutants lacking a functional NER show no significant repair, whereas rad-54(ok615) mutants exhibit a response similar to wild type mutants. (c) RAD-54::YFP foci develop slowly and persist for long in UV-C-irradiated wild-type animals. Mid–late pachytene germ cell nuclei from staged young adult wild-type animals were scored for the presence of RAD-54::YFP foci, at 5, 10, 20 min, 1, 2, 4, 8 and 16 h post 100 J/m2 of UV-C treatment
Unlike the relatively rapid appearance and repair of CPDs, RAD-54::YFP foci developed more slowly, peaking 2 h after treatment, and also persisted for much longer (Figure 4c). Their perdurance might explain the apparent conundrum that UV-C-induced apoptosis persists for much longer than the CPDs (over 36 versus 10–20 h): it is not the CPDs themselves, but rather some type of DNA intermediates, recognized or marked by RAD-54, which promote apoptosis.
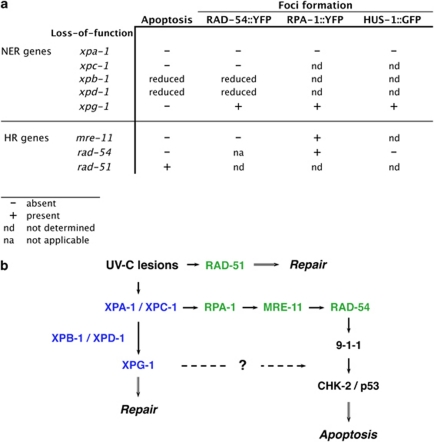
How might these two types of damage be linked? We suggest the following model: UV lesions by themselves are not pro-apoptotic. However, their processing by NER can lead, in a fraction of cases, to the generation of other types of DNA damage (e.g., DSBs, or long stretches of ssDNA). These are in turn recognized and processed by HR, leading to repair and, if necessary, apoptosis of the damaged cell (Figure 5).
Figure 5.
UV-C-induced apoptosis requires the action of both the NER and HR pathways. (a) Summary of the genetic requirements for the recruitment of NER and HR pathway components onto DNA lesions, as well as the induction of apoptosis following UV-C treatment. (b) Molecular model for UV-C-induced apoptosis in C. elegans. The NER machinery (blue) transforms a fraction of the UV-C lesions into other types of DNA damage (e.g., double-strand breaks). These, in turn, are then recognized by the HR machinery (green), leading to DNA repair and/or apoptosis of the damaged cell. Remarkably, XPG-1 defines a new branch that acts downstream of 9-1-1 to induce apoptosis
Discussion
In this paper, we took advantage of the powerful genetics in C. elegans to investigate in detail the signaling pathways that activate apoptosis in response to UV-C light. We show that apoptosis requires the coordinated action of two repair systems, the NER and HR pathways, and present a molecular model that explains this novel finding.
How does UV-C cause apoptosis in C. elegans? A simple explanation would be that UV-C directly causes double-strand breaks, which could activate the well-characterized ATM(/ATR)/CHK2/p53 pathway previously shown to respond to such lesions.27, 28 However, while mutagenic and cytotoxic, UV-C lesions are not pro-apoptotic per se: mutants in the DNA damage recognition components of the NER machinery, xpa-1 and xpc-1, are defective in UV-C-induced apoptosis.11 The requirement for these components suggests that their substrates, CPDs and 6-4PPs, are the relevant lesions, and not DSBs. Furthermore, induction of apoptosis requires both recognition and processing of these lesions by the NER machinery, as the DNA helicases XPB-1 and XPD-1 and the ssDNA endonuclease XPG-1 were also important for the activation of UV-C-induced apoptosis (Figure 3). A similar requirement for XPB and XPD has also been reported in human primary fibroblasts, as part of a p53-mediated apoptotic pathway.29
The requirement of a functional NER pathway for UV-C-induced apoptosis raises the question as to whether it is the repair process per se, or rather its output (the resulting DNA structures) that triggers apoptosis. Interestingly, the NER pathway is not the only repair pathway involved. We showed here that mutations in the HR pathway genes mre-11 and rad-54 also abrogate UV-C-induced cell death (Figures 1a and f). This lack of apoptosis in a post-mitotic cell population (mid–late pachytene germ cells), in which UV-C lesions cannot be converted into DSBs via replication, strongly suggests that the damage recognized by the HR repair system is a product of NER activity.
On the basis of these results, an attractive model for UV-C-induced apoptosis would be that the NER machinery transforms a fraction of the UV-C lesions – wittingly or unwittingly – into DSBs. These, in turn, are recognized by the HR pathway and lead to apoptosis via the pathway that we and others described previously (Figure 5).11, 30 Consistent with this model, the lesions or DNA structures positive for the presence of RAD-54 only appear progressively, over a few hours, in response to NER activity. How the latter can generate DSBs remains elusive. Might they happen accidentally when the NER machinery excises a stretch of the damaged strand? Assuming two CPDs per 104 bases at 100 J/m2 (compare Boyd et al.31), one haploid worm genome (108 bp) would initially harbor 4 × 104 lesions. Theoretically, this leaves a risk of almost 500 occurrences per cell of two lesions on opposing DNA strands within 30 nt, which, when processed simultaneously, could result in a DSB. For comparison, Vilenchik et al.32, 33 estimated that in one mammalian cell cycle, 1% of single-strand lesions are converted into 50 endogenous DSBs.
Although attractive, this model cannot fully explain a number of intriguing observations. First, we previously showed that UV-C and IR activate different sets of checkpoint kinases.11 However, we propose here that both treatments activate apoptosis via DSBs. Perhaps, the nature of the DSBs or of the associated protein complexes is different in the two cases. Second, we showed that while XPG-1 is required for UV-C-induced apoptosis, it is not needed for the generation of RAD-54-positive foci; furthermore, loss of xpg-1 function can suppress the increased apoptosis in non-irradiated worms defective for rad-54. At which level precisely XPG-1 functions to promote apoptosis is itself an intriguing topic.
Muzi-Falconi and co-workers34, 35 have suggested a functional link between NER and checkpoint activation in both yeast and mammalian cells, in which processing of UV lesions by the XPA-1 homolog enables 9-1-1 complex recruitment and activation of the ATR kinase.34 Toussaint et al.36 have characterized the co-participation of HR components in rendering G2-phase yeast cells more resistant to UV irradiation. Recent evidence might shift the concept of individual repair pathways toward that of a DNA damage response network, in which DNA repair factors interact to drive a coordinated response toward genome integrity and cell survival,37 or towards cell death.
Our findings suggest the existence of a cross-talk between the NER and HR repair pathways, and assign a role to several DNA repair components in apoptotic signaling. As repair pathways are highly conserved through evolution, the interaction between the NER and the HR pathways that we describe here for C. elegans might also occur in vertebrates, including humans.
Materials and Methods
Genetics
All strains were grown at 20°C on NGM agar seeded with Escherichia coli OP50. The Bristol N2 strain was used as the wild-type strain. Mutant alleles used are listed below: LGI: atm-1(gk186),11 hus-1(op244),38 xpg-1(tm1670) (this study), xpg-1(tm1682) (this study), xpa-1(ok698),11 rad-54(ok615) (this study), rad-54(tm1268) (this study); LGIV: xpc-1(ok734),11 spo-11(ok79);39 and LGV: atl-1(tm853),30 mre-11(ok179),12 chk-2(gk212).11 Transgenes: opIs257 (RAD-54::YFP) and opIs263 (RPA-1::YFP) (this study). Essential mutations were maintained as balanced strains: rad-54(ok615)/hT2[qIs51](IV;V), rad-54(tm1268)/hT2[qIs51](IV;V), spo-11(ok79)/nT1[n754 let](IV;V), atl-1(tm853)/nT1[qIs51](IV;V) and mre-11(ok179)/nT1[n754 let](IV;V). Additional information on mutations cited here can be obtained at www.wormbase.org.
Germline apoptosis
Staged young adult worms (12 h post the L4/adult molt) were exposed to different doses of UV-C light (254 nm) (J/m2) or X-rays (Gy). A Stratalinker UV crosslinker, model 1800 (Stratagene, Basel, Switzerland) and an Isovolt 160/225/320/450 HS X-ray machine (Rich. Seifert & Co., Ahrensburg, Germany) were used to deliver the appropriate doses. Corpses were scored in the meiotic region of one gonad arm at indicated time points using Nomarski optics. For the RNAi experiments, staged L1 larvae were transferred onto plates containing 2 mM IPTG seeded with E. coli strain HT115(DH3) expressing the respective RNAi clone, and scored as young adults for germline apoptosis.
Immunocytochemistry
For antibody staining of gonads, young adult hermaphrodites were dissected and fixed in 3% PFA/0.1 M K2HPO4 (pH 7.2) for 50 min at room temperature, followed by a 10-min incubation in 100% methanol on ice. Gonads were blocked in 5% BSA/PBS–Tween-20 0.1% for 1 h, followed by incubation with 1 : 50 anti-RAD-51 polyclonal15 antibody overnight at 4°C. Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen, Basel, Switzerland) was used as secondary antibody (1 : 500). The tissues were co-stained with DAPI before mounting. Fluorescent images were captured with a Leica DMRA2 microscope equipped with an ORCA-ER digital CCD camera and processed with an Openlab software.
CPD staining and quantification
A modified fixation protocol was applied to preserve/create denatured DNA structures for CPD staining, as this antibody specifically binds to CPDs in ssDNA.40 Adult hermaphrodites were irradiated with 100 J/m2 UVC and, at the indicated time points, were dissected in M9 (0.5 mM levamisole) to extrude the gonads and fixed in M9/3% PFA for 30 min. Fixed worms were transferred to poly-lysine-coated slides, freeze-cracked and permeabilized in PBS/Tween-20 0.1% for 3 × 10 min. For DNA denaturation, slides were incubated in PBS/0.07 N NaOH for 8 min and washed in PBS/Tween-20 0.1% for 3 × 10 min before blocking (1 h at RT) and incubation with 1 : 500 anti-CPD monoclonal antibody (MBL International, Woburn, MA, USA; product no. D194-1) for at least 6 h at 4°C. Samples were washed and incubated (1 h at RT) with Alexa Fluor 488 goat anti-rabbit (Invitrogen) as secondary antibody (1 : 500) and counterstained with DAPI (200 ng/ml) before mounting in Prolong Gold (Invitrogen). Blocking and antibody incubations were in antibody buffer/10% goat serum.
Fluorescent images were captured as described above with defined exposure settings. For quantification of CPD signal intensity, a macro was created and run in ImageJ (version 1.42q; http://rsb.info.nih.gov/nih-image/), allowing for automated measurements after definition of a region of interest (ROI) in the immunofluorescence pictures. Briefly, an ROI was drawn around approximately 100 germ cell nuclei of the transition/early meiotic region. A mask was defined from a binary threshold image of the DAPI staining to account for DNA containing areas and applied to the anti-CPD image. Average anti-CPD signal intensity over DAPI-positive areas was compared with average intensity of the background within the ROI, and expressed as difference.
Foci quantification
Staged young adult hermaphrodites were treated with either 100 J/m2 of UV-C light or 120 Gy of X-rays, and gonads were dissected at indicated time points. RAD-54::YFP and RPA-1::YFP foci formation was quantified by counting the number of bright foci present in mid–late pachytene germ cells, in one focal plane within 100 μm from the first oocyte. Fluorescent images were captured with a Leica DMRA2 microscope equipped with an ORCA-ER digital CCD camera and were processed with an Openlab software.
RNAi constructs
Fragments corresponding to exonic sequences of xpb-1 (Y66D12A.15) and xpd-1 (Y50D7A.2) were amplified by PCR from a wild-type cDNA library using primers listed in Supplementary Table S2, and cloned into the L4440 RNAi vector. The constructs were used to transform HT115(DH3) bacteria, which were subsequently used to feed worms in the RNAi experiments. For the rest of the RNAi clones in this study, we used the Ahringer RNAi library. A standard gfp sequence cloned into L4440 vector or the empty vector was used as controls for the RNAi experiments.
Transgenic worms
Genomic fragments corresponding to rpa-1 and rad-54 promoter and ORF, as well as the 3′-UTR region were separately amplified by PCR from N2 genomic DNA using primers (Supplementary Table S2) that added the appropriate restriction sites. The amplified fragments were cloned into the pLN022 expression vector upstream of yfp to generate pLS69 (RPA-1::YFP) or pLS57 (RAD-54::YFP). Low-copy transgenic worms were then generated by ballistic transformation. Sequences of all used plasmids are available upon request.
Acknowledgments
We thank Professor H Naegeli and Dr A Gartner for critical comments on our manuscript, and members of the Hengartner lab for discussions. This work was supported by the Kanton of Zurich, the Swiss National Science Foundation, the Ernst Hadorn Foundation and the Josef Steiner Cancer Research Foundation. RE was supported by an MD-PhD fellowship from the Swiss National Science Foundation. We also thank Dr A Gartner for providing us with the RAD-51 antibody, and the Caenorhabditis Genetics Center, which is funded by the National Institute of Health (NIH), National Center for Research Resources (NCRR) as well as Shohei Mitani of the National Bioresource Project in Japan for strains.
Glossary
- UV-C
ultraviolet light C (254 nm)
- IR
ionizing radiation
- CPDs
cyclobutane pyrimidine dimers
- DSB
double-strand DNA break
- NER
nucleotide excision repair
- HR
homologous recombination
- 9-1-1 complex
Rad9-Hus1-Rad1 complex (HPR-9/HUS-1/MRT-2 in C. elegans)
- XPA, XPB, XPD, XPF, XPG
xeroderma pigmentosum complementation group A, B, D, F, G
- RPA-1
replication protein A large subunit homolog in C. elegans
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Baehrecke
Supplementary Material
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie. 1999;81:127–137. doi: 10.1016/s0300-9084(99)80045-5. [DOI] [PubMed] [Google Scholar]
- Wijnhoven SW, Hoogervorst EM, de Waard H, van der Horst GT, van Steeg H. Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat Res. 2007;614:77–94. doi: 10.1016/j.mrfmmm.2005.12.018. [DOI] [PubMed] [Google Scholar]
- van der Wees C, Jansen J, Vrieling H, van der Laarse A, Van Zeeland A, Mullenders L. Nucleotide excision repair in differentiated cells. Mutat Res. 2007;614:16–23. doi: 10.1016/j.mrfmmm.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Carvalho H, Ortolan TG, dePaula T, Leite RA, Weinlich R, Amarante-Mendes GP, et al. Sustained activation of p53 in confluent nucleotide excision repair-deficient cells resistant to ultraviolet-induced apoptosis. DNA Repair. 2008;7:922–931. doi: 10.1016/j.dnarep.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Stergiou L, Hengartner MO. Death and more: DNA damage response pathways in the nematode C. elegans. Cell Death Differ. 2004;11:21–28. doi: 10.1038/sj.cdd.4401340. [DOI] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK.Germline survival and apoptosisIn: WormBook (ed). The C. elegans Research Communityvol. 145.1. 2008pp 1–20. . http://www.wormbook.org . [DOI] [PMC free article] [PubMed]
- Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14:1129–1138. doi: 10.1038/sj.cdd.4402115. [DOI] [PubMed] [Google Scholar]
- Chin GM, Villeneuve AM. C. elegansmre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G DNA damage checkpoint. Genes Dev. 2001;15:522–534. doi: 10.1101/gad.864101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Petukhova G, Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J Biol Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- Alpi A, Pasierbeck P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missura M, Buterin T, Hindges R, Hübscher U, Kaspárková J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA–RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Sung P. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J Biol Chem. 2002;277:43578–43587. doi: 10.1074/jbc.M205864200. [DOI] [PubMed] [Google Scholar]
- Stauffer ME, Chazin WJ. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J Biol Chem. 2004;279:25638–25645. doi: 10.1074/jbc.M400029200. [DOI] [PubMed] [Google Scholar]
- Solinger JA, Heyer WD. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc Natl Acad Sci USA. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, et al. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Chiganças V, Galhardo R, Carvalho H, Menck CF. The eukaryotic nucleotide excision repair pathway. Biochimie. 2003;85:1083–1099. doi: 10.1016/j.biochi.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Cann KL, Hicks GG. Regulation of the cellular DNA double-strand break response. Biochem Cell Biol. 2007;85:663–674. doi: 10.1139/O07-135. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Wang XW, Vermeulen W, Coursen JD, Gibson M, Lupold SE, Forrester K, et al. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Crocker TL, Rodriguez AM, Leung MC, Lehmann DW, Freedman JH, et al. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutat Res. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG. Radiation dose-rate effects, endogenous DNA damage, and signaling resonance. Proc Natl Acad Sci USA. 2006;103:17874–17879. doi: 10.1073/pnas.0607995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, et al. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci USA. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint M, Wellinger RJ, Conconi A. Differential participation of homologous recombination and nucleotide excision repair in yeast survival to ultraviolet light radiation. Mutat Res. 2010;698:52–59. doi: 10.1016/j.mrgentox.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde LH, Wu H. Involvement of nucleotide excision and mismatch repair mechanisms in double strand break repair. Curr Genomics. 2009;10:250–258. doi: 10.2174/138920209788488544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, et al. Caenorhabditis elegans HUS--1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Tsujino T, Suzuki T, Nikaido O, Ohtsuka E. Antigen structural requirements for recognition by a cyclobutane thymine dimer-specific monoclonal antibody. Nucleic Acids Res. 1997;25:3889–3894. doi: 10.1093/nar/25.19.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.