Abstract
Identifying the molecular mechanisms responsible for the resistance of gliomas to anticancer treatments is an issue of great therapeutic interest. Δ9-Tetrahydrocannabinol (THC), the major active ingredient of marijuana, and other cannabinoids inhibit tumor growth in animal models of cancer, including glioma, an effect that relies, at least in part, on the stimulation of autophagy-mediated apoptosis in tumor cells. Here, by analyzing the gene expression profile of a large series of human glioma cells with different sensitivity to cannabinoid action, we have identified a subset of genes specifically associated to THC resistance. One of these genes, namely that encoding the growth factor midkine (Mdk), is directly involved in the resistance of glioma cells to cannabinoid treatment. We also show that Mdk mediates its protective effect via the anaplastic lymphoma kinase (ALK) receptor and that Mdk signaling through ALK interferes with cannabinoid-induced autophagic cell death. Furthermore, in vivo Mdk silencing or ALK pharmacological inhibition sensitizes cannabinod-resistant tumors to THC antitumoral action. Altogether, our findings identify Mdk as a pivotal factor involved in the resistance of glioma cells to THC pro-autophagic and antitumoral action, and suggest that selective targeting of the Mdk/ALK axis could help to improve the efficacy of antitumoral therapies for gliomas.
Keywords: Midkine, ALK, autophagy, DNA arrays, tumor xenografts
Glioblastoma multiforme (GBM), or grade IV astrocytoma, is the most frequent class of malignant primary brain tumor and one of the most aggressive forms of cancer. As a consequence, median survival after diagnosis is usually just 12–15 months.1, 2 This dramatic behavior is mainly due to the high invasiveness and proliferation rate of GBM. In addition, GBM exhibits a high resistance to standard chemotherapy and radiotherapy. Current strategies for the treatment of GBM are only palliative, and include surgical resection (which is frequently incomplete because of the proximity of the tumor to vital brain structures) and focal radiotherapy. A large number of chemotherapeutic agents (e.g. alkylating agents such as temozolomide and nitrosureas such as carmustine) have also been tested, but they display limited efficacy.1 It is therefore essential to develop new therapeutic strategies for the management of GBM. Recently, much effort has been made to understand GBM in terms of its underlying genetic lesions.3, 4 This approach, together with the use of gene expression profile studies, may help to precisely characterize these tumors, thus allowing the identification of factors for resistance to different anti-glioma therapies and the development of specific targeted therapies.5
The hemp plant Cannabis sativa produces ∼70 unique compounds known as cannabinoids, of which Δ9-tetrahydrocannabinol (THC) is the best characterized owing to its high potency and abundance.6 THC exerts a wide variety of biological effects by mimicking endogenous substances – the endocannabinoids anandamide and 2-arachidonoylglycerol – that bind to and activate specific cannabinoid receptors.6 So far, two G protein-coupled cannabinoid-specific receptors have been cloned and characterized from mammalian tissues: CB1, abundantly expressed in the brain and at many peripheral sites, and CB2 expressed in the immune system and also present in microglia, some neuron sub-populations and glioma cells.7
One of the most active areas of research in the cannabinoid field is the study of the potential application of cannabinoids as antitumoral agents.8 Thus, cannabinoid administration has been shown to curb the growth of several models of tumor xenografts in rats and mice,8 including gliomas.9, 10, 11 On the basis of this preclinical evidence, a pilot clinical study has been conducted to investigate the effect of THC on recurrent GBM.12 The mechanism of cannabinoid antitumoral action relies on the ability of these agents to inhibit tumor angiogenesis, inhibit cell cycle progression and stimulate cancer cell death.8, 13 We have recently found that cannabinoids activate an endoplasmic reticulum (ER) stress-related signaling route that leads to the upregulation of the transcriptional co-activator p8 and its target the pseudo-kinase tribbles homolog 3 (TRB3).9, 14 The stimulation of this pathway promotes autophagy via TRB3-mediated inhibition of the Akt/mammalian target of rapamycin complex 1 (mTORC1) axis and is indispensable for the pro-apoptotic and antitumoral action of THC.15
The present work was undertaken to identify the molecular factors responsible for the resistance of glioma cells to cannabinoid antitumoral activity. By using DNA microarrays we have identified a gene expression profile characteristic of THC-resistant glioma cells. We also show that one of the genes upregulated in cannabinoid-resistant glioma cells, the growth factor Mdk, acting through the anaplastic lymphoma kinase (ALK) receptor, has a pivotal role in enhancing the resistance to cannabinoid-evoked autophagy-mediated cell death. Our results support that targeting of the Mdk/ALK axis could help to improve the efficacy of antitumoral therapies for gliomas.
Results
Identification of the gene expression profile characteristic of THC-resistant glioma cells
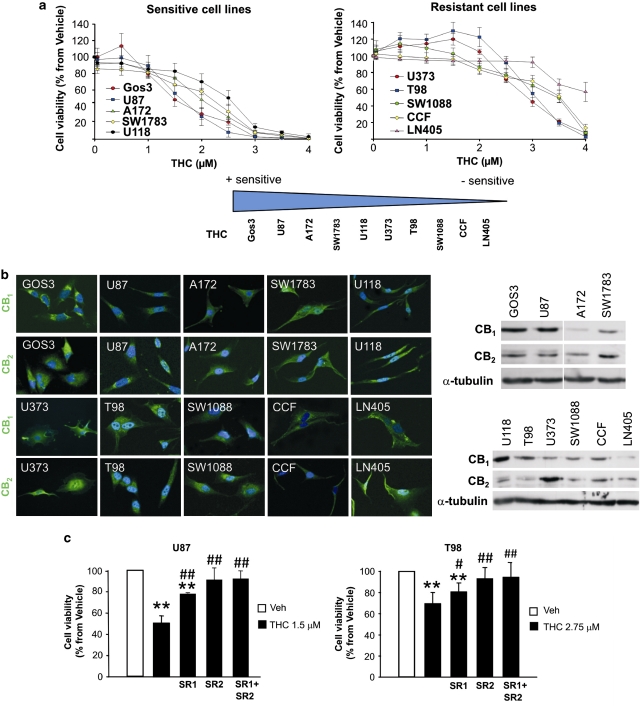
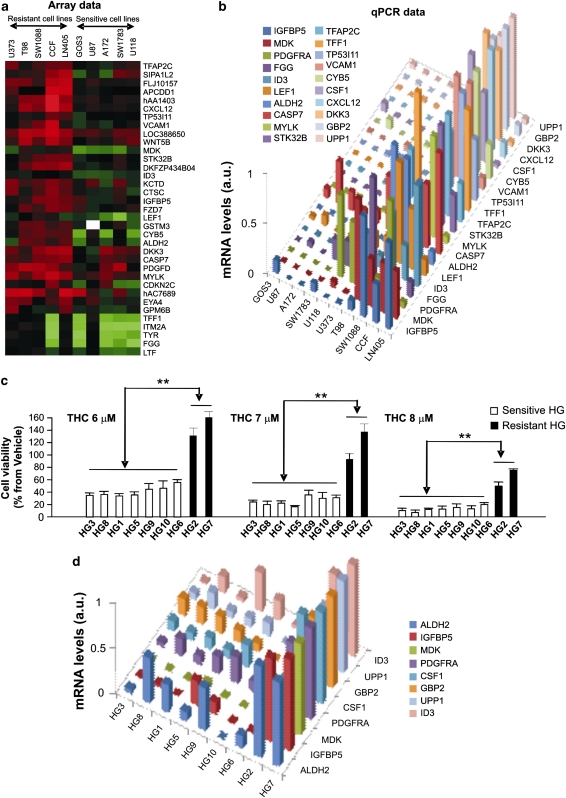
To gain insight into the molecular features associated with the resistance of glioma cells to cannabinoid action, we initially characterized 10 human glioma cell lines on the basis of their different sensitivity to THC-induced cell death (Figure 1a). Thus, cell lines were classified as cannabinoid sensitive (IC50 for THC lower than 2.5 μM) and cannabinoid resistant (IC50 for THC higher than 2.5 μM). To note, we did not find any correlation between the sensitivity of these cell lines to THC and to other two chemotherapeutic agents (carmustine and temozolomide) frequently used in glioma treatment (Supplementary Figures 1A, B and C), which indicates that the resistance to THC action is determined by specific mechanisms distinct from those promoting resistance to other drugs. In addition, differences in the levels of CB1 or CB2 receptors of glioma cells did not correlate with a higher or lower sensitivity to THC-induced cell death (Figure 1b), and pharmacological blockade of CB1 receptors with the selective antagonist SR141716 or CB2 receptors with the selective antagonist SR144528 abrogated THC-induced death in both sensitive and resistant cell lines (Figure 1c and Supplementary Figure 1D). This supports that mechanisms other than changes in the expression or the functionality of CB receptors are responsible for the differences in THC sensitivity of glioma cells. Analysis of some of the genes most frequently altered in gliomas (p16, PTEN, p53 and EGFR) did not reveal association between the presence of mutations, amplifications or deletions on those genes and sensitivity to cannabinoid treatment (data not shown). Therefore, in order to identify the molecular factors that determine the different sensitivity of glioma cells to THC, we analyzed by DNA microarrays the gene expression profile of the 10 glioma cell lines. After selection of those genes whose expression was significantly changed (FDR<0.2) at least 2.5-fold in more than three sensitive or resistant cell lines with respect to their sensitive or resistant counterparts (Figure 2a, Supplementary Figure 2A and Supplementary Tables 1 and 2) and real-time quantitative PCR validation of the array data (Figure 2b, Supplementary Figure 2B and Supplementary Tables 3 and 5), we selected 20 genes as potential candidates to predict the resistance of glioma cells to cannabinoid action. Next, we analyzed the expression of these genes in primary cultures of glioma cells obtained from tumor samples of GBM patients (in which we confirmed that THC promoted cell death via stimulation of cannabinoid receptors; Supplementary Figure 3D). Data revealed that 8 of the 20 initially selected genes were differentially upregulated in the cultures that were more resistant to THC-induced cell death (Figures 2c and d, Supplementary Figure 3A and Supplementary Table 4). These observations support that this set of genes is strongly associated with a higher resistance of glioma cells to THC-induced death.
Figure 1.
Classification of human glioma cell lines according to their sensitivity to THC-induced cell death. (a) Effect of THC on the viability (72 h) of 10 human glioma cells lines (cells with IC50<2.5 μM (left panel) and cells with IC50>2.5 μM (right panel) were considered sensitive and resistant, respectively, to THC-induced cell death; mean±S.D.; n=5). (b) CB1 and CB2 expression of the human glioma cells lines as determined by immunostaining (left panel) and western blot (right panel). One representative experiment of three is shown. (c) Effect of THC, SR1 (1 μM) and SR2 (1 μM) on the viability (72 h) of U87 (left panel) and T98 (right panel) cells (mean±S.D.; n=3; **P<0.01 from vehicle-treated cells, #P<0.05 and ##P<0.01 from THC-treated cells; Veh, Vehicle, THC, Δ9-tetrahidrocannabinol)
Figure 2.
Identification of the gene expression profile associated with resistance to THC-induced cell death. (a) Array data: selection of genes that exhibit different basal expression levels between THC-resistant and THC-sensitive human glioma cell lines. Data correspond to genes whose expression was significantly increased (FDR<0.2) at least 2.5-fold in three or more resistant glioma cell lines with respect to three or more sensitive glioma cell lines. Each square represents the fluorescence ratio (log2 scale) between sample RNA (cell lines) and reference RNA for one gene; red squares represent positive values, green squares represent negative values and white squares represent non-determined values. (b) mRNA levels, as determined by quantitative real-time PCR, of 20 genes associated with resistance to THC-induced cell death in human glioma cell lines. Data correspond to mRNA levels for each gene relative to the highest mRNA expression level for that gene detected in any of the 10 human glioma cell lines and are expressed in arbitrary units (a.u.) in a 0–1 scale. (c) Effect of THC on the viability (72 h) of primary cultures of human glioma cells (mean±S.D.; n=5; **P<0.01 from each THC-sensitive primary culture of glioma cells). (d) mRNA levels, as determined by quantitative real-time PCR, of eight genes associated with resistance to THC-induced cell death in primary cultures of human glioma cells. Data correspond to mRNA levels for each gene relative to the highest mRNA expression level for that gene detected in any of the nine primary cultures of human glioma cells and are expressed in arbitrary units (a.u.) in a 0–1 scale
The growth factor Mdk confers resistance to THC
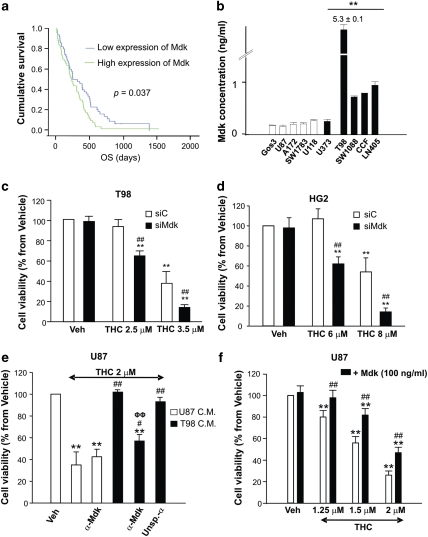
One of the genes whose upregulation was more strongly associated with the resistance of glioma cells to cannabinoid treatment was the growth factor Mdk; Mdk mRNA levels were 30- to 80-fold higher in the resistant than in the sensitive cell lines (Figures 2b and d and Supplementary Figures 4A and C). Mdk is a member of the pleiotrophin (PTN)/Mdk family and was originally described as a developmentally regulated, heparin-bound, neurotrophic factor.16, 17 Mdk has been proposed to modulate the proliferation and migration of various types of cancer cells and is often overexpressed in malignant tumors.18, 19 Of interest, analysis of samples from 201 glioma patients (including 115 GBMs) revealed that Mdk expression is associated to a lower overall survival of glioma, high-grade glioma and GBM patients (Figure 3a and Supplementary Figure 5). These analyses were validated with external data obtained from The Cancer Genome Atlas3 (Supplementary Figure 5E and Supplementary Table 6).
Figure 3.
Increased Mdk expression confers human glioma cells resistance to THC. (a) Kaplan–Meier survival curves for 115 glioblastoma tumors from patients collected at the Virgen de la Salud Hospital (Toledo) and classified according to Mdk expression. Mdk expression is associated to a lower overall survival of GBM patients (P<0.037). Mdk positive or negative expression is shown by green or blue lines, respectively. OS: overall survival. (b) Mdk concentration, as determined by ELISA, in the supernatant of 10 human glioma cell lines (mean±S.D.; n=3; **P<0.01 from each THC-sensitive glioma cell line). (c and d) Effect of THC on the viability (72 h) of T98 (c) and HG2 (d) cells transfected with control (siC) or midkine-selective (siMdk) siRNA (mean±S.D.; n=5 for each cell line, **P<0.01 from vehicle-treated, siC-transfected cells and from vehicle-treated, siMdk-transfected cells and ##P<0.01 from THC-treated, siC-transfected cells). Mdk mRNA levels (as determined by real-time quantitative PCR) were reduced in siMdk-transfected cells relative to their corresponding siC-transfected cells by 81±9% (T98 cells) and 84±9% (HG2 cells), respectively (n=5 for T98 and HG2 cells). (e) Effect of THC on the viability of U87 cells incubated with conditioned medium (C.M.) obtained from U87 or T98 cells. When indicated, the corresponding C.M. was pre-incubated with a neutralizing anti-Mdk (α-Mdk) or isotype unspecific (α-Unsp) antibody (mean±S.D.; n=4; **P<0.01 from vehicle-treated cells, #P<0.05 or ##P<0.01 from THC-treated, U87 C.M.-incubated cells and ΦΦP<0.01 from THC-treated, T98 C.M.-incubated cells). (f) Effect of THC on the viability of U87 cells incubated with exogenous Mdk (mean±S.D.; n=5; **P<0.01 from vehicle-treated cells and ##P<0.01 from THC-treated cells not incubated with Mdk)
We found that the levels of Mdk are increased in the supernatants of the cannabinoid-resistant glioma cell lines (Figure 3b). We therefore investigated whether this growth factor participates in the mechanism of resistance to cannabinoid antitumoral action of gliomas. In agreement with this possibility, Mdk silencing sensitized two cannabinoid-resistant cell lines (Figure 3c and Supplementary Figure 6A) as well as a primary culture of glioma cells (Figure 3d) to THC-induced cell death. Moreover, incubation with conditioned medium derived from the cannabinoid resistant cell line T98 (which was enriched in Mdk) (Figure 3e) or with exogenous Mdk (Figure 3f) rendered the cannabinoid-sensitive line U87 resistant to THC treatment, an effect that was abrogated by Mdk neutralization with an anti-Mdk antibody (Figure 3e and Supplementary Figure 6B). These observations strongly support that Mdk is a factor for resistance to THC-induced glioma cell death.
Of note, the cannabinoid-resistant pancreatic cancer cell line Panc1 exhibited much higher levels of Mdk than the cannabinoid-sensitive pancreatic cancer cell line MiaPaca2 (Supplementary Figures 7A and B). Furthermore, Mdk knockdown rendered Panc1 cells sensitive to THC, suggesting that a similar mechanism of resistance to cannabinoid-induced cell death also operates in pancreatic cancer cells (Supplementary Figure 7C).
Mdk promotes resistance to THC through stimulation of the ALK receptor
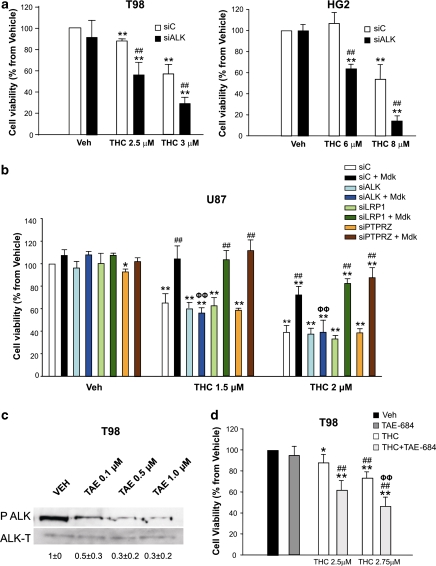
The mechanism by which Mdk mediates its different biological actions has not been completely clarified16 and may involve the stimulation of several receptors, including ALK, protein-tyrosine phosphatase, receptor-type, zeta-1 (RPTPZ), and low-density lipoprotein receptor-related protein 1 (LRP1).16 It has been proposed that the pro-survival and pro-angiogenic effect of Mdk on tumor cells relies on the stimulation of the ALK receptor.20 ALK was expressed in all the cell lines and primary cultures of human glioma cells used in this study (Supplementary Figures 8A, B and C), and therefore, we asked whether Mdk binding to ALK has a role in the resistance of glioma cells to THC-induced cell death. In agreement with this idea: (i) ALK knockdown sensitized T98 and HG2 glioma cells to THC-induced cell death (Figure 4a and Supplementary Figure 9), whereas silencing of RPTPZ and LRP1 only modestly enhanced the response to THC treatment of T98 (Supplementary Figure 9C) and HG2 (Supplementary Figure 9D) cells, respectively; (ii) incubation with T98 cell-conditioned medium or addition of exogenous Mdk increased ALK phosphorylation of U87 cells (Supplementary Figures 10C and D); (iii) silencing of ALK – but not of RPTPZ or LRP1 – abrogated the resistance to THC-induced cell death exerted by the addition of exogenous Mdk to U87 cells (Figure 4b and Supplementary Figure 10A); (iv) knockdown of ALK abolished the protective action exerted by T98-conditioned medium on U87 cells (Supplementary Figure 10B); and (v) incubation with the ALK-selective inhibitor NVP-TAE-684 reduced ALK phosphorylation (Figure 4c) and enhanced THC action in T98 cells (Figure 4d). Altogether, these observations strongly support that Mdk promotes resistance to THC-induced cell death via ALK stimulation.
Figure 4.
Mdk promotes resistance to THC-induced cell death via ALK. (a) Effect of THC on the viability of T98 (left panel) or HG2 (right panel) cells transfected with control (siC) or ALK-selective (siALK) siRNA (mean±S.D.; n=5 for T98 and HG2 cells; **P<0.01 from vehicle-treated, siC-transfected cells and from vehicle-treated, siALK-transfected cells and ##P<0.01 from THC-treated, siC-transfected cells). ALK mRNA levels (as determined by real-time quantitative PCR) were reduced in siALK-transfected cells relative to their corresponding siC-transfected cells by 77±14% (T98 cells) and 78±2% (HG2 cells), respectively (n=5). (b) Effect of THC on the viability of U87 cells transfected with siC, siALK, LRP1-selective (siLRP1) or PTPRZ-selective (siPTPRZ) siRNAs and incubated with vehicle or exogenous Mdk (mean±S.D.; n=3; *P<0.05 or **P<0.01 from vehicle-treated, siC-transfected cells; ##P<0.01 from THC-treated, siC-transfected cells not incubated with exogenous Mdk and ΦΦP<0.01 from THC-treated, siC-transfected cells incubated with exogenous Mdk). (c) Effect of the ALK inhibitor NVP-TAE-684 on ALK phosphorylation of T98 cells (a representative experiment of three is shown). Values correspond to the densitometric analysis of ALK phosphorylation and are expressed as the ratio of phosphoALK/total ALK for each condition relative to vehicle-treated cells. (d) Effect of THC and the ALK inhibitor NVP-TAE-684 (TAE 684, 0.1 μM) on the viability of T98 cells (mean±S.D.; n=5; *P<0.05 or **P<0.01 from vehicle-treated cells; ##P<0.01 from NVP-TAE684-treated cells and ΦΦP<0.01 from THC-treated cells)
Mdk inhibits the activation of the cannabinoid-evoked autophagy-mediated cell death pathway
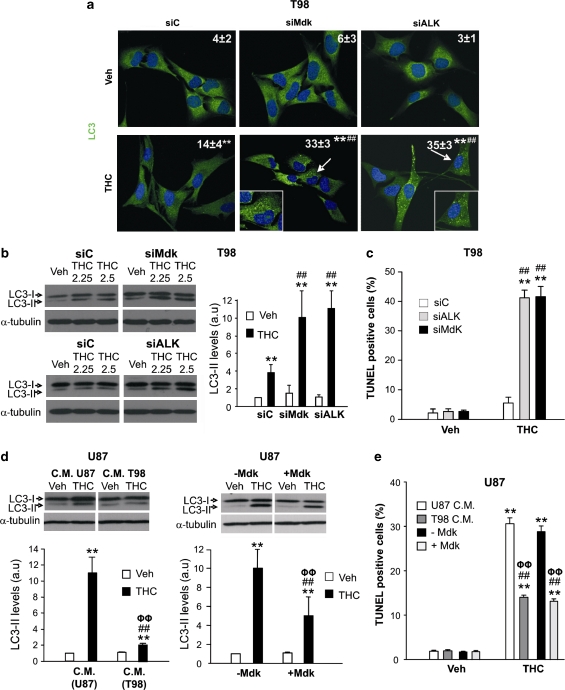
We have recently found that THC stimulates autophagy in glioma cells and that the activation of this cellular process is required for the pro-apoptotic and antitumoral action of cannabinoids.15 We therefore asked whether the mechanism by which Mdk promotes resistance to THC-induced cell death could be based on the modulation of the signaling route that regulates autophagy in response to cannabinoid treatment. In line with this hypothesis, knockdown of Mdk or ALK in T98 or HG2 cells enhanced THC-induced autophagy, as determined by the presence of cells with LC3 dots as well as by the accumulation of the lipidated form of LC3 (LC3-II; two hallmarks of autophagy;21 Figures 5a and b and Supplementary Figure 11A), as well as THC-induced apoptosis (Figure 5c and Supplementary Figure 11B). Likewise, pharmacological inhibition of ALK-enhanced THC-induced autophagy and apoptosis of T98 cells (Supplementary Figure 12). Moreover, incubation of U87 cells with T98 cell-conditioned medium or with exogenous Mdk decreased THC-induced autophagy (Figure 5d) and apoptosis (Figure 5e) in response to THC. These data strongly support that the Mdk/ALK axis modulates THC-induced autophagy.
Figure 5.
Mdk promotes resistance to THC-induced autophagy and apoptosis. (a) Effect of THC (2.5 μM, 24 h) on LC3 immunostaining of siC, siMdk or siALK-transfected T98 cells. Values in the upper right corner at each photomicrograph correspond to the percentage of cells with LC3 dots relative to the total number of cells (mean±S.D.; n=3; representative photomicrographs of each condition are shown; **P<0.01 from vehicle-treated, siC-transfected cells; ##P<0.01 from THC-treated, siC-transfected cells). Inset: high-magnification photomicrographs of the arrow-pointed cells. (b) Effect of THC (24 h) on LC3 lipidation of siC, siMdk or siALK-transfected T98 cells. A representative experiment of four is shown. Right panel: densitometric analysis of LC3-II lipidation. Data correspond to LC3-II optical density values (OD) for each condition relative to vehicle-treated, siC-transfected cells (mean±S.D.; n=4; THC 2.5 μM; **P<0.01 from the corresponding vehicle-treated cells; ##P<0.01 from THC-treated, siC-transfected cells). (c) Effect of THC (2.5 μM, 24 h) on apoptosis (as determined by TUNEL) of siC, siALK and siMdk-transfected T98 cells. Data correspond to the percentage of TUNEL-positive cells relative to the total number of cells (mean±S.D.; n=3; **P<0.01 from the corresponding vehicle-treated cells; ##P<0.01 from THC-treated, siC-transfected cells). (d) Effect of THC (1.5 μM, 24 h) on LC3 lipidation of U87 cells incubated with U87 C.M. or T98 C.M. (left panel) or with exogenous Mdk (right panel). A representative experiment of three is shown. Lower panels: data correspond to LC3-II OD for each condition relative to vehicle-treated, U87 C.M.-incubated cells (left panel) or to vehicle-treated cells (right panel; mean±S.D.; n=3; **P<0.01 from vehicle-treated, U87 C.M.-incubated cells (left panel) or vehicle-treated cells (right panel); ##P<0.01 from THC-treated, U87 C.M.-incubated cells (left panel) or THC-treated cells (right panel) and ΦΦP<0.01 from vehicle-treated, T98 C.M. cells (left panel) or vehicle-treated, Mdk-incubated cells (right panel)). (e) Effect of THC (1.5 μM, 24 h) on apoptosis (as determined by TUNEL) of U87 cells incubated with U87 C.M., T98 C.M. or with (+Mdk) or without (−Mdk) exogenous Mdk. Data correspond to the percentage of TUNEL-positive cells relative to the total number of cells (mean±S.D.; n=3; **P<0.01 from the corresponding vehicle-treated cells; ##P<0.01 from THC-treated U87 cells incubated with U87 C.M.- or from THC-treated U87 cells incubated without exogenous Mdk and ΦΦP<0.01 f from vehicle-treated U87 cells incubated with T98-conditioned medium or with exogenous Mdk)
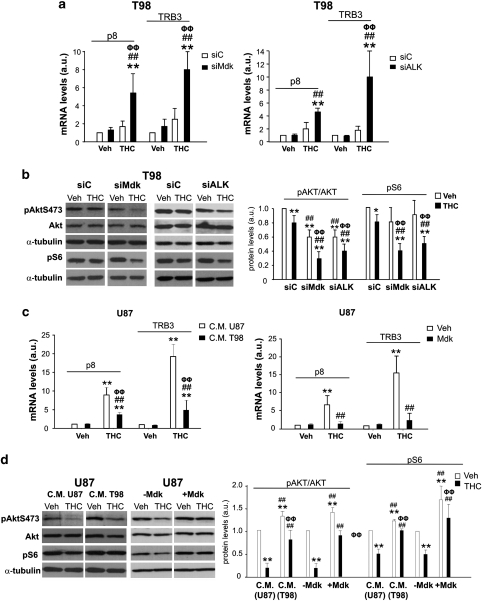
Induction of autophagy in many cellular settings relies on the inhibition of the mammalian mTORC1, which in turn regulates the activity of several autophagy proteins involved in the early triggering of this cellular process.21 Accordingly, THC stimulates an ER stress-related pathway that leads to the upregulation of the transcription factor p8 and its downstream target TRB3. This latter protein triggers the inhibition of the Akt/mTORC1 axis, thus leading to the stimulation of autophagy-mediated cell death.15 On the other hand, ALK stimulation has been shown to activate the phosphatidylinositol 3-kinase (PI3K)/Akt pathway.20 Therefore, we investigated whether the resistance to THC-induced autophagy exerted by Mdk relied on the modulation of p8/TRB3 expression and/or on the activation of the Akt/mTORC1 axis. In line with this idea, THC-triggered p8 and TRB3 induction in T98 and HG2 cells was strongly enhanced by Mdk or ALK knockdown (Figure 6a and Supplementary Figure 11C). Furthermore, a decrease in the phosphorylation of Akt and S6 (a well-established readout of mTORC1 activity) was observed upon THC treatment of Mdk- or ALK-silenced T98 and HG2 cells (Figure 6b and Supplementary Figure 11D). On the other hand, incubation of U87 cells with T98 cell-conditioned medium or with exogenous Mdk decreased THC-induced p8 and TRB3 upregulation (Figure 6c) and attenuated THC inhibition of Akt and S6 phosphorylation (Figure 6d). Taken together, these observations strongly support that Mdk (via ALK) modulates p8/TRB3 expression as well as the activity of the Akt/mTORC1 axis to prevent the stimulation of autophagy-mediated cell death by cannabinoids.
Figure 6.
Mdk promotes resistance to THC-induced autophagy by preventing p8 and TRB3 upregulation and AKT/mTORC1 inhibition. (a) Effect of THC (2.5 μM, 6 h) on p8 and TRB3 mRNA levels (as determined by quantitative real-time PCR) of siC- and siMdk- (left panel) or siC- and siALK- (right panel) transfected cells (mean±S.D.; n=5; **P<0.01 from vehicle-treated, siC-transfected cells; ##P<0.01 from vehicle- treated, siMdk-transfected (left panel) or from vehicle-treated siALK-transfected (right panel) cells, ΦΦP<0.01 from THC-treated, siC-transfected cells). (b) Effect of THC (2.5 μM, 24 h) on AKT and S6 phosphorylation of siC, siMdk or siALK-transfected T98 cells. A representative experiment of four is shown. Right panel: densitometric analysis of Akt and S6 phosphorylation. Data correspond to phospho-Akt, Akt, phospho-S6 and tubulin OD values and are expressed as the ratio of phosphoAKT/total Akt or phospho S6/tubulin for each condition relative to vehicle-treated, siC-transfected cells (mean±S.D.; n=4; *P<0.05 or **P<0.01 from vehicle-treated, siC- transfected cells; ##P<0.01 from THC-treated, siC-transfected cells; ΦΦP<0.01 from vehicle-treated, siMdk and from vehicle-treated, siALK- transfected cells). (c) Effect of THC (1.5 μM, 6 h) on p8 and TRB3 mRNA levels (as determined by quantitative real-time PCR) of U87 cells incubated with U87 C.M. or T98 C.M. (left panel) or with exogenous Mdk (right panel; mean±S.D.; n=3; **P<0.01 from vehicle-treated, U87 C.M.-incubated (left panel) or from vehicle-treated (right panel) cells; ##P<0.01 from THC-treated, U87 C.M.-incubated (left panel) or from THC-treated (right panel) cells; ΦΦP<0.01 from vehicle-treated, T98 C.M.-incubated (left panel) or from vehicle-treated, exogenous Mdk-incubated (right panel) cells). (d) Effect of THC (1.5 μM, 24 h) on AKT and S6 phosphorylation of U87 cells incubated with U87 C.M. or T98 C.M. or with exogenous Mdk. A representative experiment of three is shown. Right panel: densitometric analysis of Akt and S6 phosphorylation. Data correspond to phospho-Akt, Akt, phospho-S6 and tubulin OD values and are expressed as the ratio of phosphoAKT/total Akt or phospho S6/tubulin for each condition relative to vehicle-treated, U87 C.M.-incubated or vehicle-treated cells (mean±S.D.; n=3; **P<0.01 from vehicle-treated, U87 C.M.-incubated cells or from vehicle-treated cells; ##P<0.01 from THC-treated, U87 C.M.-incubated cells or from THC-treated cells; ΦΦP<0.01 from vehicle-treated, T98 C.M.-incubated cells or from vehicle-treated, exogenous Mdk-incubated cells)
In vivo silencing of Mdk sensitizes cannabinoid-resistant tumors to THC anticancer action
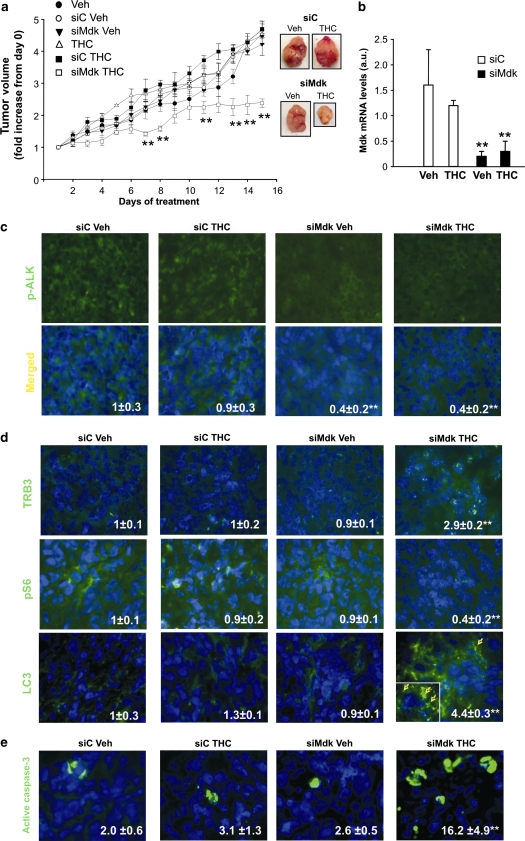
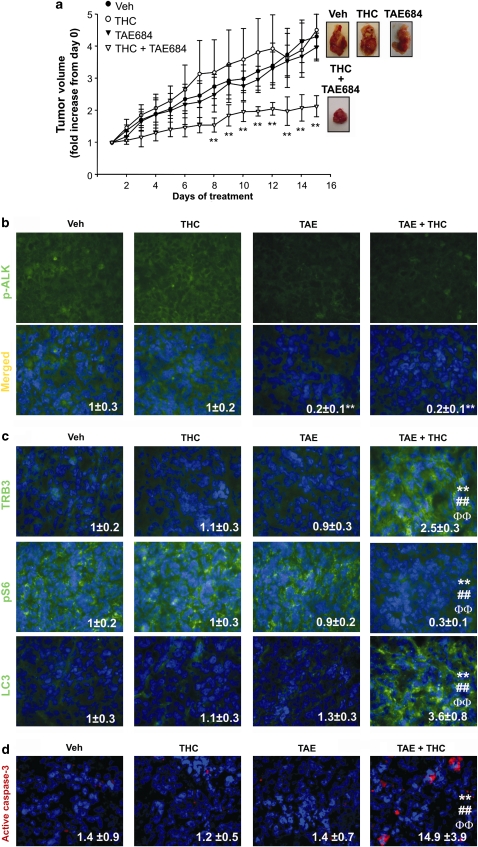
To analyze the in vivo relevance of our findings, we generated tumor xenografts by subcutaneous injection of T98 or U87 cells in nude mice. In agreement with our in vitro data, T98 cell-derived tumors exhibited higher levels of Mdk and were more resistant to THC treatment than the ones generated with U87 cells (Supplementary Figures 13A, B and C). To investigate whether Mdk is responsible for the resistance to THC antitumoral action we analyzed the effect of THC on the growth of T98 tumor xenografts in which Mdk expression had been knocked-down (Figure 7b and Supplementary Figure 13D). As shown in Figure 7a and in Supplementary Figure 13E, in vivo silencing of Mdk rendered established T98 cell-derived tumors sensitive to THC treatment, although it did not affect tumor growth by itself. Subsequent analysis of tumor samples revealed that Mdk silencing correlated with a decrease in ALK phosphorylation (Figure 7c). Moreover, THC treatment increased TRB3 expression (Figure 7d, upper panels), decreased S6 phosphorylation (Figure 7d, middle panels), enhanced autophagy (as determined by LC3 immunostaining (Figure 7d, lower panels)) and induced apoptosis (as determined by active caspase-3 immunostaining (Figure 7e)) only in those tumors in which Mdk expression had been knocked-down. Finally, we investigated the participation of ALK on the resistance of T98 tumors to THC antitumoral action. To this aim we analyzed the effect of the coadministration of THC and NVP-TAE-684 on the growth of T98 tumor xenografts. Similar to what we observed on Mdk silencing, administration of NVP-TAE-684 decreased ALK phosphorylation and sensitized established T98 cell-derived tumors to THC treatment, but did not affect tumor growth by itself (Figures 8a and b). Furthermore, THC treatment increased TRB3 expression (Figure 8c, upper panels), decreased S6 phosphorylation (Figure 8c, middle panels), enhanced autophagy (Figure 8c, lower panels) and induced apoptosis (Figure 8d) only in those tumors in which ALK had been inhibited. Altogether, these results strongly support that the Mdk/ALK axis promotes resistance to THC antitumoral action in vivo by modulating the activation of the autophagy mediated-cell death pathway.
Figure 7.
In vivo silencing of Mdk sensitizes T98 cell-derived tumors to THC antitumoral action. (a) Effect of Mdk silencing and THC (15 mg/kg per day) treatment on the growth of tumor xenografts derived from T98 cells (mean±S.D.; n=5 for each condition, **P<0.01 from the rest of the treatments). siRNAs were administered at days 1 and 7 of vehicle or THC treatment. Representative photographs of siC and siMdk, vehicle and THC-treated tumors are shown. (b) Mdk mRNA levels in T98 cell-derived tumor xenografts as determined by quantitative real-time PCR. Data correspond to Mdk mRNA levels relative to one of the siC vehicle-treated tumors (mean±S.D.; n=5 for each condition; **P<0.01 from vehicle and THC-treated, siC-transfected tumors). (c) Effect of Mdk silencing and THC treatment on phospho-ALK immunostaing of T98 cell-derived tumor xenografts. Values inside the photomicrograhs are expressed as the phospho-ALK-stained area relative to the number of nuclei in each field and correspond to 10 fields of three different tumors for each condition. Data are normalized using siC-transfected vehicle-treated tumors as a reference. Representative photomicrographs are shown (mean±S.D.; **P<0.01 from vehicle-treated, siC-transfected tumors). (d) Effect of Mdk silencing and THC treatment on TRB3 (upper panel), phospho-S6 (middle panel) or LC3 (lower panel) immunostaining of T98 cell-derived tumor xenografts. Values inside the photomicrographs are expressed as the TRB3-, phospho-S6- or LC3-stained area relative to the number of nuclei in each field and correspond to 10 fields of three different tumors for each condition. Data are normalized using siC-transfected vehicle-treated tumors as a reference. Representative photomicrographs are shown (mean±S.D.; **P<0.01 from vehicle-treated, siC-transfected tumors). Inset: high-magnification photomicrograph of the arrow-pointed cell. Arrows within the inset point to LC3 dots. (e) Effect of THC on apoptosis of T98 cell-derived tumor xenografts. Values inside the photomicrographs are expressed as the percentage of active caspase-3 positive cells relative to the total number of nuclei in each field and correspond to 10 representative fields of three different tumors for each condition. Representative photomicrographs are shown (mean±S.D.; **P<0.01 from vehicle-treated, siC-transfected tumors)
Figure 8.
Pharmacological inhibition of ALK sensitizes T98 cell-derived tumors to THC antitumoral action. (a) Effect of NPVTAE684 (TAE; 1 mg/kg per day) and and THC (15 mg/kg per day) treatment on the growth of tumor xenografts derived from T98 cells (mean±S.D.; n=5 for each condition, **P<0.01 from the rest of the treatments). Representative photographs of vehicle-, TAE-, THC- and TAE+THC-treated tumors are shown. (b) Effect of TAE and THC treatment on phospho-ALK immunostaing of T98 cell-derived tumor xenografts. Values inside the photomicrograhs are expressed as the phospho-ALK-stained area relative to the number of nuclei in each field and correspond to 10 fields of three different tumors for each condition. Data are normalized using vehicle-treated tumors as a reference. Representative photomicrographs are shown (mean±S.D.; **P<0.01 from vehicle-treated tumors). (c) Effect of Mdk silencing and THC treatment on TRB3 (upper panel), phospho-S6 (middle panel) or LC3 (lower panel) immunostaining of T98 cell-derived tumor xenografts. Values inside the photomicrographs are expressed as the TRB3-, phospho-S6- or LC3- stained area relative to the number of nuclei in each field and correspond to 10 fields of three different tumors for each condition. Data are normalized using vehicle-treated tumors as a reference. Representative photomicrographs are shown (mean±S.D.; **P<0.01 from vehicle-treated tumors; ##P<0.01 from THC-treated tumors and ΦΦP<0.01 from TAE-treated tumors). (d) Effect of THC on apoptosis of T98 cell-derived tumor xenografts. Values inside the photomicrographs are expressed as the percentage of active caspase-3 positive cells relative to the total number of nuclei in each field and correspond to 10 representative fields of three different tumors for each condition. Representative photomicrographs are shown (mean±S.D.; **P<0.01 from vehicle-treated tumors; ##P<0.01 from THC-treated tumors and ΦΦP<0.01 from TAE-treated tumors)
Discussion
GBM is one of the most aggressive forms of cancer, in part, owing to its poor response to standard radiotherapy and chemotherapy. Consequently, it is currently believed that the design of successful therapeutic strategies to manage these tumors should be based on their detailed molecular characterization, which would allow the development of more specific and personalized treatments.3, 4, 5 In line with this line of reasoning, in this study we aimed to identify the molecular features associated with the resistance of gliomas to cannabinoid antitumoral action. Our data show that the level of expression of CB1 or CB2 receptors does not determine the sensitivity of glioma cells to THC-induced cell death. This is in line with previous observations by our group and others showing that activation of either CB19, 10, 15 or CB214, 22, 23 receptors leads to tumor cell death, thus suggesting that additional factors are responsible for the differences in cannabinoid sensitivity. Detailed analysis of the gene expression profile of a series of cell lines and primary cultures of human glioma cells has led us to find a panel of genes whose upregulation is associated with differences in sensitivity to THC-induced cell death. On the one hand, this information will allow setting the basis for predicting which specific GBM patients could be more susceptible of being treated with cannabinoids. On the other hand, the genes that are selectively upregulated in the cannabinoid-resistant glioma cells become potential candidates to have a direct role in the resistance to THC. Several observations strongly support that at least one of these genes, the growth factor Mdk, is directly responsible for the resistance to cannabinoid antitumoral action: (i) Mdk levels are much higher in cannabinoid-resistant than in cannabinoid-sensitive glioma cells, (ii) incubation with Mdk-enriched conditioned medium or with exogenous Mdk renders cannabinoid-sensitive glioma cells resistant to THC-induced cell death, (iii) knockdown of Mdk sensitizes cannabinoid-resistant glioma cells to THC killing action, and (iv) in vivo silencing of Mdk renders cannabinoid-resistant tumors sensitive to THC antitumoral action.
Mdk is frequently overexpressed in malignant tumors and has been proposed to act as a protective factor against cell death,19, 24 although the precise mechanism by which Mdk mediates this effect has not been clarified. At least four receptors, namely ALK, N-syndecan, RPTPz and LRP1, have been proposed to mediate the biological activities of Mdk.16 Although controversial data have been published with respect to the notion that Mdk acts as a ligand of the ALK receptor,20, 25, 26, 27 data presented here demonstrate that the mechanism by which Mdk promotes resistance to THC-induced cell death relies on ALK. Thus, we observed that ALK silencing abolishes the protective effect exerted by an MDK-enriched conditioned medium or by the addition of exogenous Mdk on glioma cells. Likewise, genetic or pharmacological inhibition of ALK enhances THC-induced cell death in cannabinoid-resistant cells. Moreover, treatment with the ALK-selective inhibitor NVP-TAE684 sensitizes cannabinoid resistant tumors to THC anticancer action.
Of interest, alterations in ALK have been implicated in cancer progression. In particular, ALK fusion proteins associated to chromosome translocations are present in most anaplastic large cell lymphomas, a subset of non-Hodgkin lymphomas, as well as in several solid tumors including non-small cell lung cancer.28 In addition, gain-of-function mutations, mostly harbored in the kinase domain of ALK, have been associated to both familiar and sporadic cases of neuroblastoma.29, 30, 31, 32 By contrast, to the best of our knowledge no studies have been published reporting the presence of these alterations in glioma patients. Moreover, we did not find any mutation in the ALK kinase domain of the glioma cells used in this study (Supplementary Table 7). Taken together, these observations support that increased ligand production rather than mutations or amplifications of the ALK gene are responsible for the stimulation of this receptor in glioma cells. To note, no correlation between PTN (a Mdk-related protein with high homology with Mdk) expression and resistance to THC-induced cell death was found in this study (Supplementary Figure 4B), which further supports that the mechanism of resistance to THC-induced cell death in glioma cells specifically relies on the increased expression of Mdk. Our data are therefore in agreement – at least in part – with the proposal that ALK is a dependence receptor27, 33 that has an antiapoptotic role only when is activated/stimulated by its ligand. Of potential therapeutic relevance, we found that increased expression of Mdk is associated with decreased survival of GBM patients, which further supports that this ligand may have an important role in promoting resistance to cancer therapies.
We also show that Mdk, through stimulation of ALK, promotes resistance to THC-induced cell death by interfering with the induction of autophagy by cannabinoids. The mechanism by which Mdk inhibits THC-induced autophagy relies on the modulation of the Akt/mTORC1 axis. ALK, as other tyrosine kinase receptors, is coupled to PI3K/Akt.34 Accordingly, we found that Mdk stimulates Akt in human glioma cell lines. However, our data also show that, in the absence of THC, silencing of ALK or Mdk decreases Akt phosphorylation in cannabinoid-resistant glioma cells without leading to either inhibition of the mTORC1 complex or autophagy. These observations suggest that additional signals – as those triggered by THC – are necessary for the stimulation of autophagy-mediated cell death in glioma cells. Interestingly, we also found that p8 and TRB3 only become upregulated in response to THC treatment in cannabinoid-resistant cells when Mdk or ALK have been silenced. Taken together, these results support that Mdk and ALK promote resistance to THC-induced autophagy via stimulation of Akt and modulation of the expression of the pro-autophagic genes p8 and TRB3.
In summary, we have found that increased expression of the growth factor Mdk and subsequent activation of ALK are critically involved in the promotion of glioma resistance to cannabinoid antitumoral action. Furthermore, experiments with tumor xenografts demonstrate that inhibition of the Mdk/ALK axis sensitizes cannabinoid-resistant tumors to THC treatment, which correlates with an enhanced activation of autophagy and apoptosis in these tumors. Antitumoral therapies based on the selective targeting of tyrosine kinase receptors are currently being used for the management of different types of cancer.35, 36 Moreover, resistance to different chemoterapeutic agents frequently relies on a gain of function of the signaling routes activated by tyrosine kinase receptors.36, 37, 38 In line with these observations, and with the growing evidence for the participation of ALK in the origin and progression of different types of tumors,28 our findings support that selective targeting of Mdk or ALK could be used to enhance the therapeutic efficacy of antitumoral therapies in gliomas.
Materials and Methods
Reagents
THC was kindly provided by GW Pharmaceuticals (Salisbury, UK). The cannabinoid antagonists SR141716 (SR1) and SR144528 (SR2) were kindly donated by Sanofi-Aventis (Montpellier, France). Recombinant human Midkine was from Sigma (St. Louis, MO, USA). Double-stranded RNA duplexes corresponding to human Mdk, human ALK (smart-pool) and control siRNA were synthesized by Dharmacon (Lafayette, CO, USA). The RNA sequence targeting human Mdk is described in ref. 39 The ALK inhibitor NVP-TAE 684 was synthesized by Sergey A Lakatosh at the Gause Institute of New Antibiotics (Moscow, Russia). BCNU was purchased from Sigma and TMZ was kindly provided by Schering-Plough (Whitehouse Station, NJ, USA).
Cell culture and viability
The human glioma cell lines GOS3, U87 MG (U87), A172, SW1783, U118 MG (U118), U373 MG (U373), T98G (T98), SW1088, CCF-STTG1 (CCF) and LN405 were from ATCC (Middlesex, UK), DSMZ (Braunschweig, Germany) or were kindly provided by Dr. Javier Castresana (Universidad Pública de Navarra, Pamplona, Spain). Primary cultures of brain tumor cells were obtained from biopsies donated by the Tumor Bank Network coordinated by the Spanish Cancer Research Centre and from the Hospital Clínico Universitario (Madrid, Spain). Human pancreatic cancer cell lines MiaPaca2 and Panc1 were kindly provided by Dr. JL Iovanna (Marseille, France).
Human glioma or pancreatic cancer cell lines were cultured in DMEM containing 10% FBS. The preparation of adherent primary culture of brain tumor cells was as follows: tumor samples were homogenized, digested with type Ia collagenase (Sigma) for 90 min at 37°C and incubated on ice for 10 min. The supernatant was collected and, after centrifugation to discard the remaining death-floating cells, resuspended in DMEM containing 15% FBS. Finally, cells were seeded at a density of 400 000 cells per cm2 and kept in culture for 2 weeks in DMEM containing 15% FBS and 1% glutamine. Cells were transferred to a serum-free medium (human glioma cell lines), 0.5% FBS medium (primary cultures of glioma cells) or conditioned medium 18 h before performing the different treatments. When indicated, cells were preincubated with exogenous Mdk for 2 h or with NVP-TAE-684, SR1 or SR2 for 1 h before treatment with THC. Cell viability was determined after 72 h of THC treatment by using the MTT test (Promega, Madison, WI, USA) according to manufacturer's instructions. THC stock solution was prepared in dimethylsulfoxide. Control incubations contained the same amount of dimethylsulfoxide and no significant effect was observed in any of the parameters determined throughout this study at the final concentration used (0.1–0.2%, v/v).
Analysis of gene expression by cDNA microarrays
Total RNA was isolated from human glioma cells lines with RNeasy Mini Kit (Qiagen, Hilden, Germany). Double-stranded cDNA was amplified with Superscript Choice System and T7-(deoxythymidine)24 oligo-primers (Life Technologies, Inc., Gaithersburg, MD, USA) and in vitro transcription was carried out with Megascript T7 (Ambion, Austin, TX, USA). Samples were labeled with Cy3-dUTP (Amersham, Uppsala, Sweden) and Universal Human Reference RNA (Stratagene, La Jolla, CA, USA) with Cy5-dUTP (Amersham) and hybridized on an advanced version of the Spanish National Cancer Research Centre (CNIO) Oncochip (http://bioinfo.cnio.es/data/oncochip) as described in the MIAME. Slides were scanned in a Scanarray 5000 XL scanner (GSI Lumonics Kanata, Ontario, Canada) and the data matrices were processed with GenePix 6.0 software (Axon, Instruments Inc., Union City, CA, USA).
Human Mdk ELISA bioassay
Soluble Mdk levels were determined by using the Human Midkine ELISA (Biovendor, Heidelberg, Germany).
Conditioned medium experiments
U87 or T98 cells were seeded in complete medium at a density of 30 000 cells per cm2 and, after 8 h, transferred to a serum-free medium for 18 h. Media were subsequently recovered, centrifuged and incubated alone or with an anti-Mdk antibody (20 μg/ml; Ref: sc-20715, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or with an isotype antibody (rabbit IgG; 20 μg/ml; Amersham Biosciences Corp., Piscataway, NJ, USA) for 1 h at 37°C. To study the influence of the conditioned media (obtained as described above) on cell viability, U87 cells were seeded in complete medium at a density of 5000 cells per cm2 and, after 8 h, transferred to the corresponding conditioned medium 18 h before performing the different treatments. Finally, cell viability was determined as described above.
Transfections
T98, LN405, HG2, or Panc1 (75% confluent) cells were transfected with control (siC), human Mdk (siMdk) or human ALK (siALK) siRNAs using the X-tremeGENE siRNA Transfection Reagent (Roche, Basel, Switzerland) according to manufacturer's instructions. After 24 h of transfection, cells were trypsinized and seeded in complete medium at a density of 5000 cells per cm2. After 8 h, cells were transferred to a serum-free (cell lines) or 0.5% FBS (primary culture) medium for 18 h and the different treatments were performed. Transfection efficiency was monitored using a control fluorescent siRNA (Qiagen). The efficacy of transfection was higher than 80% in all the experiments.
Reverse transcription- and real-time quantitive-PCR analysis
RNA was isolated using the RNeasy Protect kit (Qiagen) including a DNase digestion step using the RNase-free DNase kit (Qiagen). cDNA was subsequently obtained using Transcriptor Reverse Transcriptase (Roche). PCR reactions were performed using the following parameters: 95°C for 5 min, 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min followed by a final extension step of 72°C for 5 min. The number of cycles (23–25 cycles for human Mdk and 22–25 cycles for GAPDH) was adjusted to allow detection in the linear range. Finally, PCR products were separated on 1.5% agarose gels. Real-time quantitive PCR assays were performed using the FastStart Master Mix with Rox (Roche) and probes were obtained from the Universal Probe Library Set (Roche). Amplifications were run in a 7900 HT-Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Each value was adjusted using 18S RNA levels as reference. Primers sequences and probes used in this study can be found in Supplementary Table 8.
Tumor xenografts
Tumors were induced by subcutaneous injection in nude mice of 10 × 106 U87 or T98 cells in PBS supplemented with 0.1% glucose. When tumors had reached an average size of 250 mm3 (an average of 2 weeks for U87 tumors and 14 weeks for T98 tumors), animals were assigned randomly to various groups and injected peritumorally for 15 days with the corresponding treatment (THC (15 mg/kg per day), NVP-TAE-684 (1 mg/kg per day) or THC (15 mg/kg per day) + NVP-TAE-684 (1 mg/kg per day)) or vehicle in 100 μl of PBS supplemented with 5 mg/ml defatted and dialyzed BSA. Tumors were measured with external caliper, and volume was calculated as (4π/3) × (width/2)2 × (length/2). At the end of the treatment, animals were killed, all tumors were excised and their weights were determined.
In vivo silencing
In vivo Mdk silencing was performed by using a 1 : 1 mixture of AteloGeneTM (Koken Co. Ltd., Tokyo, Japan) and 10 μM HPLC-purified double-stranded siRNA (synthesized by Dharmacon), according to manufacturer's instructions. Briefly, the mix of AteloGene and siRNA (1 nmol of control or Mdk-selective siRNA per injection) was administered in each tumor at days 1 and 7 of vehicle or THC treatment. RNA sequence targeting human Mdk was 5′-GUUUGGAGCCGACUGCAAGdTdT-3′.39
Western blot analysis
Western blot analysis was performed following standard methods. The antibodies used were: anti-phospho-S6 ribosomal protein Ser 235/236 and anti-total-Akt, anti-phospho-Akt Ser473 (1 : 1000; Cell Signaling Technology, Inc., Lake Placid, NY, USA), anti-α-tubulin (1 : 4000; Sigma), anti-LC3 (1 : 1000; polyclonal, MBL, Naka-Ku Nagoya, Japan, ref PM036), anti-CB1 and anti-CB2 (1 : 1000; Affinity Bioreagents, Golden, CO, USA); anti-Mdk (1 : 200; Santa Cruz Biotechnology Inc.), anti-total ALK (1 : 250; rabbit polyclonal ab75953; Abcam, Cambridge, UK) and anti-phospho ALK (Tyrosine 1604) (1 : 500; rabbit monoclonal (EP661-Y) ab51030, Abcam).
Imunofluorescence from cell cultures
Cell cultures grown on 12 mm-coverslips were washed in PBS, fixed with 4% paraformaldehyde (20 min at room temperature), permeabilized and blocked to avoid non-specific binding with 10% goat antiserum and 0.25% Triton X-100 in PBS for 45 min, and subsequently incubated with the following primary antibodies (for 1 h at room temperature): rabbit polyclonal anti-CB1 antibody and anti-CB2 antibody (1 : 500; Affinity Bioreagents), rabbit polyclonal anti-Mdk antibody (1 : 100; Santa Cruz Biotechnology), rabbit monoclonal anti-phospho ALK antibody (1/200; Abcam), rabbit polyclonal anti-cleaved caspase-3 Asp175 antibody (1 : 100; Cell Signaling Technology) and mouse monoclonal anti-LC3 antibody (1 : 100; Nanotools Antikörpertechnik GmbH & Co. Antikörpertechnik GmbH & Co., Teningen, Germany; clone 5F10). Incubation with appropriate Alexa-488 or Alexa-594-conjugated secondary antibodies (Invitrogen, Eugene, OR, USA) was performed in the dark at room temperature for 1 h. Cell nuclei were stained with Hoechst 33342 (Invitrogen). Finally, coverslips were mounted in ProLong Gold antifade reagent (Invitrogen) and visualized in a Leica TCS SP2 confocal microscope (Leica, Wetzlar, Germany).
Immunofluorescence from tumor samples
Samples from tumors xenografts were dissected and frozen. Sections (8 μm) were permeabilized and blocked to avoid nonspecific binding with 10% goat antiserum and 0.25% Triton X-100 in PBS for 45 min, and subsequently incubated with rabbit polyclonal anti-Mdk antibody (1 : 100; Santa Cruz Biotechnology; 4°C, o/n), rabbit monoclonal anti-phospho ALK antibody (1 : 200; Abcam; 4°C, o/n), mouse monoclonal anti-LC3 antibody (1 : 100; Nanotools Antikörpertechnik GmbH & Co; 4°C, o/n), rabbit polyclonal anti-cleaved caspase-3 Asp175 (1 : 100; Cell Signaling Technology; 90 min, room temperature), anti-TRB3 (1 : 800; Calbiochem, San Diego, CA, USA; 90 min, room temperature) or anti-phospho S6 (1 : 200; Cell Signaling; 90 min, room temperature) antibodies, washed, further incubated with the corresponding Alexa-488 or Alexa-594-conjugated secondary antibodies (Invitrogen; 90 min, room temperature). Nuclei were stained with Hoechst 33342 (Invitrogen; 10 min, room temperature) before montage with Mowiol (Merck, Darmstadt, Germany) was performed. Fluorescence images were acquired using an Axiovert 135 microscope (Carl Zeiss, Thornwood, NY, USA).
TUNEL
Tumor samples were fixed, blocked and permeabilized and TUNEL was performed as described.9
Tissue microarray immunohistochemistry
Tissue microarrays were constructed as described previously40 using formalin-fixed, paraffin-embedded archival tissue blocks from five non-tumoral tissues (four normal brains and one tonsil) and 201 glioma samples (115 glioblastomas WHO grade IV, GBs; 14 anaplastic astrocytomas WHO grade III, AAs; 12 diffuse astrocytomas WHO grade II, DAs; 12 pilocytic astrocytomas WHO grade I, PAs; 4 anaplastic oligoastrocytomas WHO grade III, AOAs; 8 oligoastrocytomas WHO grade II, OAs; 19 anaplastic oligodendrogliomas WHO grade III, AOGs; and 17 oligodendrogliomas WHO grade II, OGs). All samples were reviewed by means of full tissue sections stained with hematoxylin and eosin to verify tumor viability, select the representative areas of each tumor and confirm the diagnosis according to WHO guidelines.2
For immunohistochemical staining, 4-μm-thick paraffin-embedded tissue sections were subsequently dewaxed, rehydrated and subjected to antigen retrieval using EnVision FLEX Target Retrieval solution at pH10 and heated at 95°C. The slides were cooled and treated with the EnVision FLEX Peroxidase-Blocking Reagent solution (Dako, Glostrup, Denmark) for 5 min. Sections were then immunostained with Mdk mouse monoclonal antibody A-9 (1 : 50; Santa Cruz Biotechnology Inc.) using the Dako EnVision FLEX/HRP Technique (Dako), counterstained with hematoxylin and mounted. Mdk expression was considered positive in presence of cytoplasmic staining was observed.
Survival analyses
Survival curves were plotted according to the Kaplan–Meier method, and the log-rank test was used to establish the statistical significance of the differences between curves. Differences of overall survival based on MDK positive- or negative-protein expression were analyzed. Survival analysis of the MDK microarray expression TCGA (The Cancer Genome Atlas) data reported by Verhaak et al.4 was performed by categorizing patients into two groups according to the median MDK expression (high expression ≥0.12; low expression <0.12). The association between continuous MDK expression levels and overall survival was determined by a Cox regression univariate model. The threshold for statistical significance in all cases was taken to be P<0.05. Statistical analyses were carried out with SPSS v18.0 (SPSS, Chicago, IL, USA).
Statistics
Unless otherwise specified, results shown represent mean±S.D. Statistical analysis was performed by ANOVA with a post hoc analysis by the Student-Neuman–Keuls test.
Acknowledgments
This work was supported by grants from Spanish Ministry of Science and Innovation (MICINN) (PS09/01401; HF2005/0021, FR2009-0052 and IT2009-0053 to GV; SAF2006/00918 to MG), Santander-Complutense (PR34/07-15856 to GV), Comunidad de Madrid (S-SAL/0261/2006 to MG). ML was sequentially the recipient of a ‘Juan de la Cierva' contract, a postdoctoral contract from Spanish Ministry of Education and Science (MEC) and a postdoctoral contract from Comunidad de Madrid. ST was recipient of a research formation contract from Comunidad de Madrid, MS was recipient of a fellowship from MEC and of a research formation contract from Comunidad de Madrid, AC was recipient of fellowships from Gobierno Vasco, FEBS and EMBO. SH-T has a technician contract from MICINN and Fondo Social Europeo. We thank Horacio Zimman and Carmen Moreno from Hospital Clínico San Carlos as well as Leyre Urigüen from Universidad del Pais Vasco for their kind collaboration in the processing and delivery of glioma samples; Miguel Ángel Piris, Raquel Villuendas, Paloma Cueva and Rosa Pérez for technical advice in the gene expression experiments and other members of our lab for their continuous support.
Glossary
- THC
Δ9-tetrahydrocannabinol
- GBM
glioblastoma multiforme
- ER
endoplasmic reticulum
- TRB3
tribbles homolog 3
- mTORC1
mammalian target of rapamycin complex 1m
- ALK
anaplastic lymphoma kinase receptor
- Mdk
midkine
- PTN
pleiotrophin
- PI3K
phosphatidylinositol 3-kinase
- RPTP
protein-tyrosine phosphatase, receptor-type, zeta-1
- LRP1
low-density lipoprotein receptor-related protein 1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by P Mehlen
Supplementary Material
References
- Nieder C, Adam M, Molls M, Grosu AL. Therapeutic options for recurrent high-grade glioma in adult patients: recent advances. Crit Rev Oncol Hematol. 2006;60:181–193. doi: 10.1016/j.critrevonc.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purow B, Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass. Nat Rev Neurol. 2009;5:419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ruiz J, Romero J, Velasco G, Tolon RM, Ramos JA, Guzman M. Cannabinoid CB2 receptor: a new target for controlling neural cell survival. Trends Pharmacol Sci. 2007;28:39–45. doi: 10.1016/j.tips.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Guzman M. Cannabinoids: potential anticancer agents. Nat Rev Cancer. 2003;3:745–755. doi: 10.1038/nrc1188. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Lorente M, Egia A, Blazquez C, Garcia S, Giroux V, et al. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer Cell. 2006;9:301–312. doi: 10.1016/j.ccr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Galve-Roperh I, Sanchez C, Cortes ML, Gomez del Pulgar T, Izquierdo M, Guzman M. Anti-tumoral action of cannabinoids: involvement of sustained ceramide accumulation and extracellular signal-regulated kinase activation. Nat Med. 2000;6:313–319. doi: 10.1038/73171. [DOI] [PubMed] [Google Scholar]
- Velasco G, Carracedo A, Blazquez C, Lorente M, Aguado T, Haro A, et al. Cannabinoids and gliomas. Mol Neurobiol. 2007;36:60–67. doi: 10.1007/s12035-007-0002-5. [DOI] [PubMed] [Google Scholar]
- Guzman M, Duarte MJ, Blazquez C, Ravina J, Rosa MC, Galve-Roperh I, et al. A pilot clinical study of Delta9-tetrahydrocannabinol in patients with recurrent glioblastoma multiforme. Br J Cancer. 2006;95:197–203. doi: 10.1038/sj.bjc.6603236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfaraz S, Adhami VM, Syed DN, Afaq F, Mukhtar H. Cannabinoids for cancer treatment: progress and promise. Cancer Res. 2008;68:339–342. doi: 10.1158/0008-5472.CAN-07-2785. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Gironella M, Lorente M, Garcia S, Guzman M, Velasco G, et al. Cannabinoids induce apoptosis of pancreatic tumor cells via endoplasmic reticulum stress-related genes. Cancer Res. 2006;66:6748–6755. doi: 10.1158/0008-5472.CAN-06-0169. [DOI] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadomatsu K. The midkine family in cancer, inflammation and neural development. Nagoya J Med Sci. 2005;67:71–82. [PubMed] [Google Scholar]
- Kadomatsu K, Tomomura M, Muramatsu T. cDNA cloning and sequencing of a new gene intensely expressed in early differentiation stages of embryonal carcinoma cells and in mid-gestation period of mouse embryogenesis. Biochem Biophys Res Commun. 1988;151:1312–1318. doi: 10.1016/s0006-291x(88)80505-9. [DOI] [PubMed] [Google Scholar]
- Chen S, Bu G, Takei Y, Sakamoto K, Ikematsu S, Muramatsu T, et al. Midkine and LDL-receptor-related protein 1 contribute to the anchorage-independent cell growth of cancer cells. J Cell Sci. 2007;120:4009–4015. doi: 10.1242/jcs.013946. [DOI] [PubMed] [Google Scholar]
- Mirkin BL, Clark S, Zheng X, Chu F, White BD, Greene M, et al. Identification of midkine as a mediator for intercellular transfer of drug resistance. Oncogene. 2005;24:4965–4974. doi: 10.1038/sj.onc.1208671. [DOI] [PubMed] [Google Scholar]
- Stoica GE, Kuo A, Powers C, Bowden ET, Sale EB, Riegel AT, et al. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J Biol Chem. 2002;277:35990–35998. doi: 10.1074/jbc.M205749200. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Herrera B, Carracedo A, Diez-Zaera M, Gomez del Pulgar T, Guzman M, Velasco G. The CB2 cannabinoid receptor signals apoptosis via ceramide-dependent activation of the mitochondrial intrinsic pathway. Exp Cell Res. 2006;312:2121–2131. doi: 10.1016/j.yexcr.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Caffarel MM, Sarrio D, Palacios J, Guzman M, Sanchez C. Delta9-tetrahydrocannabinol inhibits cell cycle progression in human breast cancer cells through Cdc2 regulation. Cancer Res. 2006;66:6615–6621. doi: 10.1158/0008-5472.CAN-05-4566. [DOI] [PubMed] [Google Scholar]
- Tong Y, Mentlein R, Buhl R, Hugo HH, Krause J, Mehdorn HM, et al. Overexpression of midkine contributes to anti-apoptotic effects in human meningiomas. J Neurochem. 2007;100:1097–1107. doi: 10.1111/j.1471-4159.2006.04276.x. [DOI] [PubMed] [Google Scholar]
- Kuo AH, Stoica GE, Riegel AT, Wellstein A. Recruitment of insulin receptor substrate-1 and activation of NF-kappaB essential for midkine growth signaling through anaplastic lymphoma kinase. Oncogene. 2007;26:859–869. doi: 10.1038/sj.onc.1209840. [DOI] [PubMed] [Google Scholar]
- Moog-Lutz C, Degoutin J, Gouzi JY, Frobert Y, Brunet-de Carvalho N, Bureau J, et al. Activation and inhibition of anaplastic lymphoma kinase receptor tyrosine kinase by monoclonal antibodies and absence of agonist activity of pleiotrophin. J Biol Chem. 2005;280:26039–26048. doi: 10.1074/jbc.M501972200. [DOI] [PubMed] [Google Scholar]
- Mourali J, Benard A, Lourenco FC, Monnet C, Greenland C, Moog-Lutz C, et al. Anaplastic lymphoma kinase is a dependence receptor whose proapoptotic functions are activated by caspase cleavage. Mol Cell Biol. 2006;26:6209–6222. doi: 10.1128/MCB.01515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Frohling S, Luther W, II, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- Allouche M. ALK is a novel dependence receptor: potential implications in development and cancer. Cell Cycle. 2007;6:1533–1538. doi: 10.4161/cc.6.13.4433. [DOI] [PubMed] [Google Scholar]
- Powers C, Aigner A, Stoica GE, McDonnell K, Wellstein A. Pleiotrophin signaling through anaplastic lymphoma kinase is rate-limiting for glioblastoma growth. The J Biol Chem. 2002;277:14153–14158. doi: 10.1074/jbc.M112354200. [DOI] [PubMed] [Google Scholar]
- Ivy SP, Wick JY, Kaufman BM. An overview of small-molecule inhibitors of VEGFR signaling. Nat Rev Clin Oncol. 2009;6:569–579. doi: 10.1038/nrclinonc.2009.130. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- Dempke WC, Heinemann V. Resistance to EGF-R (erbB-1) and VEGF-R modulating agents. Eur J Cancer. 2009;45:1117–1128. doi: 10.1016/j.ejca.2008.11.038. [DOI] [PubMed] [Google Scholar]
- Lorente M, Carracedo A, Torres S, Natali F, Egia A, Hernandez-Tiedra S, et al. Amphiregulin is a factor for resistance of glioma cells to cannabinoid-induced apoptosis. Glia. 2009;57:1374–1385. doi: 10.1002/glia.20856. [DOI] [PubMed] [Google Scholar]
- Takei Y, Kadomatsu K, Goto T, Muramatsu T. Combinational antitumor effect of siRNA against midkine and paclitaxel on growth of human prostate cancer xenografts. Cancer. 2006;107:864–873. doi: 10.1002/cncr.22068. [DOI] [PubMed] [Google Scholar]
- Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, et al. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin Cancer Res. 2003;9:3606–3614. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.