Abstract
Several microRNA (miRNA) loci are found within genomic regions frequently deleted in primary neuroblastoma, including miR-885-5p at 3p25.3. In this study, we demonstrate that miR-885-5p is downregulated on loss of 3p25.3 region in neuroblastoma. Experimentally enforced miR-885-5p expression in neuroblastoma cell lines inhibits proliferation triggering cell cycle arrest, senescence and/or apoptosis. miR-885-5p leads to the accumulation of p53 protein and activates the p53 pathway, resulting in upregulation of p53 targets. Enforced miR-885-5p expression consistently leads to downregulation of cyclin-dependent kinase (CDK2) and mini-chromosome maintenance protein (MCM5). Both genes are targeted by miR-885-5p via predicted binding sites within the 3′-untranslated regions (UTRs) of CDK2 and MCM5. Transcript profiling after miR-885-5p introduction in neuroblastoma cells reveals alterations in expression of multiple genes, including several p53 target genes and a number of factors involved in p53 pathway activity. Taken together, these data provide evidence that miR-885-5p has a tumor suppressive role in neuroblastoma interfering with cell cycle progression and cell survival.
Keywords: 3p25.3, miRNA, p53 stabilization, senescence
MicroRNAs (miRNAs) are non-coding short (18–24 nt) RNAs, controlling gene expression post trancriptionally, via inhibiting translation or triggering degradation of multiple target mRNAs. They are vitally important regulators of proliferation, differentiation and apoptosis. Upregulated and downregulated miRNAs have been found in different malignancies, implicating miRNAs as oncogenes or tumor suppressors.1 The increasing body of evidence for miRNA involvement in tumorigenesis has prompted the search in cancer-associated genomic regions for miRNA loci.2 In a deletion region at 13q14, miR-15 and miR-16, are downregulated in B-cell chronic lymphocytic leukemia. Both miR-15 and miR-16 negatively regulate BCL2 and positively regulate apoptosis, which could explain their frequent inactivation in leukemia.3 Later, miR-34a at 1p36 emerged as a tumor suppressor gene (TSG). Several independent studies demonstrated anti-proliferative and pro-apoptotic potential of miR-34a.4, 5 The potential for involvement of as yet unreported miRNAs in cancer pathogenesis is great.
Neural crest-derived neuroblastoma, one of the most common solid tumors in children, is frequently characterized as an ‘enigmatic neoplasm'. Neuroblastoma has the highest rate of spontaneous regression of all human malignancies, however, ∼40% of neuroblastomas rapidly progress despite multimodal treatment regimes.6, 7 TP53 mutations are rare in neuroblastoma, which very often retain functional p53, suggesting that inhibitors of the p53 pathway or loss of p53 pathway positive regulators may be involved in neutralizing p53 activity.8, 9 Neuroblastomas with poor outcome are subdivided into at least two biologically distinct groups, either with or without amplification of the MYCN oncogene. The second group frequently harbors segmental 3p deletions,10 implying that neuroblastoma-suppressive transcripts are encoded from this region. Several efforts have been made to map and refine the critical region of 3p loss in neuroblastoma, currently assigned to 3p25-26.2, including the VHL gene. Low VHL mRNA expression correlated with poor prognosis, however, functional analyses could not reveal differences in pVHL protein expression or pVHL pathway impairment.11 To date, the identity of neuroblastoma-relevant tumor suppressor genes (TSGs) at 3p25-26.2 is unknown. We detected several known and novel miRNA loci within regions of neuroblastoma-relevant chromosomal aberrations in previous work.12 The only miRNA mapping to 3p25.3 is miR-885-5p. In this study, we show that miR-885-5p interferes with cell proliferation and survival, and promotes cellular senescence and apoptosis. We demonstrate that miR-885-5p downregulates cyclin-dependent kinase (CDK2) and mini-chromosome maintenance protein (MCM5), activates p53, and provide evidence that CDK2 and MCM5 are direct miR-885-5p targets. Our results suggest a tumor suppressive role for miR-885-5p in neuroblastoma.
Results
Low miR-885-5p expression in primary neuroblastomas with segmental 3p loss
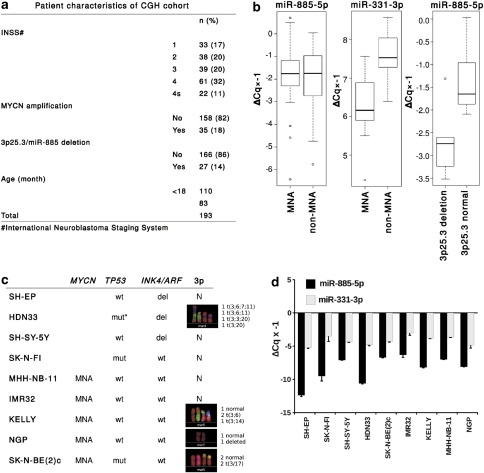
In our previous work characterizing the neuroblastoma miRNA transcriptome, we cloned miR-885-5p (MYCNSC_5_281) from a favorable tumor. miR-885-5p is a non-conserved miRNA, with no mouse or rat homologs (Supplementary Figure S1a). To evaluate rearrangements of the region encompassing miR-885-5p in primary neuroblastomas, we used array-based comparative genomic hybridization (aCGH) to analyze 193 neuroblastomas with different clinical and biological characteristics. The 3p25.3 region encompassing miR-885-5p was heterozygously deleted in the context of a segmental chromosomal 3p loss in 14% (n=27) of tumors (Figure 1a). Most 3p-deleted tumors were stage 4 (n=17). These data are consistent with previous reports on 3p25.3 status in neuroblastoma.10 We profiled miR-885-5p expression by RT-qPCR in 60 primary neuroblastomas. For comparison, we profiled miR-331-3p, a miRNA possibly repressed by MYCN with tumor suppressive functions according to our previous data.12 Assuming equal Cq-values for similar expression levels, miR-885-5p expression was lower than miR-331-3p in neuroblastomas (Figure 1b). No significant difference in miR-885-5p expression was detected between neuroblastomas with and without MYCN amplification, whereas miR-331-3p expression was lower in MYCN-amplified neuroblastomas (P<0.001, Figure 1b). Intriguingly, miR-885-5p expression was lower in aggressive tumors with segmental 3p deletions, as compared with those with intact 3p and favorable prognosis (P=0.02, Figure 1b). Expression of miR-885-5p and miR-331-3p was measured in nine neuroblastoma cell lines (both MYCN-amplified and non-amplified) using RT-qPCR and northern blotting (Figure 1c). Expression of precursor and mature miR-885-5p was low in the cell lines and did not correlate with 3p25 status, whereas expression of precursor and mature miR-331-3p was higher and did not vary significantly (Figure 1d; Supplementary Figure S1b). Although miR-331-3p data did not indicate a clear role in neuroblastoma pathogenesis as a tumor suppressor, we further included miR-331-3p as a positive control to monitor the specificity of miR-885-5p-mediated effects. The lack of correlation between miR-885-5p expression and 3p25 status in the analyzed neuroblastoma cell lines suggests that other miR-885-5p-inactivating lesions exist in these cells. Our data support a tumor suppressive role of miR-885-5p at least in a subgroup of neuroblastomas with intact 3p25.3.
Figure 1.
The miR-885-5p gene at 3p25.3 is expressed at low level in primary neuroblastomas with segmental 3p loss. (a) Patient characteristics are shown for the 193 primary neuroblastomas analyzed on either 44 or 105 K oligo microarrays. The percentage of tumors with heterozygous 3p25.3/miR-885-5p deletion is indicated in parenthesis. (b) Expression of miR transcripts was verified using RT-qPCR in 60 primary neuroblastomas, and is shown as box plots. MNA–MYCN-amplified neuroblastomas. ΔCq values were multiplied by −1 to represent relative expression of miR-885-5p and miR-331-3p. miR-885-5p: median ΔCq × −1=1.83, range −0.04–6.43; miR-331-3p: median ΔCq × −1=−7.31, range −8.57 to −4.35). (c) Neuroblastoma cell lines used in the screen. The MYCN amplification, TP53/INK4a/ARF and 3p status are listed as reported previously13, 14 or assessed in this study; An asterisk (*) denotes – age of the patient – 10 years, stage – 4. (d) Expression of miR-885-5p and miR-331-3p was assessed in nine established neuroblastoma cell lines using RT-qPCR (bar graphs). miR-885-5p ΔCq × −1 median −8.1, range −6.3 to −12.4; miR-331-3p ΔCq × −1 median −4.3, range −3 to −5.2
miR-885-5p inhibits neuroblastoma proliferation and survival
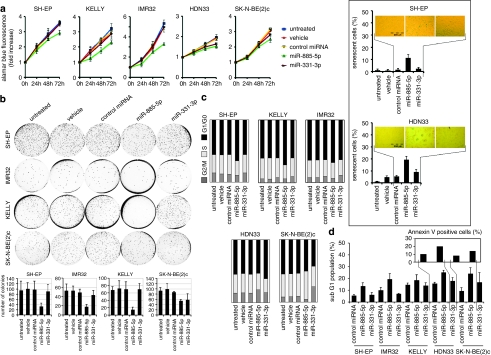
To analyze miR-885-5p function, we introduced miR-885-5p mimics into KELLY, IMR32, SK-N-BE(2)c, SH-EP and HDN33 cell lines. SH-EP, KELLY and IMR32 cell lines have wild-type TP53, while p53 in SK-N-BE(2)c is mutated in the DNA-binding domain and inactive as a transcription factor.13 Reported multinuclear cells with centrosome amplification14, 15 and a heterozygous mutation leading to a codon 15 Ser to Cys exchange (Supplementary Figure S2) indicate that the p53 pathway may not be intact in HDN33 cells. Cell proliferation was assessed using the Alamar Blue assay in cells transiently transfected with either the miR-885-5p or miR-331-3p mimics or a non-targeting control miRNA. Enforced miR-885-5p expression reduced proliferation of all cell lines, whereas miR-331-3p transfection only inhibited proliferation of HDN33 and SK-N-BE(2)c (Figure 2a). SH-EP, KELLY, IMR32 and SK-N-BE(2)c cells were also less capable of anchorage-independent growth in soft agar after enforced miR-885-5p expression (Figure 2b). Importantly, miR-885-5p introduction suppressed anchorage-independent growth more pronouncedly in cell lines with wild-type than mutant TP53. Enforced miR-331-3p expression only reduced anchorage-independent growth of SK-N-BE(2)c cells. We tested whether proliferation inhibition by miR-885-5p was associated with senescence. Senescence-associated acidic β-galactosidase (SA-β-Gal) activity was detected in miR-885-5p-transfected, MYCN-non-amplified HDN33 and SH-EP cells (Figure 2a, insert), but not in MYCN-amplified KELLY, IMR32 and SK-N-BE(2)c cells (data not shown).
Figure 2.
miR-885-5p inhibits neuroblastoma proliferation and survival. (a) TP53 wild-type IMR32, SH-EP, KELLY and TP53 mutant HDN33, SK-N-BE(2)c cells were transfected with miR-885-5p, miR-331-3p mimics, control miRNA (each 30 n), Lipofectamine 2000 (vehicle) or left untreated. Cell viability was measured by Alamar Blue assay. The data are represented as the means±S.D. (a; insert) SA-ß-Gal assays were performed 4 days after transfection; the positively stained cells were counted (mean percentages±S.D. of three experiments are presented). (b) IMR32, SH-EP, KELLY and SK-N-BE(2)c were transfected with miRNA mimics and assayed for anchorage-independent growth in soft agar after 2 weeks in culture. Colonies were fixed and stained with crystal violet and counted using ImageJ software. The graphs (right) show mean percentages of colonies number±S.D. of three experiments. (c) Cell cycle profiles of controls and cells transfected with miRNA mimics were determined using FACS analysis. (d) The sub-G1 fraction of neuroblastoma cell lines transfected with miRNA mimics or control miRNA transfectants were detected at 4-days post transfection using FACS analysis by detection of cells with a hypodiploid DNA content (bar=mean percentage of sub-G1±S.D.). Phosphatidylserine externalization was analyzed by annexin V/PI staining, the results show the percentage of annexin V-positive/PI negative and annexin V/PI double positive cells
Cell cycle profiles were analyzed after enforced miR-885-5p or miR-331-3p expression using flow cytometry (FACS). Cell lines with wild-type TP53 accumulated in G1/G0 after miR-885-5p transfection (Figure 2c), whereas enforced miR-885-5p expression in SK-N-BE(2)c and HDN33 resulted in accumulation in the S and G2/M phases, respectively. Enforced miR-331-3p expression produced no pronounced cell cycle changes (>±3%) in cells with wild-type TP53, but resulted in G1/G0 and G2/M accumulation in HDN33 and SK-N-BE(2)c, respectively. We analyzed the sub-G1 fraction to ascertain whether cell cycle changes were accompanied by cell death. Enforced miR-885-5p expression slightly increased the sub-G1 fraction in SH-EP and IMR32 and moderately increased the sub-G1 fraction in SK-N-BE(2)c and HDN33 cells, whereas miR-331-3p transfection only increased the sub-G1 fraction in SK-N-BE(2)c cells (Figure 2d). Enforced miR-885-5p expression increased the fraction of annexin V-positive cells in HDN33 and SK-N-BE(2)c cultures, but not TP53 wild-type cell lines (Figure 2d, insert; data not shown). Together, these data suggest that miR-885-5p-mediated inhibition of proliferation occurs preferentially via G0/G1 arrest in TP53 wild-type neuroblastoma cells, and apoptotic cell death in neuroblastoma cells with disabled p53 pathways.
miR-885-5p induces p53 and downregulates CDK2 and MCM5
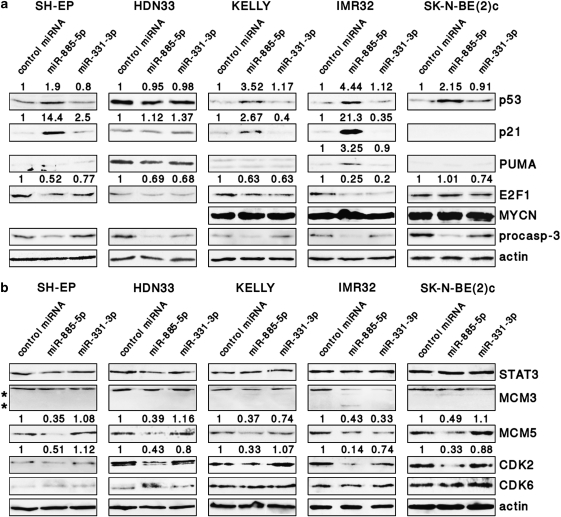
Protein levels of p53 and downstream targets were analyzed after enforced miR-885-5p expression to assess miR-885-5p influence on p53 regulation and function. The induction of p53 was marked in KELLY and IMR32 cells, moderate in SH-EP cells and also occurred in TP53 mutant, SK-N-BE(2)c cells (Figure 3a). Enforced miR-885-5p expression elevated p21waf1 levels in KELLY, IMR32 and SH-EP cells, and PUMA in only IMR32 cells. Enforced miR-885-5p expression did not alter p53 and p21waf1 protein expression in HDN33 cells or p21waf1 and PUMA protein expression in TP53 mutant SK-N-BE(2)c cells, and miR-331-3p expression had no effect on p53, p21waf1 and PUMA protein levels in any of the cell lines analyzed. Expression of miR-885-5p downregulated E2F1 in SH-EP, IMR32, KELLY and HDN33 cells, but had no effect on MYCN and p27 protein levels (Figure 3a, and data not shown). Caspase-3 activity was elevated in SK-N-BE(2)c and IMR32 after miR-885-5p introduction, although the increases were less than twofold (Supplementary Figure S3). Western blot analysis revealed reduction of procaspase-3 levels in all cell lines (Figure 3a) with no significant appearance of mature caspase-3 (data not shown). Taken together, miR-885-5p expression induces p53 and p21waf1 in TP53 wild-type neuroblastoma cells, in line with the G0/G1 arrest observed after enforced expression.
Figure 3.
miR-885-5p induces p53 and downregulates CDK2 and MCM5. (a and b) Protein expression of several cell cycle and apoptosis regulators, including potential miR-885-5p targets, is shown in western blots of neuroblastoma cells lines at 4 days after transfection of miR-885-5p, miR-331-3p or controls. MCM3 cleavage products are indicated (*). Densitometry analysis was performed using ImageJ software; the relative densitometry values are shown on top
Querying the miRNA databases at http://www.microrna.org16 and TargetScan17 (http://www.targetscan.org), and searching for candidate 3′-untranslated regions (UTRs) using the miRanda algorithm18 in the UTResource 3′-UTR database (http://www.ba.itb.cnr.it/UTR/) retrieved STAT3, CUL3, CDK2, MCM3, MCM5 and CDK6 as candidate miR-885-5p targets, and p21waf1 and E2F1 as candidate miR-331-3p targets. Cleavage of STAT3 and MCM3 by apoptosis-executing proteases has been reported, and could have interfered with our expression analysis of these proteins.19, 20 We examined candidate target protein expression after miR-885-5p and miR-331-3p transfection. CDK2 and MCM5 decreased after miR-885-5p transfection in all cell lines, supporting both as direct miR-885-5p targets (Figure 3b). Enforced miR-331-3p expression reduced E2F1 levels in KELLY, IMR32 and SK-N-BE(2)c and reduced CDK2 expression in IMR32, although to a lesser extent than miR-885-5p. MCM3 cleavage was observed after miR-885-5p transfection of IMR32 and SK-N-BE(2)c cells, which prohibited estimation of direct miR-885-5p targeting of MCM3. To test whether observed effects depended on the manner of miRNA overexpression, SH-EP and KELLY cell lines stably expressing either miR-885-5p or miR-331-3p were generated (Supplementary Figure S1c). Phenotypes of miR-885-5p-SH-EP and miR-885-5p-KELLY lines were highly similar to transiently transfected SH-EP and KELLY cells. Most importantly, CDK2 and MCM5 were downregulated and p53 and p21waf1 were upregulated in miR-885-5p-SH-EP and miR-885-5p-KELLY cells, similarly to transiently transfected SH-EP and KELLY cells (Supplementary Figure S4). Accordingly, our data from in silico prediction and western blotting support that CDK2 and MCM5 are targeted by miR-885-5p.
miR-885-5p directly targets the predicted binding sites in CDK2 and MCM5 3′-UTRs
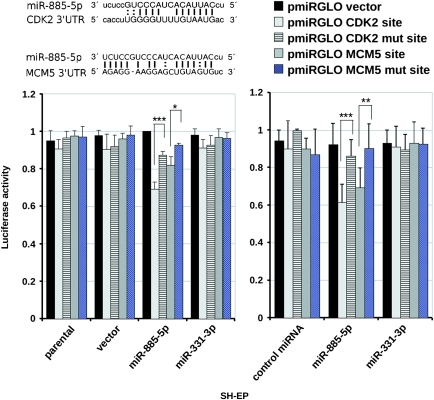
To test whether miR-885-5p binding sites in the CDK2 and MCM5 3′-UTRs are directly regulated by miR-885-5p, we performed luciferase reporter assays. Relative firefly luciferase activities of reporter plasmids containing MCM5 or CDK2 3′-UTR binding sites were inhibited by 20–40% in miR-885-5p-SH-EP cells and pre-miR-885-5p-transfected SH-EP cells but not parental, vector- or miR-331-3p-SH-EP cells and control miRNA or miR-331-3p-transfected cells. Importantly, mutation of miR-885-5p binding sites in the CDK2 and MCM5 reporter constructs restored luciferase activity in miR-885-5p-SH-EP and premiR-885-5p-transfected cells (Figure 4). These data show that miR-885-5p directly targets predicted miR-885-5p binding sites in the CDK2 and MCM5 3′-UTRs.
Figure 4.
miR-885-5p directly targets CDK2 and MCM5. Predicted miR-885-5p target sites in the CDK2 and MCM5 3′-UTRs. Lines indicate perfect matches; colons, G:U pairs. Luciferase reporter assays of stably transfected cells and miRNA-mimic-transfected cells (indicated on x-axis) transiently transfected with either empty vector or vector containing wild-type or mutant miR-885-5p binding sites (indicated as bar color). Firefly luciferase values were normalized to renilla activity. The highest normalized luciferase activity of the stable transfected cells or cells transfected with miRNA mimics was set to 1, and values are reported as fold increase (mean relative luciferase activity±S.D.). *P=<0.1; **P=<0.05; ***P=<0.01 versus vector with mutant miR-885-5p binding sites
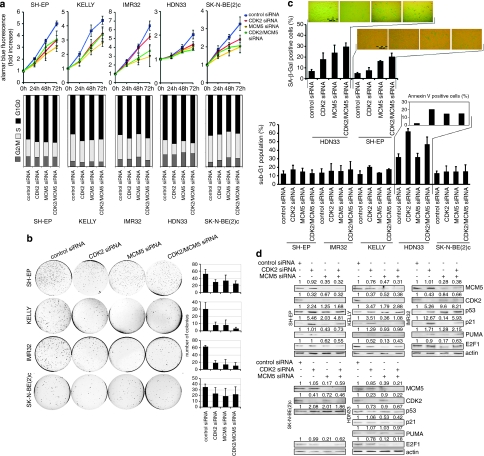
CDK2 and MCM5 knockdown mimic miR-885-5p-induced phenotype
Previous studies have linked CDK2 and MCM5 with the p53 pathway. MCM5 was identified as a negative regulator of p53-induced growth arrest,21 and CDK2 inhibition led to p53 accumulation in MYCN-amplified neuroblastoma cells.22 These suggest a link between phenotypic changes induced by miR-885-5p in neuroblastoma cells and CDK2 and MCM5 suppression. To test this possibility, we introduced short interfering RNA (siRNAs) targeting CDK2 and/or MCM5 into our neuroblastoma cell line panel, and conducted phenotype analysis using the same panel of assays used to characterize the miR-885-5p-induced phenotype. CDK2 or MCM5 knockdown alone or in combination inhibited cell proliferation and anchorage-independent growth in all cell lines (Figure 5a and b). Cell cycle analysis at 3 days after transfection of CDK2 or CDK2/MCM5 siRNAs revealed the accumulation of cells in G1/G0 phase in most cell lines (Figure 5a). MCM5 knockdown produced an inconsistent cell cycle phenotype, with G1/G0 arrest in SH-EP and KELLY and G2/M arrest in the other cell lines. SK-N-BE(2)c cells are consistently accumulated in the G2/M phase after CDK2 and/or MCM5 knockdown. The most SA-ß-Gal-positive cells were produced by combined CDK2/MCM5 knockdown in SH-EP and HDN33 cells (Figure 5c), similar to miR-885-5p overexpression in these lines. CDK2 and/or MCM5 knockdown in HDN33 increased the sub-G1 and annexin V-positive cell fractions (Figure 5c), similar to miR-885-5p overexpression in these cells. CDK2 or MCM5 inhibition significantly increased apoptosis in SH-EP, KELLY, IMR32 and SK-N-BE(2)c cells. CDK2 knockdown upregulated p53 and p21waf1 in SH-EP, KELLY and IMR32 cells, and PUMA in IMR32 cells (Figure 5d). CDK2 and/or MCM5 knockdown in HDN33 cells did not affect p53, p21waf1 or PUMA levels. Together, these data suggest that CDK2 and MCM5 inhibition is involved in miR-885-5p-mediated growth arrest and senescence in TP53 wild-type neuroblastoma cells. CDK2 and MCM5 inhibition can also has a role in miR-885-5p-triggered apoptosis and senescence in TP53 mutant HDN33 cells. Although both CDK2 and/or MCM5 knockdown and miR-885-5p transfection in SK-N-BE(2)c upregulated p53, they did not undergo apoptosis, suggesting that targets other than CDK2 and MCM5 modify the SK-N-BE(2)c response to miR-885-5p.
Figure 5.
CDK2 and MCM5 knockdown mimic miR-885-5p-induced phenotype. (a) TP53 wild-type IMR32, SH-EP, KELLY and TP53 mutant HDN33, SK-N-BE(2)c cells were transfected with anti CDK2 (5 n), anti MCM5 (5 n), anti CDK2 (2.5 n)+ anti MCM5 (2.5 n) siRNAs or control siRNA (5 n) with Lipofectamine 2000. Cell viability was measured at 24, 48 and 72 h after transfection by Alamar Blue assay. The data are represented as the mean±S.D. (b) siRNA transfected cells were assayed for anchorage-independent growth in soft agar after 2 weeks in culture. Colonies were counted using ImageJ software (mean percentages of colonies number±S.D. of three experiments are presented in the graphs (right)). (c) SA-ß-Gal assays were performed at 4 days after transfection; the positively stained cells were counted (mean percentages±S.D. of three experiments are presented). Cell cycle profiles of controls and cells transfected with siRNAs were determined using FACS analysis. The sub-G1 fraction of neuroblastoma cell lines transfected with miRNA mimics or control miRNA transfectants were detected at 4-days post transfection by FACS analysis by detection of cells with a hypodiploid DNA content (bar=mean percentage of sub-G1±S.D.). Phosphatidylserine externalization was analyzed by annexin V/PI staining, the percentage of annexin V-positive and annexin V/PI double-positive cells are indicated. (d) protein expression of MCM5, CDK2, p21waf1, p53, PUMA and E2F1 from siRNA-transfected cells is shown in western blots. The relative densitometry values are shown
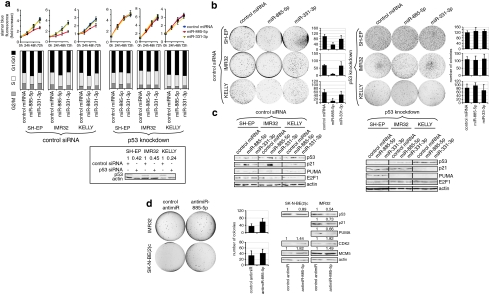
miR-885-5p-induced growth arrest is p53 dependent in TP53 wild-type neuroblastoma
To determine whether miR-885-5p-induced growth arrest is indeed p53-dependent in TP53 wild-type neuroblastoma cells, we sequentially transfected TP53 wild-type SH-EP, KELLY and IMR32 cells with siRNA targeting p53 and miRNA mimics then assessed cell proliferation. Transfection of miRNA mimic into untransfected or control siRNA-transfected cells induced the same phenotype. Expressing miR-885-5p failed to inhibit proliferation, anchorage-independent growth or induce G1/G0 arrest in p53 knockdown SH-EP, KELLY and IMR32 cells (Figure 6a and b). Consistently, p21waf1and PUMA levels were unaffected by miR-885-5p expression after p53 knockdown in these cells (Figure 6c).
Figure 6.
miR-885-5p-induced growth arrest is p53 dependent in TP53 wild-type neuroblastoma cells. (a) IMR32, SH-EP and KELLY cells were transfected with siRNA against p53 or control siRNA using Lipofectamine 2000 and transfected with miRNA mimics after 24 h. p53 knockdown efficiency was evaluated at day 4 after transfection (insert below). Cell viability was measured by Alamar Blue assay for 24, 48 and 72 h after miRNA mimic transfection (upper panel). The data are represented as the means±S.D. of three experiments. The cell cycle profiles were performed by FACS analysis (middle panel). (a, insert) p53 knockdown efficiency is shown in western blots in IMR32, KELLY and SH-EP. (b) p53 knockdown and control siRNA transfected IMR32, KELLY and SH-EP cells were transfected with miRNA mimics and assayed for anchorage-independent growth in soft agar after 2 weeks in culture. Colonies were fixed and stained with crystal violet and counted using ImageJ software. The graphs show mean percentages of colonies number±S.D. of three experiments. (c) Protein expression of p21waf1, PUMA and p53 is shown in western blots of p53 knockdown and control siRNA transfected IMR32, KELLY and SH-EP cells at 3 days after transfection of miRNA mimics. (d) IMR32 and SK-N-BE(2)c were transfected with antimiR-885-5p or control antimiR (30 n) and assayed for anchorage-independent growth for 2 weeks. Colonies were counted using ImageJ software (mean percentages of colonies number±S.D. of three experiments are presented). Protein expression of p53, p21waf1, PUMA, CDK2 and MCM5 in antimiR-transfected IMR32 and SK-N-BE(2)c cells is shown in western blots. The relative densitometry values are shown on top of the western blot panels
We thought to suppress endogenous miR-885-5p in neuroblastoma cells to further validate miR-885-p regulation of p53, CDK2 and MCM5. Only IMR32 and SK-N-BE(2)c cells expressed significant levels of miR-885-5p in RT-qPCR and northern blot analysis (Figure 1d, Supplementary Figure S1). No growth upregulation was observed upon antimiR-885-5p transfection in Alamar Blue assays (data not shown). However, antimiR-885-5p transfection of IMR32 and SK-N-BE(2)c cells produced a trend towards increased anchorage-independent growth. Importantly, antimir-885-5p transfection downregulated p53, p21waf1 and PUMA in IMR32 cells, and p53 in SK-N-BE(2)c cells (Figure 6d). MCM5 was moderately induced in SK-N-BE(2)c cells, and CDK2 was induced in SK-N-BE(2)c and IMR32 cells, after antimiR-885-5p transfection (Figure 6d). Our data show that suppressing miR-885-5p affects p53, p21waf1, CDK2 and MCM5 protein levels, consistent with miRNA mimic transfection results, and strengthening the link between miR-885-5p and p53/CDK2/MCM5.
miR-885-5p activates the p53 transcriptional program
In previous sections we provide evidence that miR-885-5p-mediated growth inhibition requires functional p53 in neuroblastoma cells. Gene expression profiles of neuroblastoma cells with wild-type TP53 were generated using oligo microarrays at 3 days after miR-885-5p mimic transfection, and compared with cells transfected with control miRNA or miR-331-3p mimic to further characterize downstream effects of miR-885-5p expression. Genes with expression changes of at least twofold in both SH-EP and KELLY cell lines were considered to be regulated. There was little overlap between genes regulated in cells expressing either miR-885-5p or miR-331-3p (8 upregulated and 28 downregulated genes, Supplementary Tables S1 and S2). Several p53 target genes (IGFBP3, ZMAT3, TNFRSF10B, BTG3, PPAP2B, PPM1D, RRM2B, PTPRE and DLC), were upregulated in miR-885-5p transfectants23 (Table 1). Only one p53 target, GDF15, was upregulated in miR-331-3p transfectants. IGFBP3 and ZMAT3 were among the top-five genes upregulated in KELLY cells after miR-885-5p expression. These data implicate p53-directed transcriptional changes downstream of miR-885-5p in neuroblastoma cells. We used the expression profiles of miR-885-5p transfectants to assess expression of genes controlling p53 transcription either positively or negatively. Several genes known to positively regulate p53, namely DYRK2, HIPK2 and GAS1, were upregulated after miR-885-5p expression. Enforced miR-331-3p expression altered expression of none of these, but did suppress the positive p53 regulator, PML (Supplementary Table S2). To validate that miR-885-5p-upregulated p53 target genes require functional p53, we profiled KELLY cells with p53 knockdown expressing either miR-885-5p or miR-331-3p. Of the nine p53 target genes identified in the first profiling series (Table 1), seven validated miR-885-5p-induced genes (IGFBP3, BTG3, ZMAT3, PPAP2B, PPM1D, RRMB2 and PTPRE) were not, or only nominally, induced after p53 knockdown (Supplementary Table S3), indicating that miR-885-5p induces these genes via increasing p53 transcriptional activity in these cells.
Table 1. miR-885-5p activates p53 transcriptional program.
| HGNC symbols | Chromosome, band | Description |
|---|---|---|
| P53 targets upregulated in miR-885-5p transfectants | ||
| IGFBP3 | 7p12.3 | Insulin-like growth factor-binding protein 3 |
| BTG3 | 21q21.1 | Protein BTG3 (abundant in neuroepithelium area protein) |
| ZMAT3 | 3q26.32 | Zinc-finger matrin-type protein 3 (p53-activated gene 608 protein) |
| PPAP2B | 1p32.2 | Lipid phosphate phosphohydrolase 3 |
| PPM1D | 17q23.2 | Protein phosphatase 1D (p53-induced protein phosphatase 1) |
| TNFRSF10B | 8p21.3 | Tumor necrosis factor-receptor superfamily member 10B |
| RRM2B | 8q22.3 | Ribonucleoside-diphosphate reductase subunit M2 B |
| DLC1 | 8p22 | Rho GTPase-activating protein 7 (Deleted in liver cancer 1 protein) |
| PTPRE | 10q26.2 | Receptor-type tyrosine-protein phosphatase epsilon |
| P53 targets upregulated in miR-331-3p transfectants | ||
| GDF15 | 19p13.11 | Growth differentiation factor 15 |
We further tested whether genes downregulated after enforced miR-885-5p expression were enriched for potential miR-885-5p targets compared with upregulated genes, based on the in silico detection of at least one miR-885-5p potential binding site in the 3′-UTR. We identified 74 downregulated genes (10.7%) compared with only 3 upregulated genes (0.9%, Supplementary Table S1). Downregulated genes identified as a miR-885-5p targets here included CDK2 (Figure 3b; Supplementary Figure S4), caspase-3 (CASP3), ß-catenin (CTNNB1), citron rho-interacting kinase (CIT), farnesyl transferase (FNTA) and microphthalmia-associated transcription factor (MITF). Genes downregulated after miR-331-3p expression similarly included putative miR-331-3p targets harboring miR-331-3p binding sites (90 genes, 15.6%, Supplementary Table S3). These included PML, the serine/threonine-protein kinase, PIM1, neurobeachin (NBEA), the nerve growth factor receptor, NGFR and ERBB2, which was also recently reported to be a miR-331-3p target.24 A total of 19 (8.2%) genes upregulated after miR-331-3p expression also contain miR-331-3p target sites (Supplementary Table S2). This distribution of prospective target genes within genes regulated after miR-331-3p expression makes less sense than those regulated after miR-885-5p expression. Together with the lack of apparent phenotype in miR-331-3p-transfected neuroblastoma cells, these results support a less specific role for miR-331-3p in directing changes in the neuroblastoma cell phenotype. Our data indicate that miR-885-5p expression activates the p53 pathway in neuroblastoma cells, including p53 induction and p53 downstream pathway activity, but that multiple pathways can be used by miR-885-5p during regulation of the neuroblastoma cell phenotype, especially in a TP53 mutant background.
Discussion
We provide evidence here that miR-885-5p is a candidate neuroblastoma tumor suppressor and involved in directing the neuroblastoma cell phenotype. Several commonly lost genomic regions have been characterized in neuroblastoma, without identification of the genes responsible for tumor suppression. Region 3p25.3 is deleted in ∼14% of neuroblastomas, and houses miR-885-5p. We show that miR-885-5p inhibits proliferation and survival, and positively regulates the p53 pathway. We present data for CDK2 and MCM5 being direct targets of miR-885-5p.
Both miR-885-5p and miR-331-3p were initially chosen as candidate tumor suppressor miRNAs in neuroblastoma. Our findings show that miR-885-5p is downregulated in neuroblastomas with heterozygous deletion of 3p25.3, and that expression in neuroblastoma cell lines is low or absent independent of 3p loss. Expression of miR-331-3p correlates inversely with amplified MYCN in neuroblastomas, but no such trend was detected in cell lines. Experimentally enforced expression of miR-885-5p, not miR-331-3p, in neuroblastoma cell lines resulted in growth inhibition, senescence and apoptosis. Clearly, miR-885-5p fulfills the criteria of a tumor suppressor gene in the context of neuroblastoma.
A master gene orchestrating cell cycle arrest, programmed cell death and senescence is TP53. We provide evidence for p53 involvement in miR-885-5p-driven cell cycle arrest, senescence and apoptosis. Wild-type TP53 neuroblastoma cells displayed G1-arrest coupled with p53 and p21waf1 induction upon miR-885-5p introduction. Among six candidates, CDK2 and MCM5 met the miR-885-5p target criteria to create this phenotype: (1) they were downregulated upon miR-885-5p introduction in all cell lines examined and (2) predicted miR-885-5p binding sites in their 3′-UTRs reduced the expression of a reporter construct in the presence of miR-885-5p.
CDK2 and MCM5 have been linked to negative regulation of p53 pathway activity.21, 22 In this study, we report prominent phenotypic similarity between enforced miR-885-5p expression and CDK2/MCM5 knockdown in TP53 wild-type neuroblastoma cells. Consistently, endogenous miR-885-5p knockdown elevated CDK2 and MCM5 levels and reduced p53 and its downstream targets. Additionally, p53 knockdown reverted changes in phenotype transcriptome and cellular proteins induced by miR-885-5p. Together, this indicates that CDK2 and MCM5 inhibition executes miR-885-5p-mediated growth inhibition in TP53 wild-type neuroblastoma cells at least in part via p53 induction. Importantly, miR-885-5p did not affect deregulated MYCN expression in MYCN-amplified neuroblastoma cells. Deregulated MYC proteins modulate p53 function by transcriptionally inhibiting p21waf1, impairing G1/G0 arrest.25 Our data indicate that miR-885-5p restores p21waf1 function leading to G1/G0 arrest in TP53 wild-type neuroblastoma cells despite high MYCN levels. This miR-885-5p-mediated effect is mainly achieved through targeting CDK2, because primarily CDK2 and not MCM5 knockdown exerts G1/G0 arrest and p21waf1 induction.
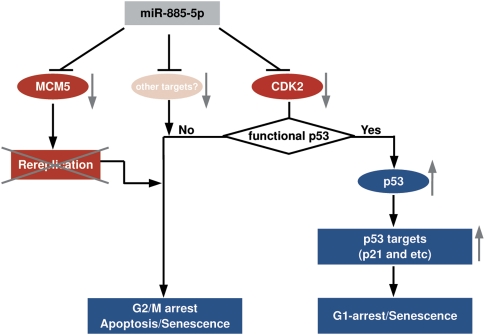
It is curious that miR-885-5p also triggers apoptosis and senescence in neuroblastoma cells with mutant TP53. Such cells cannot engage p53 in response to CDK2 inhibition, which causes G0/G1-arrest failure in these cells. However, CDK2 may still function as a pro-survival signal in a TP53 mutant background, as demonstrated by CDK2 knockdown in HDN33 cells. Alternatively, MCM5 inhibition, may prevail in TP53-mutant cells, inducing a senescence-like process and G2/M phase arrest, as evidenced by MCM5 knockdown in HDN33 cells. Other miR-885-5p targets may also influence growth and survival in p53-mutant neuroblastoma. We propose a model for miR-885-5p function targeting CDK2 and MCM5 in neuroblastoma cells in Figure 7.
Figure 7.
A model for miR-885-5p function. Proposed model for miR-885-5p function via CDK2 and MCM5 downregulation in neuroblastoma. miR-885-5p downregulates CDK2 leading to p53 activation in neuroblastoma cells with wild-type TP53. miR-885-5p-triggered CDK2 and MCM5 downregulation reduces cell renewal capacity and favors apoptosis and senescence in neuroblastoma with either wild-type or non-functional TP53
Our data provide indirect evidence for caspase-3 targeting by miR-885-5p. In neuroblastoma cells with elevated p53 protein levels, we detected only moderate upregulation of caspase-3 activity, reduction of procaspase-3 protein with no significant presence of its mature form and downregulation of CASP3 upon miR-885-5p introduction. The miRanda algorithm detected a miR-885-5p binding site in the CASP3 3′-UTR. Intriguingly, caspase-3 can be involved in p21waf1 degradation.26 We propose that caspase-3 targeting may further determine the response of miR-885-5p-transfected cells, favoring p21waf1 accumulation (Supplementary Figure S5).
Based on insights into miR-885-5p functions in neuroblastoma cells, we can speculate that miR-885-5p may withdraw normal cells from cycling via p53 activation but maintain survival. In this context, it would be interesting to examine the role of miR-885-5p in developmental processes, such as neuronal maturation. We conclude that miR-885-5p engages p53-dependent and independent regulatory layers jointly to interfere with cell cycle progression and survival of neuroblastoma cells. It negatively regulates CDK2 and MCM5 and activates the p53 pathway in TP53 wild-type neuroblastoma cells, and is able to trigger cell death and senescence in TP53-mutant neuroblastoma.
Patients and Methods
Patients
Neuroblastoma tumor samples were collected before any cytoreductive treatment, snap-frozen and stored at −80°C until RNA or DNA isolation. Written informed consent was obtained from patients' parents for tissue sampling. Genomic MYCN status was assessed in the reference laboratories of the German Neuroblastoma trial in Cologne and Heidelberg.
aCGH of primary neuroblastomas and M-FISH
Patient DNA tumor samples (n=193) were analyzed using either 44 (n=143) or 105 k (n=51) aCGH microarrays (Agilent Technologies, Santa Clara, CA, USA). A measure of 2.5 μg each of genomic tumor and reference DNA were used for each hybridization. Labeling and hybridization was performed as previously described.27 Arrays were preprocessed using the R/Bioconductor statistical software package (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). Arrays were segmented using the daglad function after lowess normalization, implementing a modified version of the methodology described in the study by Hupe et al.28 Different array platforms were matched using the chromosomal positions of the probes, and missing values were imputed only if two neighboring probes had the same segmented values. Multicolor FISH (M-FISH) analysis on neuroblastoma cell lines was performed using the 24 XCyte probe-kit (Meta-Systems, Altlussheim, Germany). Hybridization was performed according to the manufacturer's instructions. Slides were counterstained with 4,6-diamidino-2-phenylindol (DAPI) (Sigma, St. Louis, MO, USA). Images were captured with a Zeiss Axio Imager Z1 microscope (Zeiss, Thornwood, NY, USA) and analysed with Isis/mFISH imaging software (Meta-Systems, Mannheim, Germany).
Cell culture and transfection
SH-EP, KELLY, IMR32, SK-N-BE(2)c and HDN33 were grown in RPMI supplemented with 10% (v/v) fetal calf serum, 2 m -glutamine and 1% (v/v) penicillin/streptomycin in a humidified incubator at 37 °C and 5% CO2. miRNA mimics and antimiRs were purchased from Ambion (Austin, TX, USA). siRNAs to p53, CDK2 and MCM5 and control non-targeting siRNA were purchased from Qiagen (Hilden, Germany), Invitrogen (Carlsbad, CA, USA) and Santa Cruz (Santa Cruz, CA, USA). Cells were plated at 24 h before transient transfection with 30 n of miRNA mimic or antimiR or Cy3-labeled control miRNA or antimiR using Lipofectamine 2000 (Invitrogen). siRNA transfections were performed using Lipofectamine 2000 according to manufacturer's instructions. Unless otherwise stated, cells were assayed 4 days after transfection. For p53 knockdown experiments, cells were retransfected with miRNA mimics at 24 h after transfection with p53 siRNA or control siRNA. siRNA transfections were performed using Lipofectamine 2000 according to manufacturer's instructions. To construct the miR-885-5p and miR-331-3p expressing vectors, the 500 bp fragments surrounding corresponding miRNA genes were amplified and cloned into the Nhe1 and EcoR1 sites in pcDNA3.1 (Invitrogen). SH-EP and KELLY cells were plated in six-well plates for stable transfection with 400 μg pcDNA3.1-miR-331-3p, pcDNA3.1-miR-885-5p or empty pcDNA3.1 vector using Effectene (Qiagen). Individual clones were selected that grow in the presence of G418. Stable transfectants were maintained in culture no longer than 4 weeks during experiments.
Clonogenic assay, cell cycle, senescence and apoptosis assays
For cell proliferation evaluation, Alamar Blue (AbD Serotec, Raleigh, NC, USA) was used according manufacturer's instructions. Fluorescence was detected using the FluorStar Optima microplate fluorescence reader (BMG Labtech, Offenburg, Germany). The cell cycle distribution was determined as previously described.29 To assay for senescence, cells were fixed and stained using the SA-ß-Gal kit (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer's instructions. Three fields containing 100 cells were counted for each experiment and the results of three independent experiments are presented. Apoptosis was evaluated in FACS analysis by the extent of DNA fragmentation and by extent of phosphatidylserine externalization using FITC Annexin V Detection kit (BD Pharmingen, Franklin Lakes, NJ, USA).30, 31
Soft agar assay
Cells were trypsinised 18 h after miRNA mimic transfection, and 105 cells were resuspended in a top agar solution containing 0.35% noble agar, then poured onto a base layer containing 0.5% agar in six-well plates. Each well was covered with 3 ml of RPMI full medium, and incubated under standard culture conditions for 2 weeks. Colonies were fixed and stained with 10% methanol, 10% acetic acid and 0.5% of crystal violet. The colony counting was performed using ImageJ software (Rasband, WS, ImageJ, U.S. National Institutes of Health, Bethesda, MD, USA).
Luciferase reporter assay
miRNA-3′-UTR binding sites were predicted by miRanda (http://www.microrna.org) and Targetscan (http://www.targetscan.org) algorithms. 3′-UTR database was obtained from UTResource (http://www.ba.itb.cnr.it/UTR/).32 pmiRGLO vector (Promega, Madison, WI, USA) was used for reporter assay according to manufacturer' instructions. Putative miR-885-5p target sites in the CDK2 and MCM5 3′-UTRs were cloned into pmiRGLO vector downstream of the firefly luciferase gene in pmiRGLO vector to create reporter plasmids. The sequences containing miR-885-5p predicted binding sites were used to construct pmiRGLO-CDK2-site are listed in the Supplementary Table S4.
Cells were plated in 96-well plates, and transfected with 20 ng of either pmiRGLO empty, pmiRGLO-CDK2-site, pmiRGLO-CDK2-mut-site, pmiRGLO-MCM5-site or pmiRGLO-MCM5-mut-site using Effectene (Qiagen), or miRNA mimics. Cells were lysed in passive cell lysis buffer (Promega) at 24-h post transfection, and lysates were processed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's directions. Luciferase activity was detected in a FluorStar Optima microplate fluorescence reader. Firefly luciferase activity was normalized to Renilla luciferase activity as an internal transfection control. Transfections were performed in six independent experiments, and assayed in duplicate.
Real-time quantitative PCR (RT-qPCR)
TaqMan miRNA assays (Applied Biosystems) were used to quantify endogenous miR-885-5p and miR-331-3p. RNA was isolated from neuroblastoma cell lines using TRIzol, and 5 ng of total RNA was used to reverse-transcribe cDNA for miRNA assessment. Data from cell lines were normalized to U44 RNA expression. miRNA expression in primary tumor samples was analysed as previously described,33 and normalization was performed by using a set of miRNAs stably expressed in neuroblastomas. Real-time RT-PCR was performed by using the Applied Biosystems 7000 Sequence Detection system (Applied Biosystems, Foster City, CA, USA).
Northern blotting
Northern Blotting was performed as previously described.12 Total RNA was isolated with TRIZOL reagent (Invitrogen). A measure of 30 μg of total RNA was loaded per lane and separated on 15% denaturing polyacrylamide gels, transferred by electroblotting to GeneScreen+ membranes (Perkin Elmer, Boston, MA, USA). Methylene blue staining was performed before blocking with ULTRAhyb-Oligo hybridization buffer (Ambion) to assess RNA loading. Blots were incubated with 10 ng/ml 3′-biotinylated DNA probes complementary to either miR-885-5p or miR-331-3p in ULTRAhyb-Oligo buffer then visualized using the North2South Chemiluminescent Hybridization and Detection Kit (Pierce, Rockford, IL, USA).
Western blotting
Whole cells were lysed in a buffer containing 7 M urea, 1% Triton X-100, 100 m DTT, 20 m Tris-HCl, pH 8.5. Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA, USA), and 20 μg protein lysate were separated per lane on either 12 or 15% PAGE gels then transferred to nitrocellulose membranes (Protran, Whatman, Dassel, Germany). Antibody to MCM5 was purchased from Abcam (Cambridge, MA, USA). Antibody to PUMA was from ProSci Inc. (Poway, CA, USA). Antibodies to p53, MCM3, STAT3, E2F1, MYCN, CUL3 and ß-actin were from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Antibodies to p27, CDK2, CDK6 were from Dianova (Hamburg, Germany). Antibodies to precursor and mature CASP3 were from BD Transduction Laboratories (Lexington, KY, USA) and Cell Signaling, respectively. HRP-conjugated antibodies against mouse or rabbit were obtained from Jackson Immunoresearch (West Grove, PA, USA). Sheep anti-goat HRP-conjugated antibody was from Santa Cruz Biotechnologies. Membranes were incubated with the appropriate antibodies, and the bands were visualized using the ECL system (Pierce). Images were captured with a CCD camera (Vilber Lourmat, Marne-la-Vallee, France) and densitometry analysis was performed using ImageJ software.
Gene expression analysis
In the first series of experiments, RNA was isolated from cells at 3 days after transient transfection with miR-885-5p, miR-331-3p or control miRNA. In a second series of experiments, RNA was isolated from cells pretreated with p53 siRNA or control siRNA for 1 day and subsequently transfected with miR-885-5p, miR-331-3p or control miRNA for 3 days. Gene expression profiles were generated as two one-color replicates using the whole-genome oligonucleotide microarray platform from Agilent (Agilent Technologies) as previously described.34, 35 Raw microarray data were normalized using quantile normalization. Expression ratios are given as the mean of two replicates. All raw and normalized microarray data are available at the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress; Accession: E-TABM-38) or iCHIP (http://www.ichip.de/).
Statistical analysis
Statistical significance was calculated using the Student t-test. Data are presented as mean±S.D. from at least three experiments.
Acknowledgments
We thank Gabriele Becker and Yvonne Kahlert for excellent technical assistance, Filip Pattyn for critical remarks and Kathy Astrahantseff for proofreading the manuscript. This work was supported with Grants N2KR-S19T03 and 01GS0895 within NGFN-2 and NGFNPlus from the BMBF, and the EET-pipeline (No. 037260) and grant 259348 (ASSET) from the EU.
Glossary
- CDK
cyclin-dependent kinase
- MCM
mini-chromosome maintenance protein
- miRNA
microRNA
- SA-ß-Gal
senescence-associated ß-galactosidase
- siRNA
short interfering RNA
- TP53
human p53 gene
- UTR
untranslated region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of microRNA genes mir-15 and mir-16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2005;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of mir-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- Westermann F, Schwab M. Genetic parameters of neuroblastomas. Cancer Lett. 2002;184:127–147. doi: 10.1016/s0304-3835(02)00199-4. [DOI] [PubMed] [Google Scholar]
- Schwab M, Westermann F, Hero B, Berthold F. Neuroblastoma: biology and molecular and chromosomal pathology. Lancet Oncol. 2003;4:472–480. doi: 10.1016/s1470-2045(03)01166-5. [DOI] [PubMed] [Google Scholar]
- McKenzie PP, Guichard SM, Middlemas DS, Ashmun RA, Danks MK, Harris LC. Wild-type p53 can induce p21 and apoptosis in neuroblastoma cells but the DNA damage-induced G1 checkpoint function is attenuated. Clin Cancer Res. 1999;5:4199–4207. [PubMed] [Google Scholar]
- Van Maerken, Vandesompele J, Rihani A, De Paepe A, Speleman F. Escape from p53-mediated tumor surveillance in neuroblastoma: switching off the p14ARF-MDM2-p53 axis. Cell Death Diff. 2009;16:1563–1572. doi: 10.1038/cdd.2009.138. [DOI] [PubMed] [Google Scholar]
- Plantaz D, Vandesompele J, Van Roy N, Astowska M, Bown N, Combaret V, et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer. 2001;91:680–686. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1114>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Hoebeeck J, Vandesompele J, Nilsson H, De Preter K, Van Roy N, De Smet E, et al. The von Hippel-Lindau tumor suppressor gene expression level has prognostic value in neuroblastoma. Int J Cancer. 2006;119:624–629. doi: 10.1002/ijc.21888. [DOI] [PubMed] [Google Scholar]
- Afanasyeva EA, Hotz-Wagenblatt A, Glatting KH, Westermann F. New miRNAs cloned from neuroblastoma. BMC Genomics. 2008;9:52. doi: 10.1186/1471-2164-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Van de Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Carroll PE, Okuda M, Horn FH, Biddinger P, Stambrook PJ, Gleich LL, et al. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18:1935–1944. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DB. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLOS Biol. 2005;3:e264. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnowski JW, Goulette FA, Guan JJ, Chatterjee D, Yang Y-F, Cousens LP, et al. STAT3 cleavage by caspases: impact on full-length STAT3 expression, fragment formation and transcriptional activity. J Biol Chem. 2006;281:17707–17717. doi: 10.1074/jbc.M600088200. [DOI] [PubMed] [Google Scholar]
- Schwab BL, Leist M, Knippers R, Nicotera P. Selective proteolysis of the nuclear replication factor MCM3 in apoptosis. Exp Cell Res. 1998;238:415–421. doi: 10.1006/excr.1997.3850. [DOI] [PubMed] [Google Scholar]
- Agarwal MK, Amin ARMR, Agarwal ML. DNA replication licensing factor Minichromosome Maintenance Deficient 5 rescues p53-mediated growth arrest. Cancer Res. 2007;67:116–121. doi: 10.1158/0008-5472.CAN-06-2835. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Ebus ME, Geerts D, Koster J, Lamers F, Valentijn LJ, et al. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc Natl Acad Sci USA. 2009;106:12968–12973. doi: 10.1073/pnas.0901418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Qiang W, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. mir-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696–24704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le H-V, Massague J. Myc suppression of the p21 Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Levkau B, Koyama H, Raines EW, Clurman BE, Herren B, Orth K, et al. Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade. Mol Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- Spitz R, Oberthuer A, Zapatka M, Brors B, Hero B, Ernestus K, et al. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer. 2006;45:1130–1142. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- Hupé P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20:3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- Deubzer HE, Ehemann V, Westermann F, Heinrich R, Mechtersheimer G, Kulozik AE, et al. Histone deacetylase inhibitor Helminthosporium carbonum (HC)-toxin suppresses the malignant phenotype of neuroblastoma cells. Int J Cancer. 2008;122:1891–1900. doi: 10.1002/ijc.23295. [DOI] [PubMed] [Google Scholar]
- Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]
- Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, et al. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann F, Muth D, Benner A, Bauer T, Henrich KO, Oberthuer A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9:R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.