Abstract
Autophagy, an evolutionarily conserved lysosome-mediated degradation, promotes cell survival under starvation and is controlled by insulin/target of rapamycin (TOR) signaling. In Drosophila, nutrient depletion induces autophagy in the fat body. Interestingly, nutrient availability and insulin/TOR signaling also influence the size and structure of Drosophila ovaries, however, the role of nutrient signaling and autophagy during this process remains to be elucidated. Here, we show that starvation induces autophagy in germline cells (GCs) and in follicle cells (FCs) in Drosophila ovaries. This process is mediated by the ATG machinery and involves the upregulation of Atg genes. We further demonstrate that insulin/TOR signaling controls autophagy in FCs and GCs. The analysis of chimeric females reveals that autophagy in FCs, but not in GCs, is required for egg development. Strikingly, when animals lack Atg gene function in both cell types, ovaries develop normally, suggesting that the incompatibility between autophagy-competent GCs and autophagy-deficient FCs leads to defective egg development. As egg morphogenesis depends on a tightly linked signaling between FCs and GCs, we propose a model in which autophagy is required for the communication between these two cell types. Our data establish an important function for autophagy during oogenesis and contributes to the understanding of the role of autophagy in animal development.
Keywords: autophagy, Drosophila, oogenesis, starvation, insulin/TOR
Autophagy, a conserved degradation process, serves as an energy reserve in response to starvation, but also has critical roles in cellular remodeling during development, immunity and cancer.1 The central regulator of autophagy is the target of rapamycin (TOR), a downstream kinase of the insulin/insulin-like growth factor (IGF) signaling pathway (IIS).2
In Drosophila, IIS/TOR signaling regulates autophagy in the fat body,3, 4 but it remains unclear whether autophagy is also important in other nutrient-responding organs. The Drosophila ovaries are of special interest, as starvation inhibits ovarian development5 and mutations in IIS components lead to defects in oogenesis and female sterility.6, 7, 8, 9 These findings raise the question whether IIS/TOR signaling controls autophagy during oogenesis.
Notably, starvation induces programmed cell death (PCD) during Drosophila oogenesis in the germarium, in nurse cells (NCs) and follicle cells (FCs),5 and increases caspase activity during mid-oogenesis.10 At later stages, NCs also undergo developmental PCD necessary to complete oogenesis. So far, primarily the implication of apoptosis has been investigated. Only recent reports show that autophagy occurs in the germarium, during mid-oogenesis and in dying NC. Interestingly, inhibition of Atg genes prevents DNA fragmentation, suggesting that autophagy and apoptotic cell death are connected.11, 12 However, the regulatory mechanisms underlying these processes and the contribution of different ovarian cell types (GCs and FCs) are still unknown.
This motivated us to examine the crosstalk between autophagy and nutrient signaling during Drosophila oogenesis. We show that starvation induces autophagy in both GCs and FCs. Surprisingly, autophagy is specifically required in FCs, and oogenesis is unaffected when both GCs and FCs are autophagy deficient. This suggests that the incongruity between an autophagy-deficient soma and an autophagy-competent germline is responsible for the oogenesis defect. Consequently, we hypothesize that autophagy is required for proper communication between these two cell types.
Results
Starvation induces autophagy in Drosophila FCs and GCs
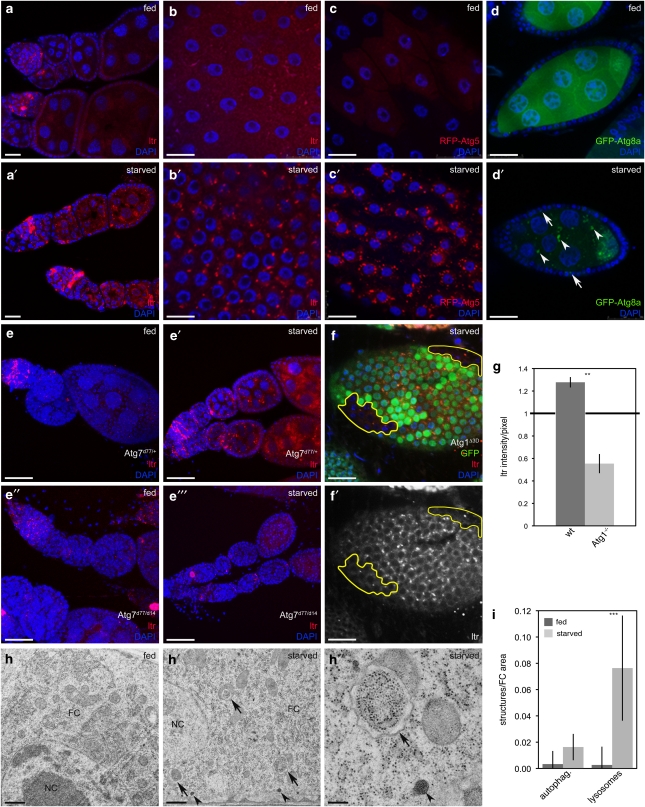
Nutrient deprivation affects Drosophila ovary size and egg production, and induces PCD in GCs and FCs.5 Thus, we tested whether autophagy is induced by starvation during Drosophila oogenesis using lysotracker (LTR). Upon starvation, LTR accumulated in region 2a/2b of the germarium and in stage 1–8 GCs (Figures 1a and a′, Supplementary Figure 1). However, LTR staining in the germarium was also visible under fed conditions (Figure 1a, Supplementary Figure 1B), but increased during starvation. Further, LTR-positive structures accumulated within FCs in stage 1–8 ovaries on starvation (Figures 1b and b′), whereas in later stages, FCs displayed starvation-independent LTR staining (Supplementary Figures 1G and G′). As reported previously,11 we also detected high levels of LTR staining in dying egg chambers (Supplementary Figures 1F and F′), whereas the staining of healthy eggs was generally more subtle, but concentrated to distinct punctae. Thus, we focused our analyses on healthy egg chambers.
Figure 1.
Starvation induces autophagy in Drosophila FCs and GCs. (a and b) LTR staining is increased in germaria, GCs (a′) and in stage 8 FCs (b′) upon starvation. (c and d) RFP-dAtg5 accumulates upon starvation in stage 8 FCs (c′) and GFP-dAtg8a in FCs (arrows) and GCs (arrowheads) (d′). (e–e″′) Atg7 mutants fail to induce autophagy. (f and f′) Atg1 mutant FC clones (marked by the lack of GFP) do not induce LTR staining. (g) LTR intensity/pixel of Atg1 mutant clones normalized to heterozygous cells. (h–h″) TEM images depict an accumulation of autophagosomes (arrows) and lysosomes (arrowheads) in starved FCs. (i) TEM quantification of FCs from fed versus starved flies (n=2). Only healthy egg chambers were considered for the analysis. Scale bars: (a, a′, d, d′, f and f′) 20 μm, (b–c′) 10 μm, (e–e″′) 50 μm, (h and h′) 500 nm, (h″) 200 nm. Error bars show S.D. of the mean, ***P<0.001, **P<0.01.Genotypes: (a–b and h–h′) y w, (c) da-Gal4/UAS-RFP-dAtg5, (d) da-Gal4/UASp-dAtg8a, (e) Atg7d14/Atg7d77, Atg7d14, (f) hs flp/+Atg1Δ3D FRT80B/FRT80B-UbiGFP
To confirm these results, we established transgenic flies expressing fluorescently tagged dAtg5 and dAtg8a proteins (Supplementary Figure 2). Starvation resulted in the formation of punctuate structures in GCs and FCs during mid-oogenesis in flies expressing UASp-GFP-dAtg8 (Figures 1d and d′), and equivalent structures were observed in FCs of flies expressing the soma-specific UASt-RFP-Atg5 (Figures 1c and c′).
Further, transmission electron microscopy (TEM) analyses revealed that lysosomes and autophagosomes are only occasionally found in FCs of fed flies (Figure 1h), whereas starvation increased the abundance of lysosomes and double membrane-bound vesicles containing undigested cytoplasmic material, indicative of autophagosomes (Figures 1h′ and h″, quantification 1i).
To confirm that these observations are truly autophagy dependent, we examined ovaries mutant for Atg7. Flies lacking Atg7 are viable, but unable to induce autophagy.13 Starvation induced LTR staining in the ovaries of Atg7 heterozygous control flies, but not in Atg7 homozygous mutant flies (Figures 1e′, and e″′). We further used the FLP/FRT system to induce FC clones homozygous mutant for Atg1, a kinase essential for autophagy.14 Although WT cells accumulated LTR-positive structures upon starvation, autophagy induction was impaired in neighboring clones lacking Atg1 (Figures 1f and f′, quantification 1g), demonstrating that ovarian autophagy requires functional ATG signaling.
Starvation induces dAtg8 conversion and Atg gene expression in Drosophila ovaries
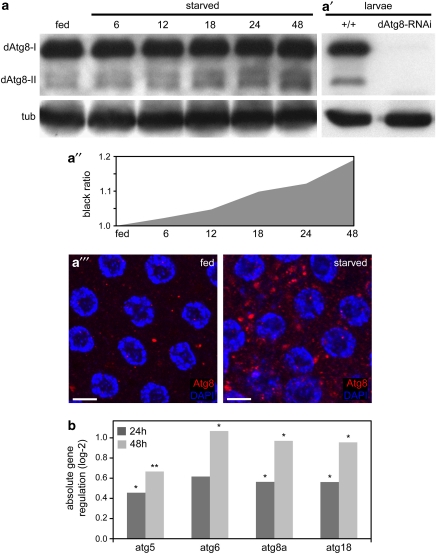
To monitor changes in cleaved dAtg8-II as an indicator of autophagy induction,15 we generated a Drosophila Atg8 antibody. An upregulation of dAtg8-II protein in the ovaries was already detectable 6 h after starvation, while the levels of dAtg8-I remained unchanged (Figures 2a and a″). Both dAtg8-I and dAtg8–II were completely vanished in protein extracts from larvae expressing UAS-dAtg8-RNAi, confirming antibody specificity (Figure 2a′). Consistently, the dAtg8 antibody detected punctuated structures in stage 8 FCs upon nutrient depletion (Figure 2a″′). Thus, starvation induces dAtg8 conversion and the accumulation of dAtg8-positive autophagosomes in Drosophila ovaries.
Figure 2.
Starvation induces dAtg8 conversion and Atg gene expression in Drosophila ovaries. (a) Western blot (WB) showing the increase of dAtg8-II in a starvation time course. (a′) Expression of dAtg8 is diminished in larvae ubiquitously expressing dAtg8a-RNAi. Tubulin served as loading control. (a″) Quantification of Atg8-II WB signals measured as grey values using ImageJ. Rising grey values represent the increase of dAtg8-II in a. (a″′) Accumulation of dAtg8 labeled autophagosomes in starved stage 8 FCs. (b) Quantitative real-time PCR of ovary RNA samples from fed flies (reference expression level), 24 and 48 h starved flies. n=5; P-values: *P<0.05, **P<0.01. Scale bars: (a″′) 10 μm. Genotypes: (a, a″′ and b) y w, (a′) y w, UAS-dAtg8-RNAi, da-Gal4
Several reports reveal that autophagy induction was accompanied by increased Atg gene expression,16, 17, 18 thus, we investigated Atg gene expression in Drosophila ovaries by quantitative real-time PCR. All genes examined showed a slight, but significant upregulation upon starvation (Figure 2b). These molecular readouts further confirm that starvation induces autophagy in Drosophila ovaries.
IIS/TOR controls ovarian autophagy
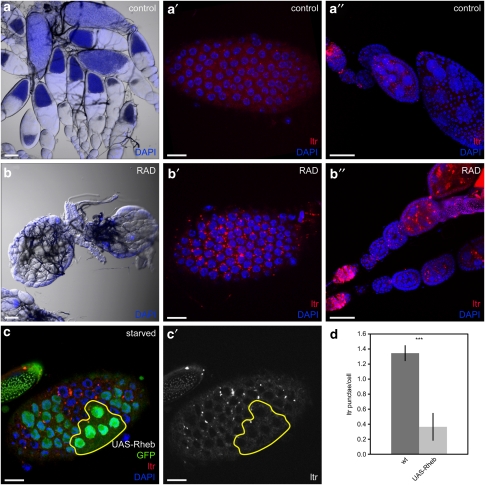
In Drosophila, autophagy is regulated by IIS/TOR signaling in the fat body3, 4 and salivary glands.19 Ovarian development is strongly affected by nutrient availability,5 and mutants in IIS/TOR pathway components are sterile,6, 7, 8, 9 suggesting that IIS/TOR signaling also regulates autophagy during oogenesis. Therefore, we investigated whether inhibition of TOR by rapamycin is able to mimic starvation-induced autophagy in the ovaries. Injection of RAD (a rapamycin derivative) into the female abdomen led to small ovaries lacking vitellogenic stages (Figure 3b), whereas control injection did not affect ovarian development and egg production (Figure 3a). RAD-treated females were fully viable, but produced 80 and 98% less offspring on day 1 and 2 after injection, respectively, compared with controls. LTR staining was dramatically increased in FCs and GCs of RAD-treated ovaries (Figures 3a′–b″), which was comparable with starvation-induced autophagy (Figures 1a–b′, Supplementary Figure 1), indicating that nutrient deprivation and TOR inhibition act on the same autophagic mechanism in Drosophila ovaries.
Figure 3.
IIS/TOR signaling controls autophagy in Drosophila ovaries. (a–b″) Injection of RAD leads to small ovaries lacking vitellogenic stages (b) and a strong accumulation of autophagolysosomes in FCs (b′) and GCs (b″). Control ovaries are of normal size (a) and barely show LTR staining in FCs (a′) or GCs (a″). (c and c′) Generation of stage 7 FC clones overexpressing Rheb using the flp-out-Gal4/UAS method results in cells with high (strong GFP signal) and low (weak GFP signal) transgene expression. Only cells with bright GFP signals and enlarged nuclei (as an indication of enhanced cell size due to Rheb overexpression) were considered for the analyses. (d) Quantification of LTR staining in Rheb overexpressing clones compared with WT cells. Error bars show S.D. of the mean, n=8, ***P<0.001. Scale bars: (a and b) 100 μm, (a′, b′, c and c′) 10 μm, (a″ and b″) 50 μm. Genotypes: (a–b) y w, (c) hs flp/+act4CD24Gal4 UAS-GFP/UAS-RhebEP50.084
Alternatively, to test whether activation of IIS/TOR signaling was sufficient to suppress starvation-induced autophagy, we generated FC clones expressing Rheb, an upstream activator of TOR.4, 8, 20 Notably, FCs overexpressing Rheb lacked LTR staining even under starvation (Figures 3c and c′, quantification 3d). Thus, IIS/TOR signaling controls starvation-induced autophagy in a cell-autonomous manner in the ovaries, and is sufficient to inhibit autophagy even under starvation.
Autophagy is required for FC development
As starvation triggers autophagy in FCs and GCs, the questions remain whether autophagy is essential for oogenesis, and whether autophagy is required in the FCs or GCs. To answer these issues, we created chimeric animals lacking Atg gene function in either the germline or the somatic FCs.
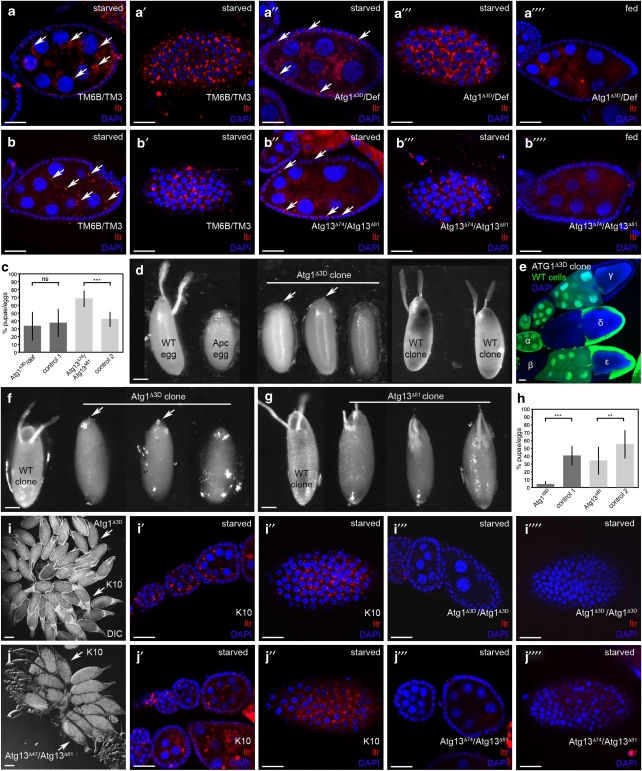
First, we generated germline chimeras by pole cell transplantations (PCT).21 Chimeric ovaries composed from an Atg1 hemizygous germline and WT FCs were defective in autophagy, as starvation did not induce LTR staining in the mutant GCs, but in the enveloping WT FCs (Figures 4a″ and a″′). This demonstrates that autophagy was induced in the chimeras, but only in WT tissue, and confirms the necessity of Atg1 for starvation-induced autophagy. In starved sibling control chimeras, in which the GCs inherited the chromosome balancers, LTR-positive structures emerged in WT FCs and GCs (Figures 4a and a′). Surprisingly, Atg1 germline chimeras developed functional ovaries, and their egg-laying behavior and hatching rates were indistinguishable from sibling control chimeras, albeit the offspring developed with a delay of 2 days (Figure 4c). When the Atg1 germline chimeras were crossed with Atg1 heterozygous males, the resulting Atg1 homozygous mutant animals died in late larval stages, similar to Atg1 homozygous mutants derived from heterozygous mothers.14 To further verify that autophagy is redundant in GCs for proper oogenesis, we created germline mosaics for Atg13. Knockout of Atg1 or Atg13 results in a similar defect in autophagy.22 Accordingly, Atg13 mutant GCs were defective in autophagy as monitored by the lack of LTR staining (Figures 4b″ and b″′), however, the chimeras were fully fertile with normal egg-laying behavior and hatching rates (Figure 4c). Further, we did not detect any defects in egg chamber development or egg morphology in Atg1 or Atg13 germline chimeras. It was recently reported that Atg1 GLCs show a partial disruption of developmental NC death.23 We occasionally observed persisting NC nuclei in stage-14 eggs; however, these events occurred with a low frequency in both the Atg1 germline chimeras as well as in control siblings (4 versus 1.3%, respectively). Atg13 germline mosaics did not show disruption of NC death, thus we conclude that developmental NC death is not affected in Atg germline chimeras. This indicates that autophagy in GCs is not required for egg development.
Figure 4.
Atg1 and Atg13 are required for FC development. (a–c) GC mosaics. Starvation induces autophagy (monitored by LTR staining) in GCs (a and b) and FCs (a′ and b′) of control TM6B/TM3 GC chimeras, whereas GC chimeras homozygous mutant for Atg1 (a″ and a″′) or Atg13 (b″ and b″′) display LTR staining only in WT FCs, which is not seen in fed control GC chimeras (a″″ and b″″). Quantification of offspring shows similar hatching rates as control TM6B/TM3 females (c). (d–h) FC mosaics. (d) X-ray induced FC clones. Shown are a normal egg deposited by a WT fly, a flaccid Apc mutant egg, missing DAs and anterior chorion structures, and eggs with Atg1 mutant FC clones, missing DAs, but showing a micropyle (arrows). Generation of WT FC clones in Apc/+ animals completely rescued the Apc phenotype. Typical examples of the resulting WT-like eggs are shown (WT clone). (e–h) Heat-shock induced clones. (e) Hs-flp induced mitotic recombination results in ovaries comprising WT (α) or completely mutant (β) egg chambers, egg chambers with all FCs mutant (γ), all GCs mutant (δ) or mosaic FCs (ɛ). (f) Eggs with Atg1 mutant clones lack DAs, but feature a micropyle (arrows). (g) Eggs with Atg13 mutant clones show variable phenotypes with reduced DAs. (h) Hatching rate of eggs containing Atg1 or Atg13 mutant FC clones. (i and j) Ovarian chimeras generated by larval ovary transplantations. After implantation, both K10 and Atg1 (i) or Atg13 (j) mutant ovaries are attached to the oviduct. Upon starvation, autophagy (monitored by LTR staining) is induced in GCs (i′ and j′) and FCs (i″ and j″) of K10 control ovaries, whereas Atg mutant ovaries are unable to induce autophagy (i″′–j″″). Error bars show S.D. of the mean, ***P<0.001, **P<0.01. Scale bars: (a–b″″, e, i′–i″″ and j′–j″″) 20 μm, (d, f and g) 100 μm, (i and j) 250 μm. Genotypes: (a–a″″) donor: Atg1Δ3D-FRT80B/TM6B, Df(3L)BSC613/TM3, host: Tm2gs/Tm2gs, (b–b″″) donor: Atg13Δ81/TM6B, Atg13Δ74/TM3, host: Tm2gs/Tm2gs, (d) w1118, Fs(3)Apc/+, Fs(3)Apc/Atg1Δ3D-FRT80B, (f and e) FRT80isogenic/FRT80-UbiGFP (WT clone), Atg1Δ3D-FRT80B/FRT80-UbiGFP, (g) FRT82isogenic/FRT82-UbiGFP (WT clone), Atg13Δ81-FRT82/FRT82-UbiGFP, (i and j) donor: Atg1Δ3D/Atg1Δ3D, Atg13Δ74/Atg13Δ81 host: fs(1)K10/fs(1)K10
To analyze the function of autophagy in FCs, we created mosaics in which only the FCs were homozygous mutant for Atg1, whereas the GCs were heterozygous. First, we made use of flies carrying the Apc mutation that disrupts FC function, leading to flaccid eggs lacking dorsal appendages (DAs) and anterior chorion structures (see Materials and Methods). The removal of Apc by irradiating +/Apc control larvae restored FC function,24 resulting in females producing eggs with normal-looking DAs and embryonic cuticle (Figure 4d), and larvae hatched and developed to adults. In contrast, females resulting from Atg1Δ3D/Apc irradiated larvae containing Atg1Δ3D/Atg1Δ3D FC clones deposited non-typical Apc eggs that were non-flaccid, but contained short and rudimentary DAs. In these eggs, embryonic cuticle never appeared, and no larvae hatched (Figure 4d), suggesting that Atg1 function is essential in FCs of Drosophila ovaries.
To confirm this, we created FC clones homozygous for Atg1 using the heatshock (hs)-flp/FRT system. Hs-induced mitotic recombination resulted in Atg1 homozygous mutant FC clones (identified by the lack of GFP) in 69% of the egg chambers with most of them being mosaic (Table 1, Figure 4e). Flies containing Atg1 mutant FC clones laid very few eggs that resembled those generated by irradiation, lacking DAs and embryonic cuticle (Figure 4f), and only 5% of the eggs hatched (Figure 4h). Quantification revealed that 89% of the eggs laid by females containing Atg1 mutant FC clones exhibited DA defects; consequently, only 11% of the chimeric eggs hatched (Supplementary Figure 3). Defective DA formation was only observed in 15% of the eggs containing control clones, and 58% of the control eggs hatched (Supplementary Figure 3). Given that 15% of the control eggs showed egg defects, the frequency of the egg phenotype that is solely due to the Atg1 deletion (74%) is in accordance with the frequency of FC clones observed in the Atg1 chimeras (69%), suggesting that almost every egg chamber containing Atg1 mutant FC clones resulted in defective eggs. These data confirm the requirement of Atg1 gene function in the FCs for proper oogenesis.
Table 1. Frequency of HS-FLP induced FC clones.
|
Percentage of FC clones |
Percentage of GC clones |
||||||
|---|---|---|---|---|---|---|---|
| Genotype | Entire clone | Mosaic clone | Total | No clone | Entire clone | No clone | Total number of egg chambers counted |
| Atg1Δ3D | 22±4 | 48±13 | 69±9 | 31±9 | 7±7 | 93±7 | 236+157 (n=2) |
| FRT80iso | 13±2 | 51±8 | 64±5 | 36±9 | 13±5 | 87±5 | 134+130 (n=2) |
| Atg13Δ81 | 38±0 | 46±9 | 84±4 | 16±9 | 30±1 | 70±1 | 40+127 (n=2) |
| FRT82iso | 51±4 | 25±6 | 76±5 | 24±10 | 40±0 | 60±0 | 48+60 (n=2) |
However, Atg7 mutants, although clearly autophagy-defective, did not exhibit a severe oogenesis phenotype. Eggs derived from Atg7 homozygous mutants showed a slight reduction in hatching rates compared with heterozygous controls (74 and 85%, respectively) with 18% of the eggs displaying eggshell defects, suggesting that Atg7 mutations have only minor effects on egg development. This may implicate that the defect in egg development caused by the lack of Atg1 is not due to autophagy, but an alternate function of Atg1. To verify that the observed phenotype is not restricted to Atg1, we created hs-induced FC clones mutant for Atg13. Clone induction was equally effective as for Atg1, with 84% of the egg chambers containing Atg13 homozygous mutant FC clones (Table 1). However, the phenotype was somewhat weaker, as we detected fewer eggs with DA defects (42%, see Supplementary Figure 3, Figure 4g) and 34% of the eggs hatched (Figure 4h, Supplementary Figure 3). The weaker effect of the Atg13 deletion is consistent with the observation that the lethality associated with Atg13 is less severe than for Atg1,22 and may be explained by differences in protein perdurance. Nevertheless, the similarity in phenotypes observed for Atg1 and Atg13 mutations confirm that autophagy in FCs is necessary for proper egg development.
Although this strongly suggests that the observed phenotype depends on autophagy, the question remains why Atg7 mutants do not show oogenesis defects, although autophagy is clearly disrupted in their FCs (Figures 1e–e″′). Apparently, the two conditions, whole animal versus mosaic, must constitute distinctive situations in which the Atg deficiency is interpreted differently. Although both GCs and FCs are autophagy defective in Atg7 mutants, the chimeras lack Atg gene function only in the FCs. This led us to propose that inputs from the cellular environment may affect the outcome of the autophagic signal. Consequently, we created chimeras lacking Atg genes specifically in the ovaries (in both GCs and FCs) by larval ovary transplantation experiments. We used host larvae carrying the fs(1)K10 mutation leading to eggs containing a mass of chorionic material instead of two DAs present in WT eggs,25, 26 therefore, the transplanted ovary can be distinguished from K10/K10 host ovaries by the appearance of DAs (Figures 4i and j). Strikingly, although autophagy was clearly disrupted in Atg1 or Atg13 homozygous mutant ovaries as monitored by the lack of LTR staining in both GCs and FCs (Figures 4i″′, i″″, j″′ and j″″), the mutant ovaries developed normally and gave rise to offspring with hatching rates comparable with those of the germline chimeras (37 and 64%, respectively). The respective K10/K10 ovary from the same animal displayed normal LTR staining in both GCs and FCs, confirming that autophagy was induced in the chimeric animals, but only in the host tissues (Figures 4i′, i″, j′ and j″). Further, we did not observe any defects in egg chamber development or DA formation in Atg1 or Atg13 mutant ovaries. This indicates that egg development is unaffected when both ovarian cell types are autophagy deficient, which is in accordance with Atg7 mutant flies not showing an oogenesis phenotype. This suggests that the oogenesis defect in the FC chimeras may be caused by an incompatibility between the mutant soma and a WT-like germline. As oogenesis requires a tight coordination of germline and soma,27 it is tempting to speculate that signaling between these tissues is dysfunctional if FCs are autophagy defective.
Discussion
This work establishes the Drosophila ovaries as an attractive model system to study autophagy. Starvation induces autophagy in FCs and GCs of Drosophila ovaries under the control of IIS/TOR signaling. Notably, IIS/TOR signaling affects various processes during Drosophila oogenesis. For example, overexpression of activated protein kinase B (Akt) disrupts the deposition of the NC cytoplasm into the oocyte (NC dumping).28 Furthermore, eliminating GCs modulates IIS, leading to prolonged lifespan and reproduction.29, 30 IIS also mediates ovarian stem cell proliferation in response to nutrients.5
Referring this to the mammalian system, the role of IIS during oogenesis is of special interest regarding one of the most common endocrine disorders, the polycystic ovary syndrome (PCOS). PCOS represents the most prevalent cause of anovulatory infertility characterized by large numbers of immature follicles. Remarkably, PCOS is often associated with type 2 diabetes and impaired IIS.31 Interestingly, apoptosis regulators are upregulated in patients affected by PCOS,32 suggesting a role for PCD in the onset of the disease. Thus, alterations in IIS could lead to a disregulation of ovarian autophagy, which might be implicated in the development of polycystic ovaries. Further investigations will reveal whether modulation of autophagy in Drosophila leads to PCOS-like phenotypes.
During Drosophila oogenesis, several cell death checkpoints have been reported. Despite the developmental PCD of NCs, starvation induces egg chamber degeneration within the germarium and during mid-oogenesis, suggesting that dying egg chambers respond to the environmental status that is monitored before investing energy into egg production.5 Interestingly, NC death, normally initiated at stage 10, is observed already at stage 8 under starvation, suggesting that those cells respond to nutrient availability as well.33, 34 Although these reports focused mainly on apoptotic cell death, the fact that PCD during mid-oogenesis strictly requires the caspase Dcp-1, which is nonessential for most other death pathways in the fly, suggests the existence of a non-redundant death mechanism during mid-oogenesis.33
Here, we show that starvation induces autophagy in the germarium and GCs during mid-oogenesis. This is consistent with recent publications indicating that autophagy contributes to PCD in the ovary.11, 12 Interestingly, this process is regulated by Dcp-1,11 suggesting that apoptosis and autophagy coordinate the progress of oogenesis.
Surprisingly, we find that autophagy is not required for germline development. This is in accordance with Atg7 mutant flies being fertile.13 However, other Atg mutant phenotypes in Drosophila suggest a role for autophagy during development. Flies mutant for Atg1 are pupal lethal,4 and Atg1 germline clones (GLCs) achieved using the OvoD technique show reduced DNA fragmentation and a partial disruption in NC death.11, 12 In the present study, we generated germline chimeras using the PCT technique where, in contrast to the OvoD system, a germline completely mutant for a certain gene is generated in a WT background. This technique excludes any perdurance and maternal contribution. Unlike the GLCs generated using OvoD, the transplanted pole cells are hemizygous mutant for the gene of interest, which excludes second site lethal effects. Further, PCT results in true germline chimeras without affecting the somatic cells, while the generation of GLC in the OvoD system also induces FC clones, which may interfere with the mutant phenotype. In germline chimeras generated by PCT, we do not observe any egg chamber defects and conclude that autophagy is not required in the germline. The discrepancy between our and the recently published data concerning reduced NC death could be explained by the different experimental setups and their limitations mentioned above.
However, as autophagy is induced upon starvation in GCs, the question remains whether oogenesis depends on autophagy in GCs when nutrients are limited. Further studies will reveal whether autophagy-deficient ovaries develop normally under such conditions.
Moreover, we demonstrate that starvation induces autophagy in FCs, confirming that FCs are involved in controlling the nutritional status to ensure germline development. Further, autophagy in FCs is essential for proper oogenesis. Thus, what could be the function of FCs during egg development, and how could autophagy contribute to this process?
FCs have a fundamental role during oogenesis. The patterning of FCs into discrete subtypes is crucial for egg development, as specialized FC sub-populations guide various steps during oogenesis. Eggshell morphogenesis further depends on the migration of different FC sub-populations to form a columnar epithelium over the oocyte, the micropyle and the DAs. FCs also secrete the chorion, a multilayered structure surrounding the oocyte essential for embryonic survival.27, 35 Interestingly, autophagy in FCs seems to be tightly associated with the spatial pattern of chorion synthesis, as autophagic death occurs at the anterior pole of the egg chamber where chorion formation is first completed.36
Notably, autophagy deficiency only affects oogenesis in a cellular context where FCs are mutant for Atg genes and GCs are WT. On the basis of this incompatibility, we hypothesize that dysfunctional signaling between soma and germline may be responsible for the oogenesis phenotype. For example, a signal arising in the WT germline may not be processed correctly in the mutant FCs and thus disrupts egg development. Alternatively, autophagy-deficient FCs may be incapable of generating a signal required in the GCs or necessary for the differentiation of specific FC sub-populations. However, if both cell types are deficient in autophagy, the absence of such a signal prevents a false interpretation by the other cell type, and egg development occurs normally. This model may be applied to explain the lack of oogenesis defects in Atg7 mutant flies.
Thus, what are the signals during oogenesis that require autophagy? Egg development depends on signaling between GCs and FCs and between sub-populations of FCs. Three signaling pathways are involved in these processes: Notch, EGFR and Jak/STAT.27 Notch is required for proliferation, differentiation and migration of FCs.27 Interestingly, loss of the cysteine protease Atg4 modulates Notch signaling in Drosophila,37 thus, it is tempting to speculate that impaired Atg signaling may lead to malfunction of the Notch receptor to affect cell fate determination during oogenesis. The identification of the signaling pathway affected by the loss of autophagy in the FCs will shed light on the yet unsolved issue on which pathways are controlled by autophagy during the development of higher organisms.
Although the lethality associated with many Atg mutations in Drosophila indicates a fundamental role for autophagy during development, the function of some Atg genes is dispensable for fly development. Thus, some Atg genes may function redundantly, or other mechanisms compensate for autophagy deficiencies during development. Alternatively, given that certain Atg mutations have cell-context specific effects, there could be factors that determine specificity. Our findings on the incompatibility between autophagy-deficient soma and autophagy-competent germline demonstrate that the generation of chimeras is crucial to elucidate the tissue-specific function of a gene in a context relevant to physiology and development.
Our data clearly indicate that autophagy is indispensable for oogenesis. The understanding of molecular events regulating PCD in the fly ovary is still incomplete, and the communication of death signals between FCs and GCs remains to be defined. The present study suggests that the nutrient response of FCs and GCs implies crosstalk between these two tissues. Further studies will aid to understand the fundamentals underlying this cell communication.
Materials and Methods
Drosophila maintenance, starvation and stocks
Flies were raised on standard yeast/cornmeal agar at 25°C. Four-day-old females were starved on 10% sucrose agar at 25°C for 24 h if not otherwise stated.
D. melanogaster stocks used: y w, w1118 (controls), Atg7Δ14, Atg7Δ77, Atg7Δ4, Atg1Δ3D, Atg13Δ74 and Atg13Δ81 (kindly provided by T. Neufeld),4, 13, 22 Atg8-RNAi 43096 (VDRC, Vienna, Austria), UAS-Rheb50.084, Fs(3)Apc,24 Tm2gs,38 fs(1)K10,39 Df(3L)BSC613, FRT80-UbiGFP, FRT82-UbiGFP and Act<CD2<Gal4 UAS-GFP (Bloomington Drosophila Stock Center, Indiana University, IN, USA).
Transgenic flies
dAtg5 (5′-CAC CAT GGC CCA CGA CCG CGA G-3′ 5′-AAC ATC CTT GTA GTC CAC CGA-3′) and dAtg8a (5′-CAC CAT GAA GTT CCA ATA CAA GGA-3′ 5′ GTT AAT TTT GGC CAT GCC G-3′) coding regions were PCR amplified and cloned into pTGW and pPGW vectors (Carnegie Institution, WA, USA) to express the transgenes either in the soma (UASt-RFP-Atg5) or in both the soma and the germline (UASp-GFP-Atg8). Constructs were injected into y w embryos for transformation according to standard procedures. Three transgenic lines on two different chromosomes were established and tested for each construct.
LTR assay, tissue preparation and confocal microscopy
Ovaries were dissected in PBS, incubated for 1 min in 100 μM Lysotracker red DND-99 (Invitrogen, Molecular Probes, Basel, Switzerland) to label acidic organelles including autolysosomes, washed three times in PBS and fixed in 4% paraformaldehyde for 20 min. Ovaries were embedded in mounting medium with DAPI (Vectashield, Vector Laboratories, Inc., Burlingame, CA, USA) and images were obtained using a confocal microscope (Leica, Wetzlar, Germany, DM5500Q, TCS-SPE; objective lenses: Leica, 20 × (0.70), 40 × (1.15), 63 × (1.30); acquisition software: LAS AF v.2.0.1, Leica, Wetzlar, Germany) at room temperature and edited using Adobe Illustrator and Photoshop CS4.
Transmission EM
Ovaries were fixed for 4 h in 2% glutaraldehyde, 1% osmium tetroxide in 0.1 m cacodylate buffer, and postfixed for 4 h in 2% osmium tetroxide. After dehydration in an acetone series, ovaries were embedded in Spurr. Sections (50 nm) were stained with uranyl acetate and lead citrate on Formvar/Carbon covered copper grids (Quantifoil, Jena, Germany) and viewed on a transmission EM (Morgani 268, FEI Europe, Eindhoven, Netherlands). Quantification of the autophagic area was performed on ovaries from two different flies for each condition. In total, 15–20 randomly chosen FCs were photographed at × 4000 magnification, and autophagic structures and lysosomes were counted. Autophagic structures were scored according to their morphology, comprising all structures that contained recognizable cytosolic material.
Antibody generation, western blotting and immunofluorescence
Rabbits were immunized with the dAtg8 peptide: H2N-MKFQYKEEHAFEKRR-CONH2 (Eurogentec, Seraing, Belgium). The serum was double affinity purified and specificity of the antibody was shown on WB (Figure 2).
For WBs, twenty ovaries per time point or alternatively, five third instar larvae were extracted in lysis buffer (120 mM NaCl, 50 mM Tris-HCl, 20 mM NaF, 1 mM Benzamidine, 1 mM EDTA, 6 mM EGTA, 15 M NA4P2O7, 1% Nonidet P-40) containing protease inhibitors. Proteins were separated on a 12% SDS-PAGE gel and blotted onto Nitrocellulose (Hybond ECL, GE Healthcare, Uppsala, Sweden). Primary antibodies were applied overnight at 4°C: anti-dAtg8 1 : 1000, anti-tubulin (T-9026, Sigma-Aldrich, Buchs, Switzerland) 1 : 10000 and secondary antibodies for 2 h at RT: anti-rabbit-HRP 1 : 10000 (Jackson ImmunoResearch Europe Ltd., Suffolk, UK), anti-mouse-HRP 1 : 10000 (Jackson ImmunoResearch Europe Ltd.). For quantification of WB signals, Image J software (National Institutes of Health, Bethesda, MD, USA) was used to calculate the grey values of Atg8-II bands in fed versus starved conditions. Grey values of ovaries from fed flies were set as one.
For immunofluorescence, ovaries were fixed for 20 min in 4% PFA in 1 : 1 PBS/Heptan, dehydrated by methanol series and blocked with 2% normal donkey serum in PBS supplemented with 0.1% Triton X-100 and 1% DMSO. Primary antibody dilution was applied overnight at 4°C (anti-dAtg8, 1 : 500), secondary antibody for 2 h at RT (anti-rabbit-TexasRed 1 : 200, Jackson ImmunoResearch Europe Ltd.).
RNA purification and quantitative real-time PCR
Total RNA from 40 ovaries per time point was extracted using the RNeasy mini kit (Qiagen, Hilden, Germany). 2 μg RNA was reverse transcribed using SuperScriptIII reverse transcriptase (Invitrogen, Basel, Switzerland), following the manufacturer's protocol. Runs were performed in duplicates for five different biological replicates with a Rotor-Gene 6000 cycler (Corbett, Qiagen, Hilden, Germany) and SYBR Green Master Mix (Roche, Basel, Switzerland) and melting curve analyses were performed. Data were analyzed using REST (relative expression software tool) and Microsoft Excel software. Relative expression ratios were normalized to rpl23 and actin5c, which showed no significant expression difference between fed and starved ovaries. mRNA levels of the respective genes of fed flies served as reference levels.
RAD treatment
RAD (Novartis, Basel, Switzerland) was dissolved in ethanol and diluted to 100 μM with Robb's minimal saline (2.6 mM NaCl, 2.0 mM KCl, 0.5 mM Glucose, 0.06 mM MgSO4. 7H2O, 0.06 mM MgCl2. 6H2O, 0.05 mM CaCl2, 0.1 mM Na2HPO4, 0.018 mM KH2PO4, pH 6.75)
RAD solution (0.2 μl per fly) or control solution containing ethanol at the same dilution was injected into the ventral mid-lateral part of the abdomen and ovaries were analyzed after 24 h. For offspring analyses, females were transferred to fresh vials every day and the number of offspring counted.
Pole cell transplantation
Pole cells (embryonic germline precursor cells) from Atg1 or Atg13 hemizygous mutant donor embryos were transplanted into host embryos derived from females homozygous for Tm2gs, a grandchildless (gs) type of mutation.38 As there is no germline in the gs-derived embryos, ovaries of gs-females are rudimentary and contain only the mesodermal components. Donor embryos were generated by crossing Atg1Δ3D/TM6B females with Df(3L)BSC613/TM3 males or Atg13Δ74/TM6B females with Atg13Δ81/TM3 males. Pole cells were collected from single blastoderm-stage donor embryos and transplanted into 2–3 host blastoderm stage embryos.21 Eclosing females were mated with WT males to determine the genotype of the progeny before LTR analysis. Germline chimeras with Atg1Δ3D/TM3, Df(3L)BSC613/TM6B or Atg13Δ74/TM3, Atg13Δ81/TM6B as well as TM3/TM6B germline cells served as internal controls. Three independent experiments with a total of 11 (Atg1) or 14 (Atg13) mutant germline chimeras and 16 (Atg1) or 23 (Atg13) control sibling females were performed.
X-ray irradiation
Atg1Δ3D/Fs(3)Apc late third instar larvae were X-ray irradiated for the induction of mitotic recombination (10 Gy; 110 kV, 1 mm Al filter, 0.31 Gy/min). Apc disrupts the function of anterior FCs, leading to the degeneration of almost all the egg primordia, with few developing to flaccid eggs lacking DAs and anterior chorion structures. Apc does not affect the function of the GCs. Removal of Apc through mitotic recombination restores FC function and allows the development of offspring from the mosaic egg primordia.24
Eclosing Atg1Δ3D/Fs(3)Apc females were mated with WT males in single vials and egg production was analyzed every day for 12 days, a time period required to identify ≥95% of the mosaics. As controls, +/Fs(3)Apc larvae were irradiated and analyzed. 17 Atg1Δ3D/Fs(3)Apc or 77+/Fs(3)Apc mosaics deposited at total of 44 or 253 non-Apc eggs, respectively, indicating that the two types of mosaics produced non-Apc eggs with a similar frequency.
FLP induced FC clones
The FLP/FRT recombination method was used to generate FC clones. FC clones overexpressing UAS-Rheb were achieved by heatshocking 4-day-old females for 20 min at 34°C. FC clones mutant for Atg1 or Atg13 were generated by heatshocking flies of the genotypes FRT80-Atg1Δ3D/FRT80-UbiGFP or FRT82-Atg13Δ81/FRT82-UbiGFP for 1 h at 37°C during larval development on five consecutive days. Resulting adults were mated with WT males in single vials and egg production was monitored every day for 5 days for egg laying analysis. Laid eggs were photographed, counted and kept on 25°C until hatching. Pupae and offspring were counted.
Larval ovary transplantation
For larval ovary transplantations,25 one mutant ovary dissected from either Atg1Δ3D homozygous or Atg13Δ74/Atg13Δ81 larvae was transplanted into fs(1)K10 homozygous host larvae. Host females were mated with WT males in single vials for identification of the egg genotype and egg laying analysis. Host females with Atg mutant eggs were starved and stained with LTR.
Acknowledgments
We thank T Neufeld, the Bloomington and the Szeged Stock Centers as well as the VDRC for fly stocks. We also thank Hugo Stocker, the members of the Hafen group, and M Matschiner for helpful discussions and technical support. This work was supported by grants from the Swiss National Science foundation (to JMIB, KK and EH) and by the Hungarian Scientific Research Fund NI 69180 (to JS).
Glossary
- Atg
autophagy-related
- DA
dorsal appendage
- FCs
follicle cells
- FRT
flp recombinase target
- GCs
germ cells
- GLCs
germline clones
- gs
grandchildless
- hs
heatshock
- IGF
insulin-like growth factor
- IIS
insulin/insulin-like growth factor signaling
- LTR
lysotracker-red
- NCs
nurse cells
- PCT
pole cell transplantation
- PCD
programmed cell death
- PCOS
polycystic ovary syndrome
- RFP
red fluorescent protein
- Rheb
Ras homologue enriched in brain
- TEM
transmission electron microscopy
- TOR
target of rapamycin
- WB
western blot
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Baehrecke
Supplementary Material
References
- Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, et al. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Richard DS, Rybczynski R, Wilson TG, Wang Y, Wayne ML, Zhou Y, et al. Insulin signaling is necessary for vitellogenesis in Drosophila melanogaster independent of the roles of juvenile hormone and ecdysteroids: female sterility of the chico1 insulin signaling mutation is autonomous to the ovary. J Insect Physiol. 2005;51:455–464. doi: 10.1016/j.jinsphys.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Werz C, Kohler K, Hafen E, Stocker H. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 2009;5:e1000596. doi: 10.1371/journal.pgen.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in drosophila oogenesis. Dev Biol. 2003;260:113–123. doi: 10.1016/s0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjorkoy G, Johansen T, et al. Cell death during Drosophilamelanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5:298–302. doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, et al. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13:358–363. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, et al. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14:1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BP, Tanner EA, Mateos San Martin D, Blute T, Kinser RD, Dolph PJ, et al. Cell-autonomous requirement for DNaseII in nonapoptotic cell death. Cell Death Differ. 2009;16:1362–1371. doi: 10.1038/cdd.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi M, Szabad J. Isolation and characterization of dominant female sterile mutations of Drosophila melanogaster. I. Mutations on the third chromosome. Genetics. 1989;122:111–127. doi: 10.1093/genetics/122.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW, Ephrussi B. Ovary transplants in Drosophila melanogaster: meiosis and crossing-over in superfemales. Proc Natl Acad Sci USA. 1937;23:356–360. doi: 10.1073/pnas.23.7.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Marsh JL, Gehring W. fs(1)K10, a germline-dependent female sterile mutation causing abnormal chorion morphology in Drosophila melanogaster. Wilhelm Roux's Arch Dev Biol. 1978;184:75–82. doi: 10.1007/BF00848670. [DOI] [PubMed] [Google Scholar]
- Poulton JS, Deng WM. Cell-cell communication and axis specification in the Drosophila oocyte. Dev Biol. 2007;311:1–10. doi: 10.1016/j.ydbio.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaliere V, Donati A, Hsouna A, Hsu T, Gargiulo G. dAkt kinase controls follicle cell size during Drosophila oogenesis. Dev Dyn. 2005;232:845–854. doi: 10.1002/dvdy.20333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, et al. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci USA. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaif A, Thomas A. Current concepts in the polycystic ovary syndrome. Annu Rev Med. 2001;52:401–419. doi: 10.1146/annurev.med.52.1.401. [DOI] [PubMed] [Google Scholar]
- Avellaira C, Villavicencio A, Bacallao K, Gabler F, Wells P, Romero C, et al. Expression of molecules associated with tissue homeostasis in secretory endometria from untreated women with polycystic ovary syndrome. Hum Reprod. 2006;21:3116–3121. doi: 10.1093/humrep/del183. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Cooley L. Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ. 2000;7:1071–1074. doi: 10.1038/sj.cdd.4400755. [DOI] [PubMed] [Google Scholar]
- Terashima J, Bownes M. Translating available food into the number of eggs laid by Drosophila melanogaster. Genetics. 2004;167:1711–1719. doi: 10.1534/genetics.103.024323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg CA. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Stravopodis DJ, Margaritis LH, Papassideri IS. Programmed cell death of follicular epithelium during the late developmental stages of oogenesis in the fruit flies Bactrocera oleae and Ceratitis capitata (Diptera, Tephritidae) is mediated by autophagy. Dev Growth Differ. 2006;48:189–198. doi: 10.1111/j.1440-169X.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- Thumm M, Kadowaki T. The loss of Drosophila APG4/AUT2 function modifies the phenotypes of cut and Notch signaling pathway mutants. Mol Genet Genomics. 2001;266:657–663. doi: 10.1007/s004380100585. [DOI] [PubMed] [Google Scholar]
- Erdelyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- Galanopoulos VK, Orr W, Szabad J, Kafatos FC. Genetic analysis of chorion formation in Drosophila melanogaster: I. The effects of one somatic-specific and seven germ-line-specific mutations. Dev Genet. 1989;10:87–97. doi: 10.1002/dvg.1020100204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.