Abstract
Hepatocellular carcinoma (HCC) is the third cause of cancer-related death worldwide. When these tumors are in advanced stages, few therapeutic options are available. Therefore, it is essential to search for new treatments to fight this disease. In this study, we investigated the effects of cannabinoids – a novel family of potential anticancer agents – on the growth of HCC. We found that Δ9-tetrahydrocannabinol (Δ9-THC, the main active component of Cannabis sativa) and JWH-015 (a cannabinoid receptor 2 (CB2) cannabinoid receptor-selective agonist) reduced the viability of the human HCC cell lines HepG2 (human hepatocellular liver carcinoma cell line) and HuH-7 (hepatocellular carcinoma cells), an effect that relied on the stimulation of CB2 receptor. We also found that Δ9-THC- and JWH-015-induced autophagy relies on tribbles homolog 3 (TRB3) upregulation, and subsequent inhibition of the serine–threonine kinase Akt/mammalian target of rapamycin C1 axis and adenosine monophosphate-activated kinase (AMPK) stimulation. Pharmacological and genetic inhibition of AMPK upstream kinases supported that calmodulin-activated kinase kinase β was responsible for cannabinoid-induced AMPK activation and autophagy. In vivo studies revealed that Δ9-THC and JWH-015 reduced the growth of HCC subcutaneous xenografts, an effect that was not evident when autophagy was genetically of pharmacologically inhibited in those tumors. Moreover, cannabinoids were also able to inhibit tumor growth and ascites in an orthotopic model of HCC xenograft. Our findings may contribute to the design of new therapeutic strategies for the management of HCC.
Keywords: cannabinoid, CB2 receptor, AMPK, autophagy, hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is one of the most common solid tumors and the third leading cause of cancer-related death worldwide.1 Its prognosis remains reserved, with a 5-year survival rate of <5%.2 It is the most common cause of death in patients with cirrhosis3 and, according to the World Health Organization, the incidence of HCC is expected to increase until 2030. The overall survival of patients with HCC has not significantly improved in the past two decades. Current treatments are only applicable at early stages of tumor development and include tumor resection, liver transplantation, chemoembolization and sorafenib administration.4 However, approximately half of the patients suffer tumor recurrence. The most important mechanism of liver cancer progression is cell proliferation. Although in recent years several clinical trials have tested the efficacy of agents that selectively target important signaling pathways involved in the control of this process, no relevant improvement in the prognostic/survival of patients with HCC has been achieved so far,5 and, therefore, it is necessary to identify novel therapeutic strategies for the management of HCC.
Cannabinoids are lipid mediators originally isolated from the hemp plant Cannabis sativa that produce their effects by activating primarily two G-protein-coupled receptors: cannabinoid receptor 1 (CB1), which is highly abundant in the brain, and cannabinoid receptor 2 (CB2), which is mainly expressed in non-neural tissues. Recently, numerous studies have evidenced the role of cannabinoids as potential anti-tumoral drugs owing to their ability to reduce tumor in different animal models, including glioma,6 breast cancer7, 8 and prostate cancer.9, 10 Recent research has also reported that the synthetic cannabinoid WIN-55 212-2 inhibits HCC growth.11, 12
It has been described that Δ9-tetrahydrocannabinol (Δ9-THC), the main active constituent of marijuana, triggers human glioma cell death through stimulation of an ER stress pathway that activates autophagy and promotes apoptosis.13, 14 Autophagy is a cellular self-digestive process whereby bulk cytoplasmic components and intracellular organelles are sequestered into double-membrane vesicles named autophagosomes and delivered for degradation to the lysosomes.8, 15, 16 In the liver, autophagy may have an important role in the regulation of energy balance for basic cell functions.17 Although functional autophagy acts as a metabolic stress buffer, many lines of evidence support a role for autophagy in antagonizing cell survival and in promoting cell death and apoptosis.18, 19, 20 Autophagy has an important role in cancer, and inhibition of this cellular process has been proposed to contribute to HCC progression,21, 22 and, therefore, it is a potentially very important target for liver cancer prevention and treatment.
This study was therefore undertaken to evaluate the potential anti-tumoral activity of cannabinoids in HCC and the mechanisms responsible for cannabinoid action in that devastating disease. We found that, both in cell cultures and in xenografted mice, Δ9-THC and the synthetic CB2 receptor-selective agonist JWH-015 promote human HCC death via autophagy stimulation. We also provide a molecular mechanism underlying CB2 receptor-mediated anti-tumoral signaling. These observations may pave the way to the design of novel therapeutic strategies for the treatment of hepatocellular carcinoma.
Results
Δ9-THC- and JWH-015-induced autophagy and apoptosis relies on CB2 receptor activation
To investigate the activity of cannabinoids on HCC cells, we first analyzed the effect of Δ9-THC (a CB1/CB2 receptor-mixed agonist that constitutes the main psychoactive ingredient of Canabis sativa) and JWH-015 (a CB2 receptor-selective agonist) on HepG2 (human hepatocellular liver carcinoma cell line) and HuH-7 (hepatocellular carcinoma cells) cells, two HCC lines that express CB1 and CB2 cannabinoid receptors (Supplementary Figure 1A). Treatment with Δ9-THC reduced the viability of HepG2 and HuH-7 cells, an event that was prevented by co-incubation with SR144528 (SR2, a CB2 receptor-selective antagonist), but not with SR141716A (SR1, a CB1 receptor-selective antagonist) (Supplementary Figure 1B). Likewise, JWH-015 decreased the viability of HCC cells, and co-incubation with SR2 abrogated this effect (Supplementary Figure 1B). These observations support that stimulation of CB2 receptors is responsible for the decrease of cell viability triggered by cannabinoids on HCC cells.
Δ9-THC and JWH-015 inhibit the growth of the human HCC lines HepG2 and HuH-7 via autophagy stimulation
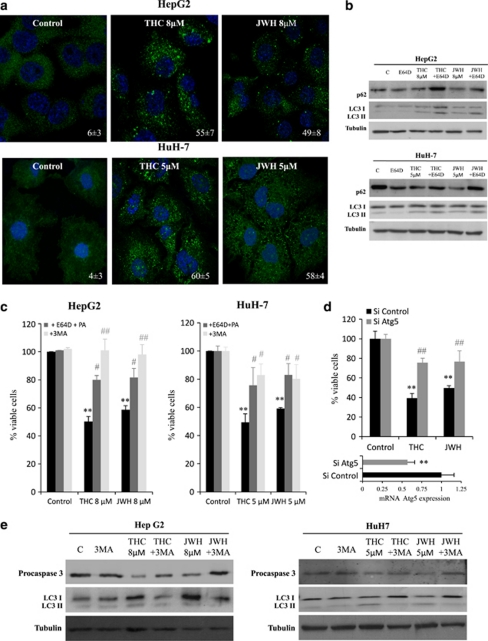
It has been recently shown that cannabinoids induce human glioma cell death via autophagy stimulation in vitro and in vivo.13, 23 We therefore examined whether Δ9-THC and JWH-015 activate a similar mechanism in HCC cells. Upon autophagy stimulation, the autophagy protein LC3 (microtubule-associated protein 1 light chain 3α) becomes conjugated to phosphatidylethanolamine (PE), which targets this protein to the membrane of the autophagosomes. The lipidated autophagosome-associated form of LC3 (LC3-II) can be monitored by immunofluorescence (autophagic cells exhibit a characteristic pattern of LC3 puncta) or western blot (LC3-II has higher electrophoretic mobility than non-lipidated LC3). Immunofluorescence analysis revealed that LC3 exhibited a punctuated distribution, consistent with its translocation to the autophagosome, in cells that had been treated with Δ9-THC or JWH-015 (Figure 1a). Likewise, incubation of HepG2 and HuH-7 cells with Δ9-THC or JWH-015 increased the levels of the lipidated form of LC3 (Figure 1b). Furthermore, pharmacological inhibition with E64d and pepstatin A (PA) of lysosomal proteases (the enzymes responsible for the degradation of the autophagosome content after fusion with the lysosome) enhanced the accumulation of LC3-II (as well as of the autophagosome cargo p62) in cells that had been treated with THC or JWH-015, thus supporting the fact that cannabinoid treatment leads to dynamic autophagy in HCC cells (Figure 1b).
Figure 1.
Δ9-Tetrahydrocannabinol (Δ9-THC) and JWH-015 treatment induces autophagy in hepatocellular carcinoma (HCC) cells. (a) HepG2 and HuH-7 cells were treated with Δ9-THC or JWH-015 for 24 h and microtubule-associated protein 1 light chain 3α (LC3) was detected by confocal immunofluorescence. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Values on the lower right corner of each panel correspond to the number of cells with LC3 dots relative to the total number of cells (n=5; mean±standard deviation (S.D.)). (b) Immunoblot analysis of p62, LC3-I and LC3-II levels after Δ9-THC or JWH-015 treatment in the presence of the lysosomal protease inhibitors E64d (2.5 μg/ml) and pepstatin A (5 μg/ml; PA) for 24 h. Tubulin levels are shown as loading control. (c) HepG2 and HuH-7 cells were treated either with 8 μM Δ9-THC or 8 μM JWH-015 in the presence of 2.5 μg/ml E64d and 5 μg/ml PA or 1 μM 3-methyladenine (3MA) for 48 h and cell viability was analyzed by 3-[4,5-dimethylthiazolyl-2] 2,5-diphenyl-tetrazolium bromide (MTT) test. Data are the mean±S.D. of three different experiments each performed in triplicate (**P<0.01 versus control; #P<0.05 and ##P<0.01 versus cannabinoid-treated cells). (d) Effect of Δ9-THC or JWH-015 on the viability – as determined by the MTT test (48 h) of HepG2 cells transfected with Atg5-selective (small interfering (si)Atg5) or control (siC) siRNA. Data correspond to the mean±S.D. of three different experiments each performed in triplicate (**P<0.01 versus control; ##P<0.01 versus cannabinoid-treated cells). Atg5 mRNA levels (mean of the three experiments) assessed by real-time polymerase chain reactions (PCRs) are shown in the lower panel. (e) HepG2 or HuH-7 cells were incubated either with Δ9-THC or JWH-015 for 30 h in the presence of 1 μM 3-MA and levels of procaspase-3 and LC3 were detected by western blot. Tubulin levels are shown as loading control. The image is representative of three different experiments
Next, we investigated whether autophagy was directly involved in the mechanism of cannabinoid-induced cell death. As shown in Figure 1c, cell death was inhibited when autophagy was pharmacologically blocked at a very early stage (by incubation with 3-methyladenine (3-MA), an inhibitor of Vps34, a class III phosphatidyl inositol-3-kinase that has a crucial role in autophagy initiation24) or at a final stage (by incubation with E64d and PA). Likewise, knock down of Atg5 (an essential autophagy gene that is part of one of the two protein conjugation systems required for autophagosome elongation25, 26) impaired THC- or JWH-015-induced cell death (Figure 1d). Taken together, these observations strongly support that autophagy is required for cannabinoid-induced HCC cell death.
Many lines of evidence indicate that there is a cross-talk between autophagy and apoptosis.27 To investigate whether cannabinoid-induced autophagy was involved in apoptosis induction, HepG2 and HuH-7 cells were incubated with either Δ9-THC or JWH-015 in the presence of the 3-MA, and levels of procaspase-3 were detected by immunoblot. As shown in Figure 1e, pre-incubation with 3-MA prevented the cleavage of procaspase-3, suggesting that autophagy induction by cannabinoids was previous to and necessary for apoptosis.
AMPK activation and TRB3 upregulation are involved in Δ9-THC- and JWH-015-induced autophagy and apoptosis of HCC cells
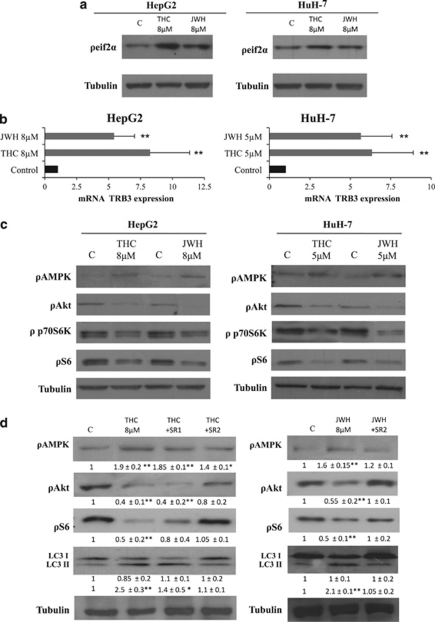
The mechanisms of autophagy stimulation by cannabinoids in glioma and other types of cancer cells relies on the stimulation of an ER stress-related pathway, which leads to the upregulation of the pseudokinase tribbles homolog 3 (TRB3). This latter protein interacts with serine–threonine kinase Akt (Akt) and promotes the inhibition of the mammalian target of rapamycin C1 (mTORC1) complex, which leads to autophagy stimulation.23 As shown in Figure 2, THC and JWH-015 increased the phosphorylation of the α-subunit of the eukaryotic translation initiation factor 2 (eIF2α, a hallmark of the ER stress response; Figure 2a), increased TRB3 levels (Figure 2b) and decreased the phosphorylation of Akt, p70S6 kinase (a well-established substrate of mTORC1) and the ribosomal protein S6 (a target of p70S6 kinase) in HepG2 and HuH-7 cells (Figure 2c). Furthermore, selective knockdown of TRB3 abrogated cannabinoid-induced inhibition of the Akt/mTOR pathway, autophagy and cell death (Supplementary Figure 2), thus supporting the fact that the mechanism by which cannabinoids promotes glioma cell death also operates in HCC cells.
Figure 2.
Δ9-Tetrahydrocannabinol (Δ9-THC) and JWH-015 upregulate tribbles homolog 3 (TRB3), inhibit the serine–threonine kinase Akt/mammalian target of rapamycin C1 (Akt/mTORC1) pathway and activate adenosine monophosphate-activated kinase (AMPK) through cannabinoid receptor 2 (CB2) receptors. (a) Effect of Δ9-THC or JWH-015 (8 h) on the phosphorylation of eIf2α in HepG2 and HuH7 cells. Tubulin levels are shown as a loading control. The image is representative of three different experiments (b) Effect of Δ9-THC or JWH-015 (24 h) on tribbles homolog 3 (TRB3) mRNA levels (as determined by quantitative polymerase chain reaction (qPCR)) of HepG2 and HuH7 cells (n=4; **P<0.01). (c) Effect of Δ9-THC or JWH-015 (24 h) in the phosphorylation of AMPK, Akt, p70S6K and S6 of HepG2 and HuH7 cells. Tubulin levels are shown as a loading control. The image is representative of three different experiments. (d) Effect of Δ9-THC (8 μM), JWH-015 (8 μM), 1 μM SR141716A (SR1) or 2 μM SR 144528 (SR2) (24 h) on AMPK, Akt and S6 phosphorylation, as well as microtubule-associated protein 1 light chain 3 alpha (LC3) lipidation of HepG2 cells. Tubulin levels are shown as loading control. The image is representative of five different experiments. Optical density (O.D.) values (mean±standard deviation (S.D.) of the five experiments; *P<0.05 and **P<0.01 versus control) are shown under the each image
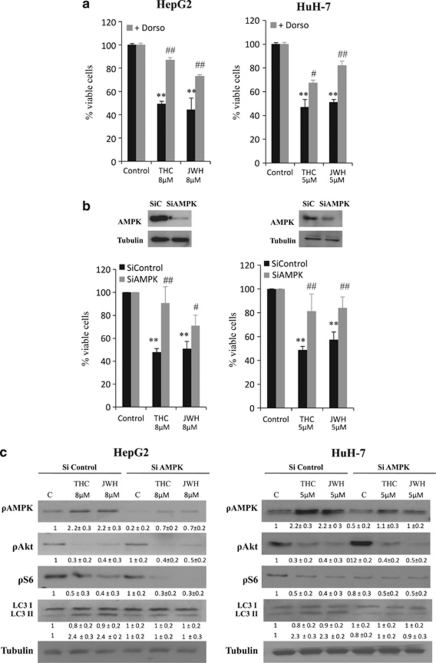
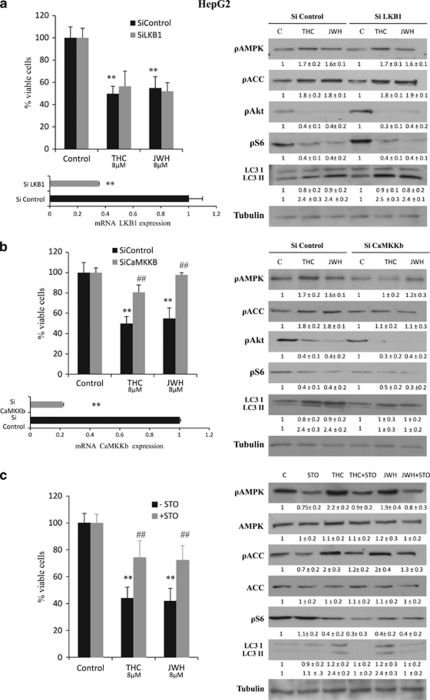
Of note, we observed that treatment of HepG2 and HuH-7 cells with THC or JWH-015 increased the phosphorylation of adenosine monophosphate-activated protein kinase (AMPK), an important intracellular nutrient status sensor that has been proposed to have a critical role in the regulation of autophagy as induced by hypoxia or nutrient deprivation (Figure 2b). In addition, pharmacological blockade of CB2 receptors with SR2 abrogated the effect of Δ9-THC and JWH-015 on Akt, S6 and AMPK phosphorylation, as well as autophagy (Figure 2d and Supplementary Figure 1C). We therefore asked whether AMPK may also have a role in the regulation of the antiproliferative effect evoked by cannabinoids in HCC cells. In line with this notion, when AMPK was pharmacologically blocked with dorsomorphin (Figure 3a) or genetically inhibited with small interfering RNA (siRNA) (Figure 3b), HepG2 and HuH-7 cells were more resistant to cannabinoid-induced cell death. Likewise, AMPK knockdown prevented LC3 lipidation (Figure 3c and Supplementary Figure 2C). These observations support the fact that activation of AMPK is necessary for the stimulation of autophagy-mediated cell death by cannabinoids in HCC cells.
Figure 3.
Δ9-Tetrahydrocannabinol (Δ9-THC) and JWH-015 induce autophagy via adenosine monophosphate-activated kinase (AMPK). (a) HepG2 and HuH-7 cells were incubated with Δ9-THC or JWH-015 for 48 h in the presence of 0.5 μM dorsomorphin and cell viability was assayed by 3-[4,5-dimethylthiazolyl-2] 2,5-diphenyl-tetrazolium bromide (MTT). Data are the mean±standard deviation (S.D.) of three different experiments, each performed in triplicate (**P<0.01 versus control; #P<0.05 and ##P<0.01 versus cannabinoid-treated cells). (b) HepG2 and HuH-7 cells transfected either with small interfering (si)C or AMPKα-selective siRNA (siAMPK) were incubated with Δ9-THC or JWH-015 for 48 h and cell viability was assayed by MTT. Data correspond to the mean±S.D. of five different experiments (**P<0.01 versus control; #P<0.05 and ##P<0.01 versus cannabinoid-treated cells). AMPKα levels of a representative experiment, assessed by western blot are shown in the upper panel. (c) Effect of Δ9-THC or JWH-015 on AMPK, Akt and S6 phosphorylation, as well as microtubule-associated protein 1 light chain 3α (LC3) lipidation (24 h) of HepG2 and HuH-7 cells transfected with siC or siAMPK . Tubulin levels are shown as loading control. A representative western blot of three different experiments is shown
AMPK and TRB3 regulate cannabinoid-induced autophagy of HCC cells through different mechanisms
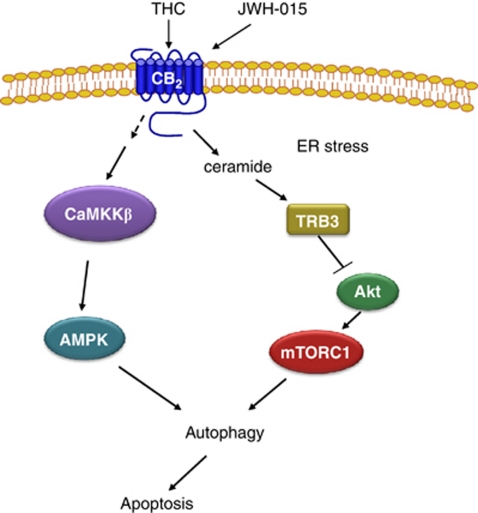
AMPK has been shown to inhibit mTORC1.28 Unlike Akt, AMPK activates tuberous sclerosis complex 2 (TSC2), a GTPase-activating protein responsible for the blockade of mTORC1.28 Therefore, we next studied whether this was the mechanism by which AMPK stimulated autophagy in our system. As shown in Figure 3c and Supplementary Figure 2C, AMPK silencing abrogated the effect of Δ9-THC and JWH-015 in the phosphorylation of AMPK and its downstream target acetyl-CoA carboxylase (ACC), as well as on autophagy, but did not modify the effect of cannabinoid treatment on the phosphorylation of Akt, TSC2 (data not shown) or the mTORC1-related substrates 4EBP1 and S6. By contrast, TRB3 knockdown did not affect AMPK or ACC phosphorylation, but did inhibit the cannabinoid-induced decrease in Akt, TSC2, 4EBP1 and S6 phosphorylation and LC3 lipidation. Likewise, pharmacological blockade of ceramide biosynthesis by using ISP-1 (a pharmacological inhibitor of serine palmitoyltransferase, one of the upstream events that trigger TRB3 upregulation in response to cannabinoid treatment29) abrogated the effect of Δ9-THC on Akt, TSC2, 4EBP1 and S6 phosphorylation, as well as LC3 lipidation, but did not affect AMPK phosphorylation. These observations support that the cannabinoid-evoked stimulation of autophagy on HCC cells relies on two different mechanisms: (i) inhibition of the Akt/mTORC1 axis via TRB3 upregulation and (ii) stimulation of AMPK. (A scheme is shown in Figure 5.)
Figure 5.
Schematic of the proposed mechanism of cannabinoid-induced hepatocellular carcinoma (HCC) cell death. Cannabinoid treatment stimulates autophagy via two different mechanism: (i) upregulation of tribbles homolog 3 (TRB3) and subsequent inhibition of the serine–threonine kinase Akt/mammalian target of rapamycin C (Akt/mTORC1) axis, and (ii) activation of adenosine monophosphate-activated kinase (AMPK) via CaMKKβ. Stimulation of autophagy by cannabinoids leads to HCC apoptosis and cell death
Activation of AMPK by cannabinoids relies on CAMKK
We next investigated the mechanism by which cannabinoids activate AMPK. Among the different kinases proposed to act as AMPKKs, the human tumor suppressor liver kinase B1 (LKB1) and the calmodulin-activated kinase kinase (CaMKK) are now widely accepted as the most relevant ones.30 Inhibition of LKB1 expression with siRNA did not have any significant effect on the viability of cannabinoid-treated HepG2 (Figure 4a) or HuH7 (Supplementary Figure 3) cells. Likewise, the effect of cannabinoid treatment on AMPK activation, Akt/mTORC1 pathway inhibition and autophagy (LC3 lipidation) was not affected by LKB1 silencing (Figure 4a), supporting the fact that LKB1 is not involved in cannabinoid-induced AMPK activation and autophagy in HCC cells. By contrast, selective knockdown of CaMKKβ or incubation with the CaMKK pharmacological inhibitor STO609 prevented the cannabinoid-evoked decrease in HCC cell viability (Figures 4b and c and Supplementary Figure 3), increase in AMPK and ACC phosphorylation (Figures 4b and c) and autophagy (Figures 4b and c), indicating that AMPK activation by cannabinoids in HCC cells relies on CaMKKβ. Of note, genetic or pharmacological blockade of CaMKKβ did not modify the inhibition of the Akt/mTORC1 pathway evoked by these agents, which again supports the fact that mTORC1 inhibition by cannabinoids in HCC occurs independently of AMPK activation (Figures 4b and c).
Figure 4.
Δ9-Tetrahydrocannabinol (Δ9-THC) and JWH-015 activate adenosine monophosphate-activated kinase AMPK via CaMKKβ. (a) Left panel: Effect of Δ9-THC (8 μM) or JWH-015 (8 μM) on the viability (48 h; as determined by the 3-[4,5-dimethylthiazolyl-2] 2,5-diphenyl-tetrazolium bromide (MTT) test) of HepG2 cells transfected with small interfering (si)C or liver kinase B1 (LKB1)-selective (siLKB1) siRNA. Data correspond to the mean±standard deviation (S.D.) of four different experiments, each performed in quadruplicate (**P<0.01 versus control). Lower panel: LKB1 mRNA levels (mean of the four experiments as determined by real-time quantitative polymerase chain reaction (PCR)) of HepG2 cells transfected with siC or siLKB1. Right panel: Effect of Δ9-THC or JWH-015 (24 h) on the AMPK, ACC, Akt, S6 phosphorylation and microtubule-associated protein 1 light chain 3α (LC3) lipidation of siC- and siLKB1-transfected HepG2 cells. Tubulin levels are shown as loading control. A representative western blot of four different experiments is shown. Optical density (O.D.) values (mean±S.D. of the four experiments) are shown under the each image. (b) Effect of Δ9-THC (8 μM) or JWH-015 (8 μM) on the viability (48 h, as determined by the MTT test) of HepG2 cells transfected with siC- or CaMKKβ-selective (siCaMKKb) siRNA. Data correspond to the mean±S.D. of four different experiments, each performed in quadruplicate (**P<0.01 versus control; ##P<0.01 versus cannabinoid-treated cells). Lower panel: CaMKKβ mRNA levels (mean of the four experiments as determined by real-time quantitative PCR) of HepG2 cells transfected with siC or siCaMKKb. Right panel: Effect of Δ9-THC or JWH-015 (24 h) on AMPK, ACC, Akt and S6 phosphorylation and LC3 lipidation of siC- and siCaMKKb-transfected HepG2 cells. Tubulin levels are shown as loading control. A representative western blot of three different experiments is shown. O.D. values (mean±S.D. of the four experiments) are shown under the each image. (c) Effect of Δ9-THC (8 μM) or JWH-015 (8 μM) on the viability (48 h, as determined by the MTT test) of HepG2 cells incubated in the presence or absence of the 10 μM STO609 (STO; a CaMKKα/β inhibitor). Data correspond to the mean±S.D. of four different experiments, each performed in quadruplicate (**P<0.01 versus control; ##P<0.01 versus cannabinoid-treated cells). Right panel: Effect of Δ9-THC, JWH-015 and STO (10 μM) on AMPK, ACC, Akt and S6 phosphorylation and LC3 lipidation (24 h). Tubulin levels are shown as loading control. A representative western blot of four different experiments is shown. O.D. values (mean±S.D. of the four experiments)
Autophagy is required for Δ9-THC and JWH-015 anti-tumoral action in human HCC xenografts
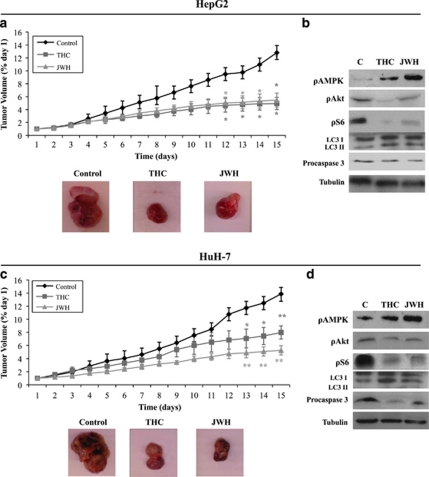
To investigate the ability of Δ9-THC and JWH-015 to inhibit HCC growth in vivo, we first generated tumor xenografts by subcutaneous inoculation of HepG2 or HuH-7 cells in nude mice. Mice were daily treated with vehicle (control), 15 mg/kg Δ9-THC or 1.5 mg/kg JWH-015 for 15 days. As shown in Figure 6a, cannabinoid administration almost totally blocked the growth of HepG2 cell-derived tumors. Moreover, treatment with Δ9-THC or JWH-015 enhanced AMPK phosphorylation and reduced Akt phosphorylation, which was accompanied by a decrease of S6 phosphorylation and an increase of LC3-II lipidation (Figure 6b). Likewise, procaspase-3 levels were also decreased in cannabinoid-treated tumors (Figure 5b). Similar results were obtained with HuH-7 cell-derived tumors, in which Δ9-THC or JWH-015 administration also decreased tumor growth (Figure 6c), increased the AMPK phosphorylation, decreased Akt and S6 phosphorylation and enhanced LC3 lipidation and caspase-3 activation (Figure 6d).
Figure 6.
Δ9-Tetrahydrocannabinol (Δ9-THC) and JWH-015 reduce the growth of HepG2- and HuH-7 cell-derived tumor xenografts. Athymic nude mice were injected subcutaneously (s.c.) in the right flank with HepG2 cells (a and b) or HuH-7 cells (c and d). When tumors reached a 150 mm3 size, mice were daily treated during 15 days with vehicle (control), 15 mg/kg Δ9-THC or 1.5 mg/kg JWH-015. Tumor volumes were measured daily. (a and c) Tumor growth curve after administration of vehicle (diamonds), Δ9-THC (squares) or JWH-015 (triangles). Results represent the mean±standard error of mean (S.E.M.) of eight mice in each group. *P<0.01 versus control compared by Student's t-test. A representative image of the dissected tumors after treatment is shown. (b and d) Immunoblot analysis of adenosine monophosphate-activated kinase (AMPK), Akt and S6 phosphorylation, microtubule-associated protein 1 light chain 3α (LC3) lipidation and active-caspase-3 levels in the dissected tumors. Western blots analyses of one representative tumor for each condition are shown
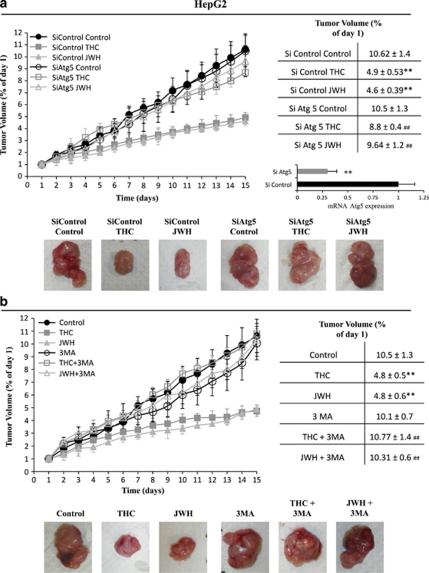
To further examine the role of autophagy on the anti-tumoral action of cannabinoids, another set of experiments was conducted to analyze the effect of cannabinoids on the growth of HepG2 tumor xenografts in which Atg5 expression had been knocked down in vivo. As shown in Figure 7a, Δ9-THC and JWH-015 failed to inhibit the growth of Atg5-silenced tumors, but not of those tumors that had been transfected with control siRNA. Furthermore, pharmacological inhibition of autophagy by using 3-MA prevented the decrease in tumor growth evoked by Δ9-THC and JWH-015 (Figure 7b). Taken together, these findings strongly support the fact that autophagy is necessary for the anti-tumoral action of cannabinoids in hepatocellular carcinoma.
Figure 7.
Autophagy is required for the anti-tumoral action of Δ9-tetrahydrocannabinol (Δ9-THC) and JWH-015 on hepatocellular carcinoma (HCC) tumor xenografts. (a) Athymic nude mice were injected subcutaneously (s.c.) in the right flank with HepG2 cells. When tumors reached a 150 mm3 size, mice daily treated during 15 days with vehicle (control), 15 mg/kg Δ9-THC or 1.5 mg/kg JWH-015. Tumors were injected with atelocollagen complexed with control RNA or atelocollagen complexed with small interfering (si)Atg5 in days 1 and 7 of the treatment. Tumor volumes were measured daily. Tumor growth curves and final tumor volumes after administration of the treatments are shown. Results represent the mean±standard error of mean (S.E.M.) of eight mice in each group. **P<0.01 versus control and ##P<0.01 versus siControl compared by Student's t-test. Expression levels of Atg5 in siC and siATG5 tumors at the end of the treatment was examined by real-time polymerase chain reaction (PCR). A representative image of the dissected tumors after the treatments is shown. (b) Athymic nude mice injected s.c. in the right flank with HepG2 cells were daily treated during 15 days with vehicle (control) (filled circles), 15 mg/kg Δ9-THC (filled squares), 1.5 mg/kg JWH-015 (filled triangles), vehicle plus 1 mg/kg 3-MA (open circles), 15 mg/kg Δ9-THC plus 1 mg/kg 3-MA (open squares) or 1.5 mg/kg JWH-015 plus 1 mg/kg 3-MA (open triangles). Tumor growth curves and final tumor volumes after administration of the treatments are shown. Results represent the mean±S.E.M. of eight mice in each group. **P<0.01 versus control and ##P<0.01 versus cannabinoid-treated tumors compared by Student's t-test. A representative image of the dissected tumors after the treatments is shown
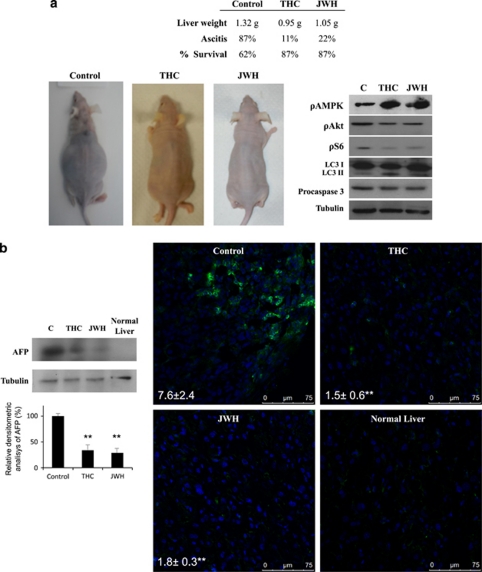
Finally, we tested the anti-tumoral efficacy of cannabinoids in an orthotopic HCC model. HepG2 cells were inoculated in the liver of nude mice, and after 1 week, mice were treated intraperitoneally with vehicle (control), 15 mg/kg Δ9-THC or 1.5 mg/kg JWH-015 for 10 days. As shown in Figure 8a, cannabinoid treatment almost completely prevented hepatomegaly and ascites. Moreover, levels of the HCC tumor marker α-fetoprotein (AFP) were dramatically reduced in the livers of animals treated with Δ9-THC or JWH-015 (Figure 8b). Analysis of tumor samples revealed that cannabinoid treatment enhanced AMPK phosphorylation and inhibited Akt and S6 phosphorylation. Furthermore, THC and JWH-015 enhanced autophagy and apoptosis in these tumors (Figure 8a).
Figure 8.
Anti-tumoral effect of Δ9-tetrahydrocannabinol (Δ9-THC) and JWH in an orthotopic transplantation tumor model of hepatocellular carcinoma (HCC). The orthotopic transplantation HCC model was established by intrahepatic implanting of HepG2 cells. At 1 week after injection, mice were daily treated intraperitoneally (i.p.) with vehicle (control), 15 mg/kg Δ9-THC or 1.5 mg/kg JWH-015 for 10 days. (a) Effect of the different treatments on liver weight and ascites development. Representative images of mice at the end of the treatment are shown. Immunoblot analysis of adenosine monophosphate-activated kinase (AMPK), Akt and S6 phosphorylation, microtubule-associated protein 1 light chain 3α (LC3) lipidation and active-caspase-3 levels in the dissected tumors. Western blots analyses of one representative tumor for each condition is shown. (b) Effect of the different treatments on α-fetoprotein levels (as determined by western Blot – left panel) and immunofluorescence (right panel) of the dissected livers are shown. A normal liver is shown for comparison
Taken together, these observations robustly support the fact that cannabinoid anti-tumoral action in HCC relies on AMPK stimulation, Akt inhibition and activation of autophagy in HCC cells both in vitro and in vivo.
Discussion
In this study, we show that the natural cannabinoid Δ9-THC and the CB2 receptor-selective agonist JWH-015 inhibit HCC cell growth via stimulation of autophagy. Importantly, although the human HCC cell lines used (HepG2 and HuH-7) expressed both CB1 and CB2 receptors, only CB2 activation was involved in the pro-autophagic and antiproliferative effect induced by cannabinoids on these cells. This is in line with the recent observation that the synthetic cannabinoid WIN-55, 212-2-induced apoptosis in HepG2 cells in a process that was partially inhibited by the CB2 receptor-selective antagonist AM630.11 Moreover, it has been previously shown that CB2 receptors are overexpressed in HCC and correlate with good prognosis.31 Those findings, together with ours, support the fact that stimulation of CB2 receptors could be a new therapeutic strategy to promote HCC death.
Our study shows that the mechanism of cannabinoid anti-tumoral action in HCC relies on the stimulation of autophagy and the subsequent activation of apoptosis. Depending on the physiopathological setting, autophagy has been proposed to protect from apoptosis, act as an apoptosis-alternative pathway to induce cell death or act together with apoptosis as a combined mechanism for cell death.32 However, very little is known about the role that the interchange between these two cellular processes have in the control of tumor growth in response to anticancer agents. Our observations are in line with previous results obtained in human glioma cells13 and support the fact that stimulation of autophagy in response to cannabinoid treatment leads to apoptosis. Nevertheless, further research is still necessary to clarify the precise mechanisms linking both cellular processes upon cannabinoid treatment.
Stimulation of autophagy in many cellular settings relies on the inhibition of the mTORC1 complex, which have a central role in the control of protein synthesis, cell growth and cell proliferation through the regulation of several downstream targets. As a result of its central position in the control of cellular homeostasis, mTORC1 integrates signals from different inputs. One of the most important upstream regulators of mTORC1 is the pro-survival kinase Akt, which phosphorylates and inactivates TSC2 (an inhibitor of the mTORC1 activator Rheb) and PRAS-40. Thus, Akt activation stimulates mTORC1 and inhibits autophagy. In this work, we found that cannabinoid treatment of HCC cells leads to Akt and mTORC1 inhibition, which is in agreement with our recent studies in glioma cells.13 Thus, it had been previously shown that inhibition of Akt/mTORC1 pathway by cannabinoids relies on the stimulation of an ER stress-related pathway, which leads to the upregulation of the pseudokinase TRB3, the inhibition of the Akt/mTORC1 axis and the induction of autophagy.13, 23 In this study, Δ9-THC and JWH-015 promoted ER stress and increased TRB3 expression. In addition, Akt/mTORC1 inhibition and autophagy were abolished when ceramide biosynthesis was inhibited or when TRB3 expression was silenced, thus suggesting that this could be a general mechanism of cannabinoid anti-tumoral action. Of importance, we also found that Δ9-THC and JWH-015 activate AMPK in HCC cells and that pharmacological or genetic inhibition of this kinase has a similar inhibitory effect on cannabinoid-induced cell death and autophagy. AMPK has been shown to negatively regulate mTORC1 via TSC2 activation, which also leads to autophagy stimulation,33 and therefore we asked whether cannabinoids also inhibit mTORC1 through this mechanism in HCC cells. In disagreement with this possibility, our data show that – unlike TRB3 silencing – AMPK knockdown does not prevent cannabinoid-induced mTORC1 inhibition. In addition, knock down of TRB3 does not affect the stimulation of AMPK by cannabinoids. These observations suggest that TRB3 and AMPK (i) are activated by different mechanisms in response to cannabinoid treatment, and (ii) regulate autophagy acting at different stages.
Two converging pathways have been described for AMPK regulation: one directed by LKB1, dependent on a change in cellular AMP, and another one directed by CaMKKs, dependent on changes in intracellular Ca2+.34 The dramatic reduction in phospho-AMPK and phospho-ACC obtained upon silencing of CaMKKβ indicates that this latter kinase rather than LKB1 is the dominant AMPKK enzyme in HCC cells in response to cannabinoids. Thus, cannabinoids induce autophagy in HCC cells, possibly by a two-pronged mechanism, one prong (similar to that operating in glioma cells) involving ER stress, TRB3 and Akt/mTORC1 inhibition, and another one reliant on AMPK stimulation via CaMKKβ. A model of this mechanism of cannabinoid action in HCC cells is depicted in Figure 5.
In addition, it has been recently shown that AMPK binds to and directly phosphorylates the Ser/Thr kinase ULK1, the mammalian ortholog of the yeast protein kinase Atg1, and that this phosphorylation is required for ULK1-mediated autophagy.35, 36 Thus, under certain cellular settings, mTORC1 inhibition and AMPK activation may cooperate to trigger autophagy.36, 37, 38 Although future research is needed to completely clarify this point, our data suggest that this could be the mechanism by which cannabinoids trigger autophagy in HCC cells.
To note, it has been recently described that mTOR signaling has a critical role in the pathogenesis of HCC and that mTOR inhibitors have antineoplastic activity in experimental models of HCC.39 Moreover, decreased autophagy in HCC correlates with a more aggressive cancer cell phenotype and poor prognosis.21, 40 Here we found that cannabinoid treatment reduces the growth of two different models of HCC subcutaneous xenografts in concert with decreased mTORC1 activation, enhanced AMPK phosphorylation and increased autophagy and apoptosis in those tumors. Moreover, knock down of the autophagic gene Atg5 as well as pharmacological inhibition of autophagy dramatically abolished the anti-tumoral activity of cannabinoids against subcutaneous HCC xenografts. Furthermore, Δ9-THC and JWH-015 efficiently reduced ascites development and AFP expression in an orthotopic model of HCC, which also paralleled mTORC1 inhibition, AMPK activation and autophagy stimulation in those tumors. Our data represent the first evidence for the antiproliferative action of cannabinoids in HCC cells in vivo and support that the ability of cannabinoids to inhibit mTORC1, stimulate AMPK and enhance autophagy could be therapeutically exploited for the management of HCC.
Materials and Methods
Reagents
Δ9-THC was obtained from GW Pharm GmbH (Frankfurt, Germany) and JWH-015 was purchased to Sigma (St. Louis, MO, USA). The CB1 antagonist SR-141716 and the CB2 antagonist SR-144528 were kindly provided from Sanofi-Synthelabo (Paris, France). The anti-LC3 polyclonal antibody was obtained from MBL International (Woburn, MA, USA) and the anti-pS6, pAKT-ser473, pACC, ACC, peIF2α, pAMPK and AMPK polyclonal antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA). The anti-caspase-3 antibody and 3-MA were purchased to Sigma. The inhibitors E64d and PA were purchased to Roche Diagnostics (Mannheim, Germany). The CaMKKα/β inhibitor STO609 was purchased from Sigma. Atelocollagen (AteloGene) was purchased to Cosmo Bio Co. (Tokyo, Japan). All the other chemicals were obtained from Sigma.
Cell cultures
Human HCC HepG2 cells (ATCC, HB-8065) (Rockville, MD, USA) were cultured according to suppliers. The human hepatoma cell line HuH-7 was kindly supply by Dr. Lisardo Boscá (IKnstituto de Investigaciones Biomédicas Alberto Sols, Madrid, Spain). Cells were routinely growth in DMEM/10% FBS supplemented with 1% non-essential amino acids and 100 IU/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, 0.25 μg/ml amphotericin B (Invitrogen, Paisley, UK). One day before the experiments, the medium was changed to 0.5% FBS medium. Experiments were carried out when cell monolayers were 80% confluent.
RT-PCR analysis
Total RNA was isolated from cells by Trizol Reagent from Gibco (Invitrogen, Carlsbad, CA, USA) according to manufacturer's protocol. One microgram total RNA was retrotranscribed to cDNA with the M-MLV Reverse transcriptase kit (Life Technologies, Carlsbad, CA, USA). Two microliters of RT reaction were then PCR amplified with specific primers for CB1: sense primer, 5′-TATATTCTCTGGAAGGCTCACAGCC-3′ antisense primer, 5′-GAGCATA CTGCAGAATGCAAACACC-3′(for amplification of a 270-bp product for human CB1); and CB2: sense primer, 5′-TTTCCCACTGATCCCCAATG-3′ antisense primer, 5′-GAGCATACTGCAGAATGCAAACACC-3′ (for amplification of a 333-bp product for human CB2). PCR products were analyzed by electrophoresis on ethidium bromide-stained 2% agarose gels and DNA was detected by exposure under UV light.
Western blot
After different treatments according to the experiments cells were lysed in ice-cold lysis buffer (50 mM Tris (pH 7.4), 0.8 M NaCl, 5 mM MgCl2, 0.1% Triton X-100, 1 mM PMSF, 10 μg/ml soybean trypsin inhibitor, 1 μg/ml aprotinin and 5 μg/ml leupeptin), and cleared by microcentrifugation. Equivalent protein amounts of each sample were separated on SDS-PAGE gels and blotted to PVDF transfer membrane. After blocking with 5% skim dried milk, immunoblot analysis was performed followed by enhanced chemo luminescence detection.
Cell viability assay
Cells in logarithmic phase were cultured at a density of 5000 cells per cm2 in a 12-well plate. The cells were exposed to various concentrations of Δ9-THC and JWH-015 for indicated times. The 3-[4,5-dimethylthiazolyl-2] 2,5-diphenyl-tetrazolium bromide (MTT) cell viability assay was used to evaluate the effects of cannabinoids on cell growth and to determine the IC50.
Confocal microscopy
After 48 h in culture, the cells were fixed in 4% paraformaldehyde in PBS and incubated with 0.1% Triton X-100 for permeabilization. Immunolabeling with the anti-LC3 polyclonal antibody was performed by incubation at room temperature for 1 h. Secondary labeling was performed with Alexa Flour 594, conjugated to anti-rabbit IgG and Alexa Flour 488 (Invitrogen). Imaging was with a Leica TCS SP5 laser-scanning confocal microscope with LAS-AF imaging software, using a 63 × oil objective.
siRNA transfections
Cells were seeded at 2 × 105 cells/35 mm well the day before transfection. Cells were then transfected in 1 ml OPTIMEN containing 4 μg Lipofectamine 2000 (Invitrogen), with 100 nM siRNA duplexes or control scrambled RNA according to the manufacturer's protocols. At 24 h after transfection, the medium was removed and replaced for DMEM containing 10% fetal bovine serum. Cells were then treated with cannabinoids for 48 h and used for MTT cell viability assays.
For each transfection, the following sequences were used: Atg 5 sense sequence, 5′-GUGAGAUAUGGUUUGAAUAdTdT-3′ α1 subunit AMPK first sense sequence, 5′-CCCAUAUUAUUUGCGUGUAdTdT-3′ second sense, sequence 5′-GAATCCTGTGACAAGCACAdTdT-3′ sense sequences of the Dharmacon Smart Pool, 5′-CCAUACCCUUGAUGAAUUAUU-3′ 5′-GCCCAGAGGUAGAUAUAUGUU-3′ 5′-GAGGAUCCAUCAUAUAGUUUU-3′ and 5′-ACAAUUGGAUUAUGAAUGGUU-3′. TRB3 sense sequence 5′-GUGCGAAGCCGCCACCGUAdTdT-3′ LKB1 sense sequence, 5′-GUACUUCUGUCAGCUGAUUdTdT-3′ CaMKKβ sense sequence, 5′-GCUCCUAUGGUGUCGUCAAdTdT-3′ (Sigma).
Real-time quantitative PCR
cDNA was obtained from cells using Transcriptor (Roche Applied Science, Mannheim, Germany). Real-time quantitative (qPCR) assays were performed using the FastStart Universal Probe Master mix with Rox (Roche Applied Science), and probes were obtained from the Universal ProbeLibrary Set (Roche Applied Science); Atg 5 sense primer, 5′-GACGCTGGTAACTGACAAAGTGA-3′ Atg 5 antisense primer, 5′-TAGGAGATCTCCAAGGGTATGCA-3′ TRB3 sense primer, 5′-GCCACTGCCTCCCGTTCTTG-3′ TRB3 antisense primer, 5′-GCTGCCTTGCCCGAGTATGA-3′ LKB1 sense primer, 5′-GGCATGCAGGAAATGCTGGACAGC-3′ LKB1 antisense primer, 5′-GTGTCCAGGCCGTTGGCAATCTCG-3′ CaMKKβ sense primer, 5′-TCGAGTACTTGCACTGCCAGAAGATC-3′ CaMKKβ antisense primer, 5′-GGGGTTCTTGTCCAGCATACGGGT-3′ 18S sense primer, 5′-GCTCTAGAATTACCACAGTTATCCAA-3′ 18S antisense primer, 5′-AAATCAGTTATGGTTCCTTTGGTC-3′. Amplifications were run in a 7900 HT-Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Each value was adjusted by using 18S RNA levels as a reference.
Animal care and handling
Athymic nude (nu/nu) 5-week-old male mice were obtained from Harlan Iberica Laboratory (Barcelona, Spain) and maintained under specific pathogen-free conditions with the approval of the Institutional Animal Care and Use Committee of Alcala University. All animal studies were conducted in accordance with the Spanish institutional regulation for the housing, care and use of experimental animals and met the European Community directives regulating animal research. Recommendations made by the United Kingdom coordinating Committee on Cancer Research (UKCCCR) have been kept carefully.
In vivo studies
To study the in vivo antitumor activity of cannabinoids, hepatocarcinoma tumors were induced in athymic mice by subcutaneal injection or by liver implantation. Mice were injected subcutaneously in the right flank with 10 × 106 HepG2 or HuH-7 cells in 0.1 ml of PBS+0.5% BSA. At 2 weeks after transplantation, tumors had grown to an average volume of 150 mm3. Mice were then divided into different experimental groups of eight animals each, which received the following treatments as subcutaneous injections according to the experiment: saline (control); 15 mg/kg b.w. Δ9-THC; 1.5 mg/kg b.w. JWH-015; and 1 mg/kg b.w. 3-MA. The injection was repeated every day and treatment was continued for 15 days. Tumor volumes were monitored every day using calliper measurements and were calculated by the formula: (4π/3) × (w/2)2 × (l/2). The body weight of the animals was recorded daily. For in vivo Atg5 knockdown, xenograft tumors were induced as indicated and 1 nmol specific Atg5 atelocollagen-complexed siRNA or control siRNA was injected peritumorally on days 1 and 7 of the treatment. Mice were treated daily for 15 days with saline, 15 mg/kg b.w. Δ9-THC or 1.5 mg/kg b.w. JWH-015.
When tumor cells were implanted in the liver, the treatments were initiated 1 week after cells injection and administered intraperitoneally. Eight animals were used in each experimental group. The study was performed for 10 days to minimize the trauma of the host animals according to UKCCCR recommendations. At the end of the treatment, the animals were killed and xenografted tumors and livers weighted and frozen.
Statistical analysis
Cell viability data were expressed as the mean±S.D. and evaluated by Student's t-test. Differences were considered significant when the P-value was less than 0.05.
Acknowledgments
This work was supported by Ministerio de Ciencia e Innovación (Grant SAF2008-03220 to ID, PS09/01401 to GV and SAF2006/00918 to MG), Comunidad de Madrid (Grants CAM/UAH CCG08-UAH/BIO-3914 and CAM S-SAL-0261-2006), Comunidad Castilla-LaMancha (Grant PII1/09-0165-0822) and Santander-Complutense (Grant PR34/07-15856 to GV). NO-H and DV received fellowships from the University of Alcalá. MS was recipient of a fellowship from MEC and of a formation contract from Comunidad de Madrid. We thank Sonia Hernández, Mar Lorente and Sofía Torres for their technical advice as well as other members of our laboratories for their continuous support.
Glossary
- ACC
acetyl-CoA carboxylase
- AFP
alpha-fetoprotein
- Akt
serine–threonine kinase Akt
- AMPK
adenosine monophosphate-activated kinase
- CaMKK
calmodulin-activated kinase kinase
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- HCC
hepatocellular carcinoma
- HepG2
human hepatocellular liver carcinoma cell line
- HuH-7
hepatocellular carcinoma cells
- LC3
microtubule-associated protein 1 light chain 3α
- LKB1
liver kinase B1
- 3-MA
3-methyladenine
- mTOR
mammalian target of rapamycin
- SR1
SR141716A
- SR2
SR144528
- Δ9-THC
Δ9-tetrahydrocannabinol
- TRB3
tribbles homolog 3
- TSC2
tuberous sclerosis complex 2
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Piacentini
This article has been corrected since Advance Online Publication and a corrigendum is also printed in this issue
Supplementary Material
References
- Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353–367. doi: 10.1586/egh.09.35. [DOI] [PubMed] [Google Scholar]
- Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- Josephs DH, Ross PJ. Sorafenib in hepatocellular carcinoma. Br J Hosp Med (Lond) 2010;71:451–456. doi: 10.12968/hmed.2010.71.8.77669. [DOI] [PubMed] [Google Scholar]
- Duffy A, Greten T. Developing better treatments in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2010;4:551–560. doi: 10.1586/egh.10.58. [DOI] [PubMed] [Google Scholar]
- Velasco G, Carracedo A, Blazquez C, Lorente M, Aguado T, Haro A, et al. Cannabinoids and gliomas. Mol Neurobiol. 2007;36:60–67. doi: 10.1007/s12035-007-0002-5. [DOI] [PubMed] [Google Scholar]
- Qamri Z, Preet A, Nasser MW, Bass CE, Leone G, Barsky SH, et al. Synthetic cannabinoid receptor agonists inhibit tumor growth and metastasis of breast cancer. Mol Cancer Ther. 2009;8:3117–3129. doi: 10.1158/1535-7163.MCT-09-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarel MM, Sarrio D, Palacios J, Guzman M, Sanchez C. Delta9-tetrahydrocannabinol inhibits cell cycle progression in human breast cancer cells through Cdc2 regulation. Cancer Res. 2006;66:6615–6621. doi: 10.1158/0008-5472.CAN-05-4566. [DOI] [PubMed] [Google Scholar]
- Sarfaraz S, Afaq F, Adhami VM, Mukhtar H. Cannabinoid receptor as a novel target for the treatment of prostate cancer. Cancer Res. 2005;65:1635–1641. doi: 10.1158/0008-5472.CAN-04-3410. [DOI] [PubMed] [Google Scholar]
- Olea-Herrero N, Vara D, Malagarie-Cazenave S, Diaz-Laviada I. Inhibition of human tumour prostate PC-3 cell growth by cannabinoids R(+)-methanandamide and JWH-015: involvement of CB2. Br J Cancer. 2009;101:940–950. doi: 10.1038/sj.bjc.6605248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano M, Pellerito O, Portanova P, Calvaruso G, Santulli A, De Blasio A, et al. Apoptosis induced in HepG2 cells by the synthetic cannabinoid WIN: involvement of the transcription factor PPARgamma. Biochimie. 2009;91:457–465. doi: 10.1016/j.biochi.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Pellerito O, Calvaruso G, Portanova P, De Blasio A, Santulli A, Vento R, et al. The synthetic cannabinoid WIN 55,212-2 sensitizes hepatocellular carcinoma cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by activating p8/CCAAT/enhancer binding protein homologous protein (CHOP)/death receptor 5 (DR5) axis. Mol Pharmacol. 2010;77:854–863. doi: 10.1124/mol.109.062257. [DOI] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente M, Torres S, Salazar M, Carracedo A, Hernandez-Tiedra S, Rodriguez-Fornes F, et al. Stimulation of the midkine/ALK axis renders glioma cells resistant to cannabinoid antitumoral action Cell Death Differ 2011. e-pub ahead of print 14 January 2011; doi: 10.1038/cdd.2010.170 [DOI] [PMC free article] [PubMed]
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- Liu YL, Yang PM, Shun CT, Wu MS, Weng JR, Chen CC. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6:1057–1065. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009;1793:1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Shi YH, Ding ZB, Zhou J, Qiu SJ, Fan J. Prognostic significance of beclin 1-dependent apoptotic activity in hepatocellular carcinoma. Autophagy. 2009;5:380–382. doi: 10.4161/auto.5.3.7658. [DOI] [PubMed] [Google Scholar]
- Shi M, Wang HN, Xie ST, Luo Y, Sun CY, Chen XL, et al. Antimicrobial peptaibols, novel suppressors of tumor cells, targeted calcium-mediated apoptosis and autophagy in human hepatocellular carcinoma cells. Mol Cancer. 2010;9:26. doi: 10.1186/1476-4598-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Egia A, Lorente M, et al. TRB3 links ER stress to autophagy in cannabinoid anti-tumoral action. Autophagy. 2009;5:1048–1049. doi: 10.4161/auto.5.7.9508. [DOI] [PubMed] [Google Scholar]
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC. Atg5 and Bcl-2 provide novel insights into the interplay between apoptosis and autophagy. Cell Death Differ. 2007;14:1247–1250. doi: 10.1038/sj.cdd.4402149. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, Granata R, Meijer AJ, Codogno P. Does autophagy have a license to kill mammalian cells. Cell Death Differ. 2009;16:12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- Memmott RM, Dennis PA. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal. 2009;21:656–664. doi: 10.1016/j.cellsig.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Lorente M, Egia A, Blazquez C, Garcia S, Giroux V, et al. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer cell. 2006;9:301–312. doi: 10.1016/j.ccr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Jaattela M. AMP-activated protein kinase: a universal regulator of autophagy. Autophagy. 2007;3:381–383. doi: 10.4161/auto.4240. [DOI] [PubMed] [Google Scholar]
- Xu X, Liu Y, Huang S, Liu G, Xie C, Zhou J, et al. Overexpression of cannabinoid receptors CB1 and CB2 correlates with improved prognosis of patients with hepatocellular carcinoma. Cancer Genet Cytogenet. 2006;171:31–38. doi: 10.1016/j.cancergencyto.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. An expanding role for mTOR in cancer. Trends Mol Med. 2005;11:353–361. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Witters LA, Kemp BE, Means AR. Chutes and Ladders: the search for protein kinases that act on AMPK. Trends Biochem Sci. 2006;31:13–16. doi: 10.1016/j.tibs.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Klionsky DJ. AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metab. 2011;13:119–120. doi: 10.1016/j.cmet.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS One. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma Gastroenterology 20081351972–1983.e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.