Abstract
As neuronal differentiation of embryonic stem cells (ESCs) recapitulates embryonic neurogenesis, disturbances of this process may model developmental neurotoxicity (DNT). To identify the relevant steps of in vitro neurodevelopment, we implemented a differentiation protocol yielding neurons with desired electrophysiological properties. Results from focussed transcriptional profiling suggested that detection of non-cytotoxic developmental disturbances triggered by toxicants such as retinoic acid (RA) or cyclopamine was possible. Therefore, a broad transcriptional profile of the 20-day differentiation process was obtained. Cluster analysis of expression kinetics, and bioinformatic identification of overrepresented gene ontologies revealed waves of regulation relevant for DNT testing. We further explored the concept of superimposed waves as descriptor of ordered, but overlapping biological processes. The initial wave of transcripts indicated reorganization of chromatin and epigenetic changes. Then, a transient upregulation of genes involved in the formation and patterning of neuronal precursors followed. Simultaneously, a long wave of ongoing neuronal differentiation started. This was again superseded towards the end of the process by shorter waves of neuronal maturation that yielded information on specification, extracellular matrix formation, disease-associated genes and the generation of glia. Short exposure to lead during the final differentiation phase, disturbed neuronal maturation. Thus, the wave kinetics and the patterns of neuronal specification define the time windows and end points for examination of DNT.
Keywords: stem cell, development, neurotoxicity, neuron, astrocyte, electrophysiology
Ultimately, the entire complexity of the mammalian central nervous system (CNS) is generated during ontogenesis from a few single cells. Neuronal generation and differentiation can be recapitulated by embryonic stem cells (ESCs) under appropriate culture conditions.1, 2, 3, 4, 5, 6 ESC-based studies of neurodevelopment allow investigations, which are not easily possible in vivo.7 However, known differentiation protocols differ in their suitability for toxicological studies. For instance, older protocols involve a step of embryoid body formation.8 Frequently, only a small number of the initially present ESCs form neurons, and the observation of individual cells is hardly possible. Other protocols use co-cultures with stromal cell lines to differentiate ESCs towards neurons, and would therefore introduce additional complexity into models for developmental neurotoxicity (DNT). A recently developed monolayer differentiation protocol allows monitoring of the differentiation procedure and of possible effects of different chemicals during the whole period of differentiation on a single cell level.9
DNT is the form of toxicity least examined and hardest to trace, as it is not necessarily related to cell loss. In all, <0.1% of frequently used industrial chemicals have been examined, and for the few known toxicants, the mechanism of action is still elusive (reviewed in Makris et al.10; Grandjean and Landrigan11; Bal-Price et al.12). Behavioral pathology in the absence of cell loss is also known from disease models, for example, for Huntington's disease13 or schizophrenia.14 Toxicants, such as mercury or lead, may trigger behavioral or cognitive deficits without histophathological hallmarks.11 Cellular physiology may be affected during the period of exposure.15 This may eventually lead to changes in differentiation and patterning in the CNS, which is the basis for long-term effects that are observed after the exposure to toxicants has ceased.
CNS development is assumed to be orchestrated by waves of gene expression16, 17 that determine different intermediate cell phenotypes. Some periods may be more sensitive to certain toxicants than others. Epidemiological proof for such ‘windows of sensitivity' in organ development with long-term consequences for the organism comes from thalidomide exposure in man3 and various animal models.18
Current test systems based on the differentiation of stem cells to either cardiomyocytes19 or neural cells12 neither yield mechanistic information, nor do they account for the complexity of CNS development, that is, the establishment of a balance between multiple neuronal cell types.3, 20 The ‘toxicology for the 21st century' initiative21, 22 suggests the identification of pathways as opposed to the current black-box test systems. In the case of ESC-based models of DNT, this requires a detailed understanding of the developmental process leading to multiple different cell types. Detailed knowledge on the waves of gene induction controlling different developmental steps would be an essential prerequisite. However, CNS development is proceeding at different paces. For instance, the anterior and posterior parts of the neural tube follow different kinetics, and some regions of the CNS continue neurogenesis, whereas in other regions, cells have already reached fully post-mitotic stages.20
Our study was undertaken to analyze the wave-like expression pattern of mESC neurodevelopment as a basis for the definition of test windows and markers. This knowledge should help to identify non-cytotoxic, but neuroteratogenic compounds that are able to shift neuronal composition or phenotypes. Finally, the markers should distinguish multiple cell types and differentiation stages, and be able to indicate subpopulations of cells.
Results
Monolayer differentiation of mESCs to neurons
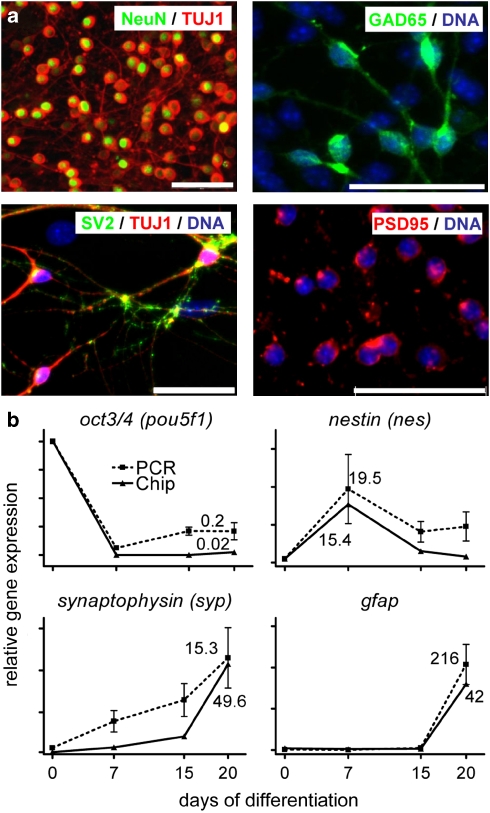
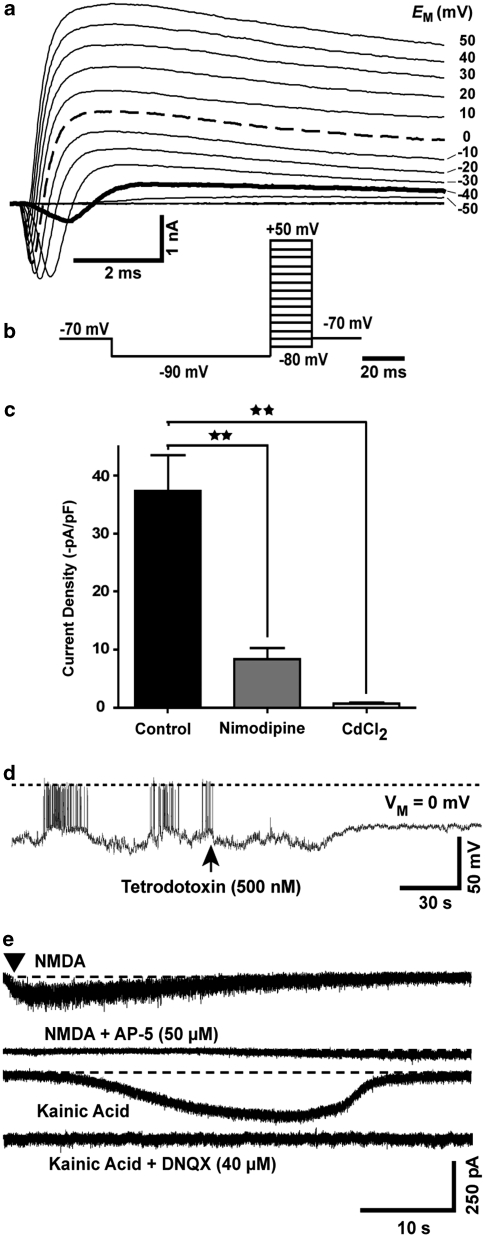
On day of differentiation 20 (DoD20), the majority of cells was positive for the pan-neuronal markers Tuj1 and NeuN. Many cells also expressed the synapse-associated markers SV2 and PSD95 (Figure 1a). As a more quantitative overall measure for the robustness of the differentiation protocol, we chose mRNA expression, which we followed over time. The kinetics for different markers was highly reproducible across experiments (Figure 1b). Differentiation to mature, electrophysiologically active neurons was shown by the presence of voltage-dependent Na+ and K+, and Ca2+ channels in individual patch-clamped neurons (Figure 2a–c, Supplementary Figure S1). Further experiments also identified spontaneous neuronal electrical activity (Figure 2d) and action potentials (Supplementary Figure S1). Currents were also evoked by exposure to N-methyl--aspartate or kainic acid and blocked by the respective selective antagonists (Figure 2e). Thus, our differentiation protocol yielded bona fide neurons.
Figure 1.
Protein and mRNA-based markers of robust neuronal differentiation of mESCs. (a) Cultures of mESCs were fixed and stained on day 20 of differentiation. DNA (blue) was stained with H-33342. Proteins are indicated as text on the micrograph in the same color as used for the display of their staining pattern. Tuj1: neuronal form of beta-III tubulin; NeuN: neuron-specific nuclear antigen, encoded by fox3;40 GAD65: glutamate decarboxylase 2; SV2: synaptic vesicle glycoprotein 2a; PSD95: post-synaptic density protein 95. Scale bars: 20 μm. (b) mESC cultures (n=5 biological experiments) were differentiated towards neurons, and RNA was prepared at the indicated days of differentiation. Gene expression of the stemness factor Oct4, the neural stem cell marker Nestin (nes), the mature neuronal marker synaptophysin and the glial marker gfap was quantified by quantitative RT-PCR. The mean±S.D. of the relative expression compared with day 0 (set to 1 on each diagram) was calculated and displayed (dotted lines). Relative gene expression data were also obtained by chip analysis and the means (n=2) are displayed (solid line). Note the different scaling of the axes for chip or RT-PCR analysis, which was chosen for reasons of better comparability of the overall curve shapes. The figures in the diagram indicate the relative expression level on DoD20 (DoD7 for nestin) versus DoD0, and thus define the axis scaling
Figure 2.
Electrophysiological evidence for successful neuronal development. Cells were differentiated on glass coverslips towards the neuronal lineage for 20–24 days and then placed into a temperature-controlled recording chamber for whole-cell patch-clamp studies. (a) Representative example for the currents observed during the 20 ms voltage steps of the whole-cell voltage clamp recording protocol displayed in (b). Note that Na+ currents (downwards deflection) are observed at voltages ⩾−40 mV (solid line). Strong depolarizing and repolarizing (K+ currents; upwards deflection) are observed at depolarization to 0 mV (dashed line). (c) For voltage clamp recording (voltage step from −80 to 0 mV) of Ca2+ channels, Na+ and K+ channels were blocked by addition of tetrodotoxin, tetraethylammoniumchloride (5 mM), 4-aminopyridine (10 mM) and substitution of intracellular K+ ions by 120 mM Cs+. Moreover, the measurement of Ca-currents was favoured by a bath solution containing barium ions (10 mM) instead of calcium ions. Current traces were obtained without Ca2+-channel blocker, or with the blockers nimodipine (1 μM) or Cd2+ (1 mM) added. Current data at 15 ms after the voltage step were corrected for cell capacitance (indirect measure for cell size) and displayed. Data represent means±S.D. **P<0.01. (d) Spontaneous action potentials were recorded in current clamp mode (0 pA). At the time indicated by an arrow, tetrodotoxin was added. The dashed line indicates 0 mV membrane potential. The scale bars indicate the dimensions of the membrane potential and the time domain. (e) Recordings at individual neurons excited with specific glutamate receptor agonists in the presence or absence of blockers. Current traces were recorded after application of N-methyl--aspartate (NMDA) or kainic acid. All agonists were also tested in the presence of their respective specific antagonist (traces with 5-aminophosphovalerate (AP-5), 6,7-dinitroquinoxalin-2,3-dione (DNQX)). The scale bars represent the current and time dimensions of the experiment. Data are representative for n⩾10 neurons (for agonists) and n=3 for antagonists (on neurons with positive agonist response)
Transcription-based end points to identify disturbed neuronal differentiation
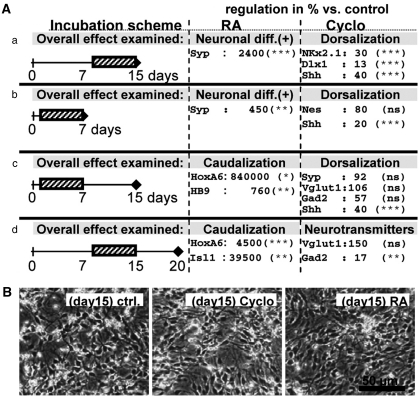
We next investigated whether subtle perturbations of the differentiation process below the cytotoxicity threshold would be detectable by mRNA-based readouts. Parallel mESC cultures were differentiated for 7, 15 and 20 days and mRNA was prepared for quantitative RT-PCR analysis. These cells were treated during two different time windows (DoD1–7, DoD8–15) with two neuroteratogens (Figure 3A). With the concentrations used here, cell death was not detectable (data not shown), and cells looked viable and were morphologically indistinguishable from untreated cells (Figure 3B). We used the morphogen retinoic acid (RA) as a known in vivo and in vitro reproductive toxicant and cyclopamine for its ability to alter sonic hedgehog (Shh) signaling, resulting in the disruption of patterning gradients responsible for floor plate and ventral neurons.2, 20 As expected from the literature,23 RA induced accelerated neuronal differentiation (increased synaptophysin expression) whereas cyclopamine reduced the expression of markers typical for more ventrally located neurons like Shh, Nkx2.1 and Dlx1, but not overall neuronal differentiation (Figure 3A). Thus, marker genes can indicate subtle shifts in differentiation patterns not visible morphologically. When cultures were exposed to cyclopamine from DoD1–7 and immediately analyzed thereafter, treatment did not affect the overall formation of neural precursor cells (NPCs), but the reduced Shh expression suggested a reduced ventral development. In cells left to differentiate further without the compound, reduced Shh expression was still observable on DoD15. A shift of neurotransmitter phenotype from GABAergic (Gad2 as marker) to glutamatergic (Vglut1 as marker)2 was not observed after treatment for the first 7 days, but a significant decrease in Gad2 (more ventrally prominent) was observable when the cells were treated between DoD8 and 15.
Figure 3.
Detection of non-cytotoxic developmental disturbances by transcriptional analysis. Cultures of mESCs were neuronally differentiated for 7, 15 or 20 days as indicated in a–d. They were exposed to RA or cyclopamine (Cyclo) for the time periods indicated by the hatched boxes. (A) RNA was isolated at the indicated days (diamond) and used for quantitative RT-PCR analysis of selected differentiation and patterning markers. Headings indicate the overall biological effect, such as accelerated neuronal differentiation (e.g. neuronal diff. (+)) or altered patterning (e.g. caudalization). Names are the official gene names, apart from the following: Vglut1=Slc17a7, HB9=Mnx1. The data indicate relative expression levels in % compared with untreated controls at the same time point and are means±S.D. from two to three independent experiments for each treatment and exposure schedule. Significance levels (by ANOVA within a given experimental condition) are indicated (*P<0.05, **P<0.01, ***P>0.001). The complete data set with S.D. is given in Supplementary Figure S2. (B) Representative images of cultures on DoD15 in condition a. RA- and cyclopamine-treated cultures were viable, indistinguishable from controls (ctrl.)
In the case of RA, the acceleration of development (synaptophysin) was already significant at early stages and we found upregulation of markers usually expressed in caudal parts of the neural tube (hoxa6, hb9), and associated with the development of motor neuron precursors (isl1; Figure 3).
We also examined whether inhibited differentiation was detectable by RNA markers. Early exposure to 3i, a kinase inhibitor mix known for inhibiting differentiation of mESCs,24 resulted in cultures with retarded neural differentiation indicated by a decreased expression of hes5, nestin and Tubb3, and an increased expression of Oct4 (Supplementary Figure S2). Treatment of cells with 3i after DoD7 (after neural differentiation had been initiated) did not return them to the stem cell state, but was cytotoxic. These examples demonstrated the usefulness of transcript profiling for detection of patterning disturbances.
It may be necessary to measure the impact on differentiation at different DoD, and specific markers need to be selected for each stage.
Identification of clusters of genes regulated during neuronal differentiation of mESCs
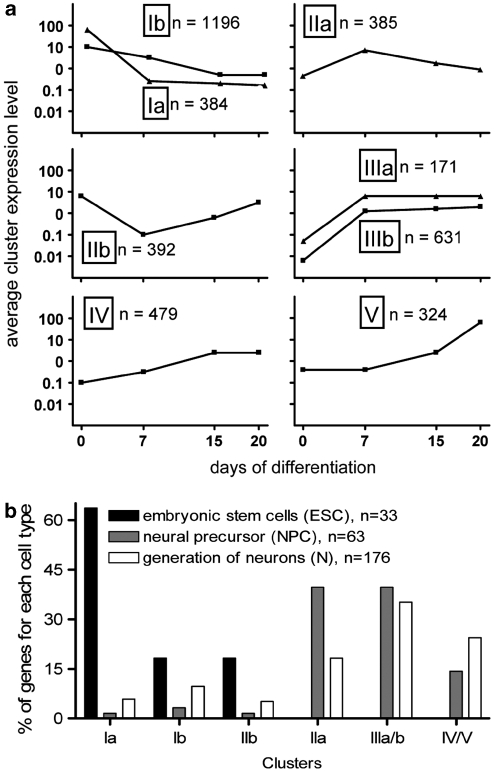
Using oligonucleotide microrarrays, we analyzed changes in the transcriptome over time to identify toxicity markers. The differentiation kinetics identified in the microarray analysis matched the ones observed during many other well-controlled experiments (Figure 1b). The kinetics of expression of each gene represented on the chip was used as input for an unbiased clustering analysis, which yielded eight regulation profiles (Figure 4a, Supplementary Figure S3), besides the genes not regulated at all. Cluster Ia was characterized by rapid downregulation, and cluster Ib by slow downregulation. These two clusters exemplify the principle of superimposed gene regulation waves with different amplitudes. Clusters IIa and IIb contained genes that were transiently regulated at DoD7 (IIa: up, IIb: downregulated). Cluster IIIa and IIIb were characterized by a rapid increase in transcripts between day 0 and DoD7, maintained then at high levels. Cluster IV contained genes, which remained low until DoD7 and then reached high levels on DoD15. The final cluster V comprised transcripts that were hardly upregulated until DoD15, and reached their maximum on DoD20 (Figure 4a).
Figure 4.
Cluster analysis of mRNA time course profiles, and their association with distinct phases of differentiation. (a) Gene expression kinetics was determined for all genes represented on the chip. An unbiased clustering analysis of the kinetic profiles of all regulated genes was performed. For each cluster (named Ia, Ib, IIa, IIb, IIIa, IIIb, IV, V), the means of the absolute expression level of all genes in the respective cluster, for each analysis time point, are displayed and plotted on a logarithmic scale; n: number of genes in the cluster (b). Number of genes expressed in mESCs (ESCs=40 evaluated, 33 found), NPCs (73 evaluated, 63 found) and developing neurons (N) were analyzed by extensive literature search (mESCs, NPCs) or GO-analysis (N). The relative distribution of these genes across the different clusters was calculated (in %) and displayed (e.g. 65% of all ESC markers was found in cluster Ia, 35% of all N markers in cluster III)
The genes were subjected to a more detailed analysis. Of 40 genes that characterize the initial mESC stage,3 33 were identified and all were downregulated (Figure 4b). Most NPC markers were found in clusters IIa and IIIa/b, containing genes upregulated early. In contrast to this, most neuronal markers were found in the clusters with increasing gene expression (III–V), whereas about 20% were found in cluster IIa (transient upregulation on DoD7; Figure 4b). The clusters identified by unbiased bioinformatics methods may therefore correspond to waves of real biological processes underlying the differentiation of mESCs to neurons. To explore this working hypothesis, we continued with an analysis of the biological significance of genes in individual clusters.
Loss of pluripotency is accompanied by progressive changes in transcripts responsible for chromatin organization and DNA/cell cycle functions
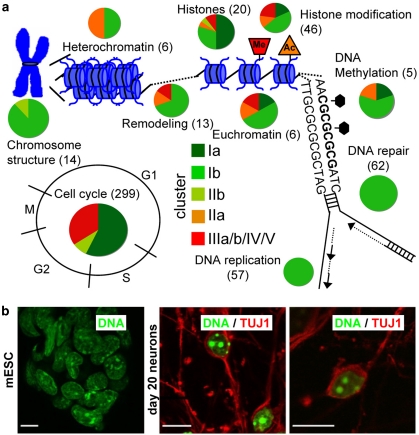
Genes in cluster I were analyzed for gene onthology (GO) categories significantly overrepresented. Besides the cell cycle, we found chromatin structure and epigenetic processes to be affected (Figure 5a, Supplementary Figure S4). All genes known to be associated with chromosome structure, DNA replication, DNA repair and DNA methylation were downregulated. Also, most of the genes coding for histones, histone modifiers, chromatin remodeling and chromatin substructuring were found in clusters Ia/b. Confocal microscopy showed that chromatin distributed relatively homogeneously over the nucleus in mESCs, but was organized entirely differently after 20 days of differentiation (Figure 5b).
Figure 5.
Indication of a progressive change in chromatin organization and epigenetic factors in waves of fast and slow downregulation. Gene lists of relevant processes were assembled both with the help of the GO database and extensive literature search. The clusters were then queried for the presence of these genes. (a) Processes linked to chromatin or DNA repair and DNA replication are displayed, and for each of them, the number of genes found to be regulated during neuronal differentiation of mESCs is displayed in brackets. The individual genes are listed in Supplementary Figure S4. Among the identified genes, four (Smarca1, Myst4, Jmjd3 and Hdac11) are known to be neurospecific and five (Suz12, Ezh2, Bmi1, Cbx2 and Cbx8) are components of the polycomb repressor complexes, which play an important role in differentiation-related control of gene promoters. These genes could serve as sensitive markers to detect negative effects of compounds on early developmental processes. For each process, the percentage of genes present in the different clusters is indicated by colour-coded pie charts. All green shades represent clusters of genes downregulated from DoD0 to DoD20. (b) Changes in chromatin structure during differentiation were visualized by DNA staining with DAPI (green) and confocal microscopic analysis. Left panel: undifferentiated mESCs; right panels: neuronally differentiated cells on DoD20 that were stained with neuron-specific beta-III tubulin antibody (red). Scale bar: 10 μm
Correlation of neural precursor formation with a strong, transient change in gene expression levels
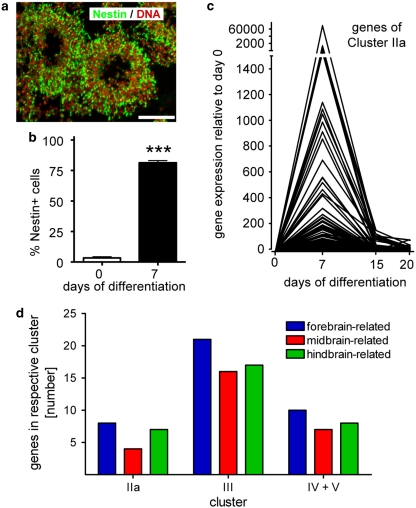
We examined whether genes of cluster II were specifically linked to the process of NPC formation. Nestin was expected, and found, in cluster IIa. Nestin-positive cells were often arranged in ring-like structures, reminiscent of rosettes or two-dimensional neural tubes25, 26 (Figure 6a). Quantification by flow cytometry analysis confirmed the immunocytochemical finding that about 80% of all cells in the culture became nestin positive (Figure 6b). High synchronization of differentiation was suggested by the sharp expression profile of genes in cluster IIa (Figure 6c). Besides nestin, many other genes typically associated with neuroepithelial precursors (NPCs) and neurogenesis were found in cluster IIa (Supplementary Figure S3). Also some genes associated with early, but definitive neuronal development were identified (Dll1, Hes3). Cluster IIa also contained apparently unspecific genes (e.g. Jak2, Foxd4, Bcl-2, Kif21a, Agtr1a, Moxd1, Aacs, Arl2bp, Scd2). We examined which GO categories were statistically overrepresented by cluster IIa genes. The GO ‘nervous system development' emerged with a P-value <10−13, and only neuronal/neurodevelopmental GOs were identified with the exception of ossification (eight weakly significant genes; Table 1). Thus, genes of cluster IIa represent an important end point for testing the disturbed proliferation and differentiation during the neuroectodermal/neuronal development time window.
Figure 6.
Correlation of neural precursor formation with a transiently upregulated group of genes. (a) On DoD7, cultures were immunostained for the neural stem cell marker nestin (green) and DNA (red). Scale bar: 100 μm. (b) For quantification of nestin-positive NPCs, cells were immunostained for nestin on DoD7 and analyzed by flow cytometry. Data are means±S.D. of seven independent differentiations. ***P<0.001. (c) Relative expression profiles of genes from cluster IIa were calculated by normalization of expression of each gene to DoD0 expression, which was arbitrarily set to 1. The expression kinetics for each gene within that cluster is displayed. (d) Genes upregulated during neuronal differentiation of mESCs were analyzed for their role in regional specification of the brain and classified accordingly (colour-coding). The number of genes associated with each of the three chosen subregions of the brain are displayed separately for each regulation cluster. A detailed list of genes with their regional assignment is given in Supplementary Figure S5
Table 1. GO categories significantly overrepresented in cluster IIa.
| Biological process (GO)a | No. of genes in IIa | P-value | Examples of upregulated genes listed in the GO |
|---|---|---|---|
| Nervous system development | 51 | 3e−14 | Neurod4, Nes (nestin), Cdh2 (N-cadherin), Fgf5, Sema5b, Efnb2 |
| Regulation of nervous system development | 17 | 7e−09 | Nefm (neurofilament M), Chrna3 (cholinergic R), Ntrk3, Isl1, Foxg1 |
| Regulation of neurogenesis | 16 | 9e−09 | Hoxa2, Smo, Dll1 (delta-like 1), Hes3, Metrn, Ntrk3 (=Trkc) |
| Neuron projection morphogenesis | 13 | 2e−05 | Epha7, Mtap1b, Myh10 (myosin heavy chain), Egr2, Epha7, Isl1 |
| Central nervous system development | 21 | 1e−06 | Mtap1b (microtubule-associated protein), Bmi, Foxg1, Isl1, Fgfr3 |
| Neuron projection regeneration | 5 | 2e−06 | Mtap1b, Bcl2, Smo, Chst3 (carbohydrate sulfotransferase) |
| Parasympathetic nervous system development | 4 | 4e−06 | Hoxb2, Egr2, Smo (smoothened), Hes3 (hairy and enhancer of split) |
| Neuron development | 20 | 5e−06 | Mtab1b, Foxg1, Epha7 (Eph receptor A7), Isl1, Ulk2, Bmpr1b |
| Cranial nerve development | 5 | 1e−05 | Gli3, Hoxb2, Egr2 (early growth response), Smo, Hes3 |
| Dorsal/ventral pattern formation | 9 | 7e−07 | SP8, Foxg1, Bmpr1a, Bmpr1b (bone morphogenic protein R.), Hoxa2 |
| Tissue development | 28 | 1e−06 | Homer1, Prox1 (prospero-related homeobox 1), Fzd2, Sdc1 (syndecan) |
| MAPKKK cascade | 12 | 2e−06 | Mapk8, Fgf13, Jak2, Nrg1, Fgfr3, Tgfbr1, Mapk8 (=Jnk) |
| Anterior/posterior pattern formation | 12 | 4e−06 | Hoxb2, Hoxa2, Tgfbr1 (transforming growth factor, beta receptor) |
| Regulation of ossification | 8 | 3e−05 | Smad5, Calca (calcitonin), Sfrp1 (secreted frizzled-rel. protein 1), Egr2 |
All categories are identified by gProfiler bioinformatics analysis, with their P-values indicated after correction by removal of ‘nervous system development' genes from non-neuronal GOs.
Markers of regional fate decisions in the CNS
We examined the expression of regional markers in cluster IIa. Amongst the few markers expressed, those for forebrain (Foxg1) and hindbrain (Hoxa2/b2) were evenly distributed (Figure 6d). Also, in the clusters containing continuously upregulated genes (IIIa/b, IV+V), forebrain (Reln), midbrain (En1/2) and hindbrain (Lmx1a or Hoxa1) markers were evenly distributed (Supplementary Figure S5). Accordingly, our experimental model appears to reflect several parallel lines of in vivo neural specification, and the ratios of expression of different patterning markers may provide sensitive indicators of disturbed neurodevelopment.
Specificity for neuronal induction with respect to glial cells
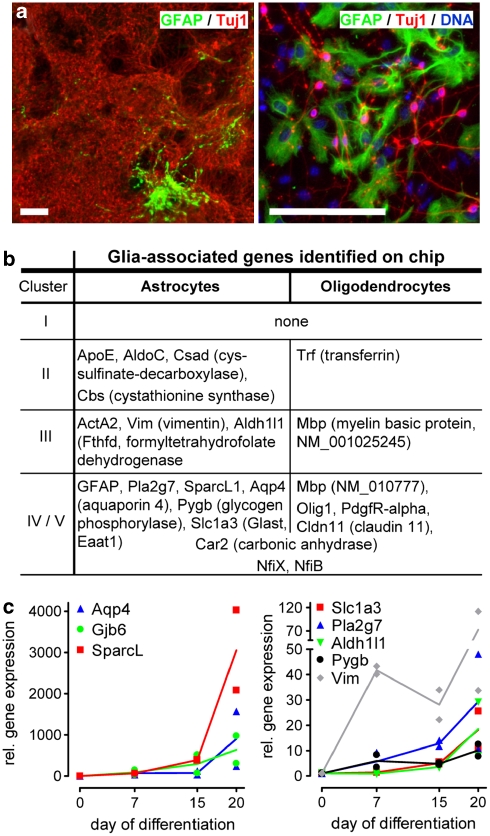
The transcriptional profile allowed us a detailed analysis of potentially contaminating non-neuronal cells. Some small GFAP-positive cell areas were reproducibly (1–2 small islands/cm2) identified by immunocytochemistry (Figure 7a). As an unbiased search for overrepresented GO categories did not result in any hits related to gliogenesis (Table 2), we used a list of 25 astrocyte-related genes3 and found 11 of them to be upregulated on DoD20 compared with DoD0, with four additional astrocytes-related genes transiently upregulated on DoD7 (Figure 7b). This early upregulation of apparent astrocytic markers (e.g. vimentin) may be owing to the generation of radial glia-like NPCs at DoD7. This cell type, as exemplified by the upregulation of Fabp7 in cluster IIIb27 or Ascl1 (=Mash1),28 shares many markers with astrocytes.5 The late upregulation of astroglial markers was corroborated by qPCR (Figure 7c). Small increases in this astrocytic population may affect toxicity testing during the later differentiation phases. Microglia appeared to be absent. The contribution of oligodendrocytes appears to be negligible.
Figure 7.
Analysis of glia-associated genes. (a) DoD20 cultures were fixed and stained for GFAP (green; to identify astrocytes) and Tuj1 (red; to identify neurons). The left image shows a representative overview with large neuronal areas and one typical astrocytic island. The right image shows an astrocytic island in greater detail. Scale bars=100 μm. (b) The table indicates the glia-related genes identified in this study, sorted by the cluster of expression kinetics they fell into. Astrocyte-related genes searched for, but not identified here were glutamine synthetase (Glul), S100b, Slc1a2 (Glt-1, Eaat2), Connexin 30/43 (Gjb6/Gja1), NfiA (also found in oligodendrocytes). Oligodendrocyte-related genes not found here were ATP-binding cassette, sub-family A (Abca2), CNPase (Cnp1), a microtubule-associated protein (Mtap4), myelin-glycoproteins (Omg and Mog), Olig2/3 (Olig2, Olig3), myelin protein zero (Mpz), Ng2 (Cspg4), NfiA. (c) Expression of selected astrocyte-related genes was monitored by qPCR on day 0, 7, 15 and 20 of two differentiations. Data for each differentiation are given individually. The lines indicate the resulting mean values
Table 2. GO categories that are overrepresented in the clusters comprising genes upregulated during differentiation.
| Cluster | Biological process (GO)a | No. of genes | P-value | Examples of upregulated genes |
|---|---|---|---|---|
| IIIa/b | Nervous system development | 107 | 2e−32 | Hes5, Notch3, Otx1, FoxA2, Nkx2.2, Ntrk3, Nrxn2 (neurexin) |
| Generation of neurons | 69 | 6e−23 | Sox5, Shh (sonic hedgehog), Wnt3a, Dcx (doublecortin), Nog (noggin) | |
| CNS development | 49 | 1e−16 | Zic1, Wnt7a, Fgf8, Pitx2 | |
| Neuron development | 39 | 7e−13 | Gap43, Gprin2 (inducer of neurite outgrowth), App (Aβ precursor protein), Reln | |
| Axogenesis | 28 | 3e−12 | Cdk5r1 (kinase), EfnB1 (ephrin), Ntng1 (netrin), Stxbp1 (syntaxin-binding protein) | |
| Axon guidance | 19 | 3e−10 | Apbb1 (APP-binding), Cxcr4, Slit2, Kif5C (kinesin), Ephb1 (ephrin-R) | |
| Neuron projection | 32 | 4e−7 | Grik5 (glutamate-R), Gria3 (glutamate-R), Cacna1g (Ca2+ channel), Mtap2 (map2) | |
| IV | Vesicle | 33 | 2e−7 | Sv2a (synaptic vesicle glycoprotein), Syn2 (synapsin), Syt1 (synaptotagmin) |
| Nervous system development | 43 | 4e−7 | Neurog2 (neurogenin), Unc5b (netrin-R), Bai2, FoxD1, Egfr, Dner, En1 (engrailed) | |
| V | Extracellular matrix | 24 | 10e−11 | Dcn (decorin), Col1a1 (collagen), Spon2 (spondin), Lum (lumican), Tnc (tenascin) |
| Lipid storage | 5 | 4e−6 | Apoa1 (apolipoprotein), Gm2a (ganglioside activator), Enpp1, Cav1 (caveolin) | |
| Response to stress | 42 | 1e−5 | Hspa2 (heat shock protein), Fas (fas), Fos, Pparg (PPAR-gamma), Pink1, Snca | |
| IV+V | Extracellular matrix | 39 | 3e−11 | Col1a2 (collagen), Col3a1 (collagen), Ecm1 (extracellular matrix), Efemp2 (fibulin) |
| Response to hormone stimulus | 38 | 7e−10 | Rbp4 (retinol BP), Rxra, Thra, Rgs9, Igfbp7 (insulin binding) | |
| Nervous system development | 70 | 2e−10 | Nrxn1 (neurexin), Mapt (tau), Tgfbr2, Dlx1 | |
| Blood vessel development | 29 | 5e−9 | Cdh13 (cadherin-H), Prrx1, SphK1 (sphingosine kinase), Cul7 (cullin) | |
| Neuron projection | 33 | 5e−7 | Tubb4 (tubulin), Syt1 (synaptotagmin), Psd2, Syt4, Ttyh1 (tweety homolog) | |
| Neurogenesis | 44 | 9e−7 | Myo6 (myosin), Nrn1 (neuritin), En2 (engrailed), Hoxa1, Lhx5 | |
| Synaptic vesicles | 14 | 1e−6 | Syp (synaptophysin), Slc17a6, Rabac1 (rab acceptor) | |
| Muscle organ development | 22 | 3e−6 | Gata6, Des (desmin), Myl2 (myosin light chain), Vamp5 (vesicle-associated protein) | |
| Transmission of nerve impulse | 23 | 2e−5 | Gria2 (glutamate-R), Slc17a6 (vGlut), Chrnb1 (ACh-R), Kcnmb4 (K+ channel) |
All categories are identified by gProfiler bioinformatics analysis, with their P-values indicated after correction by removal of ‘nervous system development' genes from non-neuronal GOs.
Specificity for neuronal induction with respect to other germ layer lineages
All GO categories significantly overrepresented by the genes of clusters III–V (upregulation on DoD20 versus DoD0) were determined bioinformatically, and were searched for evidence of non-neural cell type formation. Individual clusters did not indicate any non-neural cell types while representation of neuronal GOs was highly significant (Table 2). Upon pooled analysis of clusters IV and V, the GOs ‘blood vessel development' and ‘muscle organ development' emerged as significant. Thus, a subpopulation of cells present on DoD20 may display smooth muscle features.
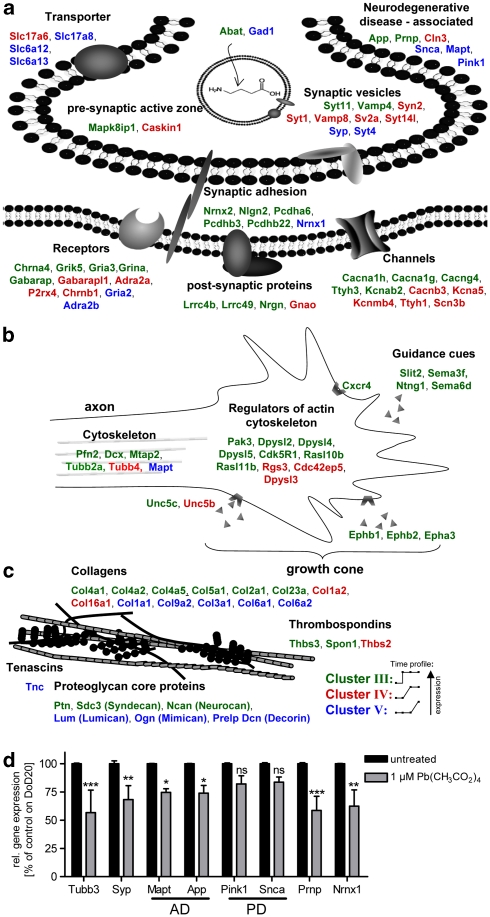
Waves of clustered genes related to neuronal induction
For characterization of the cultures, we used non-biased bioinformatics methods to identify overrepresented GOs (Table 2). In a complementary approach, based on literature and expert judgement, we hand-picked interesting groups of genes (Figure 8). The major result from this combination of strategies was our finding that the differentiation did not proceed as sequence of sequential steps, but rather involved strongly overlapping processes with one underlying large wave (cluster IIIa/b) superseded by shorter waves (cluster IV and V). For instance, generation of neurons and axogenesis/growth cone formation seemed to be ongoing in the entire period from DoD7 to DoD20, as indicated by groups of neuroreceptors and growth cone/axon guidance-related genes in cluster III (Figure 8a and b). A larger group of genes associated with synaptic vesicles or the transmission of nerve impulse only appeared later (cluster IV/V). In the latest phase, genes associated with ‘responses to stress' and ‘hormonal stimuli', ‘regulation of extracellular matrix components' and genes known to be ‘associated with hereditary neurodegenerative diseases' were strongly upregulated (Table 2, Figure 8c). Analysis by PCR confirmed the latter finding. The upregulation for disease-associated genes from DoD0 to DoD20 (n=2 differentiations) was: 92- and 16-fold for the Alzheimer's disease associated genes App and Mapt, 273-fold for the schizophrenia-associated gene Nrnx1, 91-fold for the prion protein Prnp and 19-fold/56-fold for the Parkinson's disease-related genes Pink1/Snca.
Figure 8.
Functional assignment of neuronal genes upregulated in different waves. A combination of bioinformatics tools and literature information was used to search all upregulated clusters for conspicuous biological themes and for genes associated with them. Themes are displayed and corresponding genes (with original NCBI gene names) are colour-coded according to the clusters they were found in (displayed graphically besides the legend, with dots on the lines representing DoD0, DoD7, DoD15 and DoD20). (a) Core neurochemical themes. Note a relatively early induction of receptors and channels, compared with late emergence of genes coding for transporters and synaptic vesicles, and those related to neurodegenerative disease. (b) Themes related to neurite growth indicate an early focus on growth cone formation and guidance. (c) Genes related to extracellular matrix are displayed. (d) The cells were treated with a non-cytotoxic concentration (assessed by resazurin reduction and LDH release, data not shown) of lead (1 μM) only during the last phase of differentiation (DoD14–DoD20). RNA was isolated on DoD20 and used for quantitative RT-PCR analysis of genes associated with neurodevelopment and known to be involved in neuronal disease. Pink1 and Snca were not affected. Also, their relative increase with respect to the pan-neuronal marker synaptophysin was not significant. Data indicate relative expression levels in % compared with the untreated controls on DoD20 and are means±S.D. (n=2). Significance levels (ANOVA) are indicated (*P<0.05, **P<0.01, ***P<0.001)
We wondered whether toxicants with a purported role in the developmental origins of neurodegenerative diseases (see Supplementary Figure S6), such as lead, affect this very late phase of neuronal differentiation. The transcript levels of two neuronal markers and the set of disease-associated genes were used to examine differences in differentiation. Lead exposure had a dampening effect on the expression of App, Mapt, Nrnx1 and Prnp (Figure 8d). Thus, the knowledge on markers together with that of the expected timing of their expression provides an ideal toolkit for fine-mapping of subtle developmental disturbances.
Discussion
We have here demonstrated a concept of overlapping waves of gene regulation and suggested its use to define protocols, test windows and end points for DNT testing. Our findings should be helpful to close a gap between two highly developed, but isolated disciplines: experimental developmental neurobiology and toxicology. The former has been highly successful in defining the functional importance, regional expression and cell type association of genes. The latter has an urgent need for robust and sensitive marker genes to identify disturbances of development. We showed that subtle changes in the speed of differentiation, or in dorso–ventral or anterior–posterior patterning owing to toxicants can be detected by using the right choice of mRNA markers. Such changes may be considered in vitro correlates of known teratogenic effects of the chosen compounds. For instance, cyclopamine causes dramatic patterning disturbances (holoprosencephaly) in a defined period of brain development; RA causes shifts in the anterior–posterior axis organization, favouring the more posterior parts, as found here by transcript markers. Lead affects multiple neuronal types, which is in agreement with the broad pattern of disturbances found here (see Supplementary Figure S6 for references). The data also suggest some warning on the limitations of in vitro–in vivo correlations. Although our cyclopamine data suggest a disturbance in patterning, they would not indicate a problem in the separation of the forebrain hemispheres, as observed in animal studies. Thus, observations from stem cell systems will have a major value for raising alerts on certain compounds and pinpointing potential mechanisms, while complementary data from other systems may be required to predict specific effects on humans.
Transcriptional profiling studies, relying predominantly on bioinformatics analysis, suffer from the weakness and errors of databases and algorithms. For example, assignment of genes to GO categories is not always perfect. For instance, the GO for gliogenesis contains ubiquitous signaling and metabolic molecules as well as highly specific transcription factors. On the other hand, typical astrocyte markers such as Gfap and glutamine synthetase are not members of this GO. Moreover, the equal weight given to ubiquitous versus specific genes in statistical analysis results in biological skewing. An additional problem is the visualization of the large amount of data in a form that generates meaningful knowledge. With these considerations in mind, we chose to combine bioinformatic analysis with classical knowledge-based approaches. During this procedure, the entire hit list of several thousand genes was manually screened, sorted and annotated. A consortium of experts was consulted, and results were compiled in an open access review format.3 We strongly advocate such combined approaches for toxicological systems biology, which is at present driven too strongly by computational methods.22, 29
Electrophysiology studies have a rather qualitative character, as the cells that were patched may not represent the entire culture. However, our results fully corroborate earlier findings that functional neurons can be generated from mESCs.8, 30 Immunostaining and quantitative RT-PCR were used as classical and established methods to link chip-based transcript profiling to other experiments that have been performed with much higher replicate number. In the future, extensive studies, involving RT-PCR controlled by internal standards, will be necessary for a quantitative definition of a final set of markers. Notably, we did not use differences in absolute values of regulation in the present study as basis for any of our conclusions, and all major conclusions are built on groups of co-regulated and biologically linked genes as opposed to speculations based on the presence or absence of a single gene. Even though mRNA correlated well with protein levels, as for example, in brain inflammation studies,31 our approach should not be interpreted as phenotype definition on single cell resolution. The genes grouped within the clusters described here are not necessarily expressed in the same cell and therefore do not automatically describe a single biological entity. However, with these caveats, we feel that indicators of disturbances of the default development can be selected with confidence on the basis of our study.
In the area of developmental toxicology and especially in DNT, cause–effects relationships are still mostly unknown, and human epidemiological data are only available for a handful of industrial chemicals.11 Rodent data based on the OECD test guideline 426 are available for about 200 substances.10 With this lack of human-relevant information and the better animal database, it appears reasonable for us to perform proof-of-principle experiments for the usefulness of a new approach in rodent cells first, and to validate human cells against these in case of a positive outcome.
At present, DNT studies are based on, for example, behavioral, cognitive or neuropathological end points, and the next step towards mechanistic information would be an understanding of changes on the level of cells and in gene expression. The overlapping waves defined here would provide a conceptual framework for this. Such waves (i.e. temporally and spatially shifting activation) of gene expression are known from many pioneering studies of mammalian in vivo CNS development20 and are, for instance, well characterized in high density and resolution in the hippocampus.32 Waves have also been defined in vitro in mESCs17, 33 or differentiating embryonic carcinoma cells.16, 34 Here, we extended this concept by relating regulation clusters to underlying biological processes important for toxicity testing. This translation from developmental biology to the toxicological perspective defines the windows of sensitivity relevant for test protocols.
In the field of cardiac development, the mESC-based embryonic stem cell test has been frequently applied.19 Exposure of cells during the entire test period is confounded by relatively unspecific toxicity. Therefore, separation of exposure into the proliferation and differentiation phase has been suggested.35 We want to expand this principle here by suggesting four relevant test periods. DoD1–7: testing of lineage commitment, efficiency of NPC formation and of epigenetic changes associated with the transition from pluripotent cells to more committed NPCs. DoD8–15: major phase of neuronal patterning and vesicle development. DoD15–20: a more unexpected, but highly interesting and relevant phase, when most proliferation has ceased and maturation becomes evident by expression of matrix components, important transporters and disease-associated genes. Our data on lead exposure during this phase show that it will be of high importance for future testing. DoD20+ has not been explored here. It requires further investigation to determine whether this period can be used as stable reference for neurotoxicity versus DNT, or whether new processes such as synaptogenesis, gliogenesis or myelination take a dominant role here.
The major task for the future will be the validation of a larger set of such markers, first with known specific and mechanistically defined disruptors of developmental pathways, then with known DNT compounds, to select the smallest group of final markers useful for a comprehensive description of toxicities triggered by the test compounds.
Materials and Methods
Materials
Unless otherwise mentioned, cell culture media and reagents were from Invitrogen (Carlsbad, CA, USA) and accessory reagents from Sigma (Munich, Germany). Antibodies used were anti-Tuj1 (cat. no. MMS-435P; Covance, Munich, Germany), anti-NeuN (cat. no. MAB377; Chemicon, Schwalbach/Ts, Germany), anti-GAD65 (GAD-6; DSHB, Iowa City, IA, USA), anti-SV2 (SV2; DSHB), anti-PSD95 (cat. no. 51-6900; Zymed, Darmstadt, Germany), anti-Nestin (cat. no. MAB353; Chemicon), anti-GFAP (clone: G-A-5; Sigma) and anti-Nestin-647 (clone: 25/NESTIN; BD Biosciences, Franklin Lakes, NJ, USA). mRNA primers used were Pou5f1-forward: 5′-CTCTTTGGAAAGGTGTTCAGCCAGAC-3′, Pou5f1-reverse: 5′-CGGTTCTCAATGCTAGTTCGCTTTCTC-3′ Nestin-forward: 5′-CTGGAAGGTGGGCAGCAACT-3′, Nestin-reverse: 5′-ATTAGGCAAGGGGGAAGAGAAGGATG-3′ Synaptophysin-forward: 5′-GGGTCTTTGCCATCTTCGCCTTTG-3′, Synaptophysin-reverse: 5′-CGAGGAGGAGTAGTCACCAACTAGGA-3′ Gfap-forward: 5′-GCCCGGCTCGAGGTCGAG-3′, Gfap-reverse: 5′-GTCTATACGCAGCCAGGTTGTTCTCT-3′ Shh-forward: 5′-CAGCGGCAGATATGAAGGGAAGATCA-3′, Shh-reverse: 5′-GTCTTTGCACCTCTGAGTCATCAGC-3′ Hes5-forward: 5′-CCCAAGGAGAAAAACCGACTGCG-3′, Hes5-reverse: 5′-CAGCAAAGCCTTCGCCGC-3′ Tubb3-forward: 5′-GACAACTTTATCTTTGGTCAGAGTGGTGCTG-3′, Tubb3-reverse: 5′-GATGCGGTCGGGGTACTCC-3′ Nkx2.1-forward: 5′-TACCACATGACGGCGGCG-3′, Nkx2.1-reverse: 5′-ATGAAGCGGGAGATGGCGG-3′ Dlx1-forward: 5′-TCACACAGACGCAGGTCAAGATATGG-3′, Dlx1-reverse: 5′-AGATGAGGAGTTCGGATTCCAGCC-3′ HoxA6-forward: 5′-CTGTGCGGGTGCCGTGTA-3′, HoxA6-reverse: 5′-GCGTTAGCGATCTCGATGCGG-3′ Hb9-forward: 5′-CGAACCTCTTGGGGAAGTGCC-3′, Hb9-reverse: 5′-GGAACCAAATCTTCACCTGAGTCTCGG-3′ Vglut1-forward: 5′-GGTCACATACCCTGCTTGCCAT-3′, Vglu1-reverse: 5′-GCTGCCATAGACATAGAAGACAGAACTCC-3′ Gad2-forward: 5′-AAGGGGACTACTGGGTTTGAGGC-3′, Gad2-reverse: 5′-AGGCGGCTCATTCTCTCTTCATTGT-3′ Isl1-forward: 5′-ACCTTGCGGACCTGCTATGC-3′, Isl1-reverse: 5′-CCTGGATATTAGTTTTGTCGTTGGGTTGC-3′ Tubb3-forward: 5′-GACAACTTTATCTTTGGTCAGAGTGGTGCTG-3′, Tubb3-reverse: 5′-GATGCGGTCGGGGTACTCC-3′ Mapt-forward: 5′-ACACCCCGAACCAGGAGGA-3′, Mapt-reverse: 5′-GCGTTGGAC GTGCCCTTCT-3′ App-forward: 5′-TCAGTGAGCCCAGAATCAGCTACG-3′, App-reverse: 5′-GTCAGCCCAGAACCTGGTCG-3′ Pink1-forward: 5′-GGGATCTCAAGTCCGACAACATCCT-3′, Pink1-reverse: 5′-CTGTGGACACCTCAGGGGC-3′ Snca-forward: 5′-ATGGAGTGACAACAGTGGCTGAGA-3′, Snca-reverse: 5′-CACAGGCATGTCTTCCAGGATTCC-3′ Prnp-forward: 5′-ACCATCAAGCAGCACACGGTC-3′, Prnp-reverse: 5′-GACAGGAGGGGAGGAGAAAAGCA-3′ Nrnx1-forward: 5′-GTGGGGAATGTGAGGCTGGTC-3′, Nrnx1-reverse: 5′-TCTGTGGTCTGGCTGATGGGT-3′ Aqp4-forward: 5′-GCTCAGAAAACCCCTTACCTGTGG-3′, Aqp4-reverse: 5′-TTCCATGAACCGTGGTGACTCC-3′ Gjb6-forward: 5′-CGTACACCAGCAGCATTTTCTTCC-3′, Gjb6-reverse: 5′-AGTGAACACCGTTTTCTCAGTTGGC-3′ SparcL-forward: 5′-CCCAGTGACAAGGCTGAAAAACC-3′, SparcL-reverse: 5′-GTAGATCCAGTGTTAGTGTTCCTTCCG-3′ Slc1a3-forward: 5′-CTCTACGAGGCTTTGGCTGC-3′, Slc1a3-reverse: 5′-GAGGCGGTCCAGAAACCAGTC-3′ Pla2g7-forward: 5′-GGGCTCTCAGTGCGATTCTTG-3′, Pla2g7-reverse: 5′-CAACTCCACATCTGAATCTCTGGTCC-3′ Aldh1l1-forward: 5′-CTCGGTTTGCTGATGGGGACG-3′, Aldh1l1-reverse: 5′-GCTTGAATCCTCCAAAAGGTGCGG-3′ Pygb-forward: 5′-GGACTGTTATGATTGGGGGCAAGG-3′, Pygb-reverse: 5′-GCCGCTGGGATCACTTTCTCAG-3′ Vim-forward: 5′-GAGATGGCTCGTCACCTTCGTG-3′, Vim-reverse: 5′-CCAGGTTAGTTTCTCTCAGGTTCAGG-3′. The toxicants used were RA (cat. no. R2625; Sigma), cyclopamine: (cat. no. 239803; Calbiochem, Darmstadt, Germany), PD184352: (cat. no. Axon 1368; Axon Medchem, Groningen, The Netherlands), SU5402 (cat. no. 572631; Calbiochem) and CHIR99021 (cat. no. Axon 1386; Axon Medchem).
Cell culture and differentiation
CGR8, a widely available murine ESC (mESC) line suitable for feeder-free culture maintenance and with established potential to develop along the neuroectodermal and neuronal lineage36, 37 was kindly provided by K-H Krause (Geneva). Cells were cultured in complete Glasgow's modified Eagles medium (GMEM), supplemented with 10% heat inactivated fetal bovine serum (FBS; PAA, Pasching, Austria), 2 mM glutamax, 100 μM non-essential amino acids, 50 μM β-mercaptoethanol, 2 mM sodium pyruvate and 1000 U/ml leukemia inhibitory factor (Chemicon). Cells were kept at 37 °C in 5% CO2 on tissue culture plates coated with 0.1% gelatin, and were routinely passaged every 48 h.
The mESCs were differentiated towards the neural lineage according to the protocol developed by Ying and Smith.9 At critical steps, we used the following parameters: cells were plated in the priming phase at 1.2 × 105 cells/cm2 in complete GMEM on 0.1% gelatin-coated Nunclon culture dishes (Nunc, Langenselbold, Germany). Next day, for neural induction, cells were plated on gelatin-coated Nunclon dishes at 104 cells/cm2 in N2/B27 medium (composition as described in Ying and Smith9, for a detailed description of B27 see http://www.paa.com/cell_culture_products/reagents/growthsupplements/neuromix.html). On day 7 of differentiation (DoD7) for neuronal generation and maturation, cells were replated at 104 cells/cm2 on poly--ornithin (10 μg/ml) and laminin (10 μg/ml)-coated Nunclon dishes in N2/B27 medium. Cells were fed every other day with complete medium change with N2/B27 medium.
Immunostaining and FACS analysis
For immunocytochemical analysis, cells were fixed with methanol (−20 °C) or 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 10% FBS, cells were incubated with primary antibodies (Tuj1 1 : 1000, NeuN 1 : 200, GAD65 1 : 200, SV2 1 : 200, PSD95 1 : 500, Nestin 1 : 500, Nestin-647 1 : 40, GFAP 1 : 800) overnight. After incubation with appropriate secondary antibodies, nuclei were counterstained with Hoechst H-33342 dye. Images were taken on the original cell culture dishes using an IX81 inverted microscope (Olympus, Hamburg, Germany) equipped with a -40X, NA 0.6 long-range lens and processed using CellP imaging software (Olympus). For confocal microscopy, cells were grown on four-well chamber slides (Nunc), fixed with 4% PFA/2% sucrose in PBS and permeabilized with 0.6% Triton X-100 in PBS. After blocking with 5% BSA/0.1% Triton X100 in PBS, cells were incubated with Tuj1 antibody in blocking buffer for 1 hour at room temperature. After incubation with appropriate secondary antibodies, nuclei were counterstained with DAPI. Confocal images were taken using a Zeiss LSM 510Meta confocal microscope equipped with a Plan Apochromat -63X, NA 1.4 oil DIC lens. Images were analyzed and processed using ImageJ.
For flow cytometry, cells were dissociated on DoD7 with accutase, fixed and permeabilized in Cytofix Buffer followed by Perm Buffer I (both BD Biosciences), and stained with anti-nestin antibody conjugated to Alexa-647, or isotype control. Cells were analyzed with an Accuri C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI, USA) and data were processed with CFlow Plus (Accuri Cytometers).
Quantitative PCR and quality control of differentiation
Total RNA of five independent differentiation experiments, performed at different times, with different CGR8 cell batches, and by different operators was isolated at indicated time points for marker gene expression analysis using Trizol, the RNA was retro-transcribed with SuperScript II reverse transcriptase, and the resultant cDNAs were amplified in a Biorad Light Cycler (Biorad, München, Germany) with primers specific for the genes of interest and designed for a common melting temperature of 60 °C. Real-time quantification for each gene was performed using SybrGreen and expressed relative to the amount of gapdh mRNA using the 2(−ΔΔ C(T)) method.38 For each run, the consistency of conditions and constancy of gapdh amounts in the samples were controlled by assessment of its absolute cycle number (=18±0.5).
Gene expression analysis
Cells were used for RNA preparation as undifferentiated mESCs before the priming phase (day 0), on DoD7 (before replating), DoD15 and DoD20. RNA was extracted from Trizol preparations and purified using RNeasy Mini prep columns (Qiagen, Hilden, Germany). The total RNA harvested was quantified using a Nanodrop device (Thermo Scientific, Waltham, MA, USA) and its integrity was assessed using Agilent Bioanalyser (Agilent, Santa Clara, CA, USA). Illumina TotalPrep RNA Amplification Kit (Ambion, Foster City, CA, USA) and 500 ng total RNA of each sample was used according to the manufacturer's protocol to produce biotin-labelled cRNAs. For hybridization onto Sentrix Mouse Ref.8 V2 mRNA microarray beadchips (Illumina, San Diego, CA, USA), 750 ng of labeled cRNA were incubated for 16 h at 58 °C. After hybridization, chips were washed, blocked and streptavadin-Cy3 stained. Fluorescence emission by Cy3 was quantitatively detected using BeadArray Reader Scan. Statistical analysis data are based on duplicate samples. Each of the samples contained pooled RNA from two differentiations to further increase robustness of results. Technical variation of the chip was minimal as tested by rerun of the same sample on two different arrays and by comparison of results from two beadchips within one array.
Data analysis
Original and processed data have been deposited for public access in the EBI Arrayexpress database (accession no. E-TABM-1068, specified release date; 30 September 2010). For initial processing, data were uploaded to Beadstudio (Illumina) for background subtraction. Further processing (baseline transformation and normalization to 75 percentile) and analysis was performed with Genespring 9.0 (Agilent), and all normalized expression kinetics data sets were used as input for an unsupervised non-hierarchical clustering with relation to the average of expression of all genes on the chip, using the K-means algorithm. The eight major clusters were selected for further analysis. Within these, significant gene expression profiles were selected, based on a minimum regulation of twofold on any of the time points and on two-way ANOVA, taking into account the regulation range and the variation between different arrays.
Patch-clamp recording
For functional characterization, neurons from at least three independent differentiations were tested for electrophysiological activity. Electrodes with a resistance of 2–5 MΩ were pulled of borosilicate glass (Clark, G150F; Warner Instruments, Hamden, CT, USA) on a Sutter Instruments (Novato, CA, USA) P-97 horizontal micropipette puller. All experiments were carried out using a custom built recording chamber (800 μl volume) made of Teflon within a temperature-controlled microscope stage (37 °C). Whole-cell voltage and current clamp recordings were obtained from cells at DoD20–24. Cells were grown on coated glass cover slips (10 mm) from DoD7. Whole-cell currents were recorded using an L/M-EPC-7 amplifier (List Medical Electronic, Darmstadt, Germany), digitised at sampling frequencies between 10–50 kHz using a DigiData 1320A AD/DA converter (Axon Instruments Inc.). The patch pipettes for spontaneous and evoked action potential measurements as well as for the neurotransmitter responses were filled with (in mM) 90 K+-gluconate, 40 KCl, 1 MgCl2, 10 NaCl, 10 EGTA, 4 Mg-ATP, 10 HEPES/KOH (pH 7.4 at 37 °C), whereas the bath solution contained (in mM) 155 NaCl, 1 CaCl2, 3 KCl, 10 -(+)-glucose, 10 HEPES/NaOH (pH 7.4 at 37 °C). The protocol for recording of Na+ and K+ channels was as follows: cells were hyperpolarized to −90 mV, and subsequently stepped to a defined voltage as indicated and returned to −70 mV, before the next cycle, with a different voltage step, was run. Each cycle took 120 ms. For the neurotransmitter response measurements, the different substances were directly added as concentrated stock solutions to the recording chamber in amounts of 1–10 μl. Antagonists were added at least 1 min before the agonists. Recordings were initiated within 100 ms after addition of agonists. For the measurement of barium currents through calcium channels, the pipette filling solution contained (in mM) 110 CsF, 10 NaCl, 20 TEA-Cl, 10 EGTA, 4 Na2-ATP, 10 HEPES/CsOH (pH 7.4 at 37 °C), whereas the bath solution contained (in mM) 130 NaCl, 10 BaCl2, 10 -(+)-glucose, 5 tetraethylammonium chloride, 10 4-aminopyridine, 0.5 tetrodotoxin, 10 HEPES/NaOH (pH 7.4 at 37 °C).
All current signals were normalized against the individual cell capacitances (as a surrogate measure for cell size) and are expressed in current densities (current divided by cell capacitance). Liquid junction potentials were measured and corrected, using the method described by Erwin Neher (1992), except for barium current measurements. Stimulation, acquisition and data analysis were carried out using pCLAMP 10.2 (Axon Instruments Inc., Sunnyvale, CA, USA) and ORIGIN 8.0 (OriginLab Corp., Northampton, MA, USA). Fast and slow capacitive transients were canceled online by means of analog circuitry. Residual capacitive and leakage currents were removed online by the P/4 method. Series resistance compensation was set to at least 50%. For analysis, traces were filtered offline at 5 kHz. Cells for measurements were chosen with respect to their morphological phenotype (small, round, highly elevated (phase-bright) cell bodies with projections of at least five times the cell body diameter, growing in network-like clusters containing at least 20–30 similar cells). The patch pipette was approached to these cells perpendicular to the plane formed by the cell membrane in the patch region.
Statistics and data mining
The numbers of replicates of each experiment are indicated in figure legends. Data were presented, and statistical differences were tested by ANOVA with post-hoc tests as appropriate, using GraphPad Prism 4.0 (Graphpad Software, La Jolla, CA, USA). Assignment of significantly overrepresented GO categories to different clusters and calculation of probabilities of a false-positive assignment was performed by G-profiler (http://biit.cs.ut.ee/gprofiler/39). For coverage of biological domains without appropriate and well-controlled GO category, relevant genes were assembled from the literature and cross-checked by 2–3 independent specialists. The number of genes within these groups identified in this study was indicated in relation to the overall number of possible hits or in relation to their distribution over different clusters. The genes defined in this study as embryonic stem cell markers or neural stem cell (NPC) markers were derived from recent literature.3 Neuronal (N) differentiation markers (n=574) were defined as all members of GO GO:0048699 (generation of neurons) corrected for those genes used as NPC markers. The graphical representation of identified genes (or groups) within their biological context is based on the major gene function as indicated on the NCBI-gene website and the literature. Importantly, members of each identified group were scored according to their suitability as markers for a PCR-based quality control of the differentiation pattern in toxicity experiments. Several selection rounds were run to identify the final set of markers displayed as example genes in the tables and some of the figures.
Toxicity experiments
Cells were exposed to chemicals during different phases of differentiation to test the suitability of the model system for neurotoxicity testing, and for testing of DNT during defined time windows. RA (1 μM), ‘3i' (a mixture of 0.8 μM PD184352, 2 μM SU5402, 3 μM CHIR99021)24 or cyclopamine (1 μM) were added to cultures from DoD1–DoD7 or from DoD8–DoD15. Then, the experiment was ended, or incubation was continued in the absence of chemicals for additional 6 days. On the final day, RNA was prepared by the Trizol method for PCR analysis. For morphological observations, the monolayer regions within the culture wells were imaged. Genes were preselected before the analysis as end points for initial proof-of-concept experiments, and results from all genes chosen are presented.
Acknowledgments
The work was supported in part by the Doerenkamp-Zbinden Foundation, the DFG, the EU FP7 project ESNATS (ML, SK), an IRTG1331 fellowship (BZ) and a fellowship from the KoRS-CB (PBK). We are grateful to Giovanni Galizia and Sabine Kreissl for help with the electrophysiological recordings and indebted to Bettina Schimmelpfennig for invaluable experimental support. The monoclonal antibodies Gad-6 developed by DI Gottlieb and SV2 developed by KM Buckley were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA 52242, USA. We thank KH Krause for the CGR8 mESC-line, and J Vilo and S Ilmjärv for help with bioinformatics analysis.
Glossary
- CNS
central nervous system
- DNT
developmental neurotoxicity
- DoD
day of differentiation
- ESC
embryonic stem cells
- GO
gene onthology
- mESC
murine embryonic stem cell
- N
gene onthology neuronal differentiation
- NPC
neural precursor cell
- RA
all-trans retinoic acid
- Shh
sonic hedgehog
- GMEM
Glasgow's modified Eagles medium
- FBS
fetal bovine serum
- PFA
paraformaldehyde
- PBS
phosphate-buffered saline
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JC Marine
Supplementary Material
References
- Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities. Nat Rev Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- Kuegler PB, Zimmer B, Waldmann T, Baudis B, Ilmjarv S, Hescheler J, et al. Markers of murine embryonic and neural stem cells, neurons and astrocytes: reference points for developmental neurotoxicity testing. Altex. 2010;27:17–42. doi: 10.14573/altex.2010.1.16. [DOI] [PubMed] [Google Scholar]
- Barberi T, Klivenyi P, Calingasan NY, Lee H, Kawamata H, Loonam K, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotech. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, et al. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS One. 2009;4:e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Bremer S, Brundin P, Hescheler J, Kirkeby A, Krause KH, et al. The biological and ethical basis of the use of human embryonic stem cells for in vitro test systems or cell therapy. Altex. 2008;25:163–190. [PubMed] [Google Scholar]
- Strübing C, Ahnert-Hilger G, Shan J, Wiedenmann B, Hescheler J, Wobus AM. Differentiation of pluripotent embryonic stem cells into the neuronal lineage in vitro gives rise to mature inhibitory and excitatory neurons. Mech Dev. 1995;53:275–287. doi: 10.1016/0925-4773(95)00446-8. [DOI] [PubMed] [Google Scholar]
- Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Makris SL, Raffaele K, Allen S, Bowers WJ, Hass U, Alleva E, et al. A retrospective performance assessment of the developmental neurotoxicity study in support of OECD test guideline 426. Environ Health Perspect. 2009;117:17–25. doi: 10.1289/ehp.11447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Landrigan PJ. Developmental neurotoxicity of industrial chemicals. Lancet. 2006;368:2167–2178. doi: 10.1016/S0140-6736(06)69665-7. [DOI] [PubMed] [Google Scholar]
- Bal-Price AK, Hogberg HT, Buzanska L, Lenas P, van Vliet E, Hartung T. In vitro developmental neurotoxicity (DNT) testing: relevant models and endpoints. Neurotoxicology. 2009. in press. [DOI] [PubMed]
- Hansson O, Petersén A, Leist M, Nicotera P, Castilho RF, Brundin P. Transgenic mice expressing a Huntington's disease mutation are resistant to quinolinic acid-induced striatal excitotoxicity. Proc Natl Acad Sci USA. 1999;96:8727–8732. doi: 10.1073/pnas.96.15.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penschuck S, Flagstad P, Didriksen M, Leist M, Michael-Titus AT. Decrease in parvalbumin-expressing neurons in the hippocampus and increased phencyclidine-induced locomotor activity in the rat methylazoxymethanol (MAM) model of schizophrenia. Eur J Neurosci. 2006;23:279–284. doi: 10.1111/j.1460-9568.2005.04536.x. [DOI] [PubMed] [Google Scholar]
- Rossi AD, Larsson O, Manzo L, Orrenius S, Vahter M, Berggren PO, et al. Modifications of Ca2+ signaling by inorganic mercury in PC12 cells. FASEB J. 1993;7:1507–1514. doi: 10.1096/fasebj.7.15.8262335. [DOI] [PubMed] [Google Scholar]
- Wei Y, Harris T, Childs G. Global gene expression patterns during neural differentiation of P19 embryonic carcinoma cells. Differentiation. 2002;70:204–219. doi: 10.1046/j.1432-0436.2002.700409.x. [DOI] [PubMed] [Google Scholar]
- Aiba K, Sharov AA, Carter MG, Foroni C, Vescovi AL, Ko MS. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells. 2006;24:889–895. doi: 10.1634/stemcells.2005-0332. [DOI] [PubMed] [Google Scholar]
- Jongen-Relo AL, Leng A, Luber M, Pothuizen HH, Weber L, Feldon J. The prenatal methylazoxymethanol acetate treatment: a neurodevelopmental animal model for schizophrenia. Behav Brain Res. 2004;149:159–181. doi: 10.1016/s0166-4328(03)00228-6. [DOI] [PubMed] [Google Scholar]
- Marx-Stoelting P, Adriaens E, Ahr HJ, Bremer S, Garthoff B, Gelbke HP, et al. A review of the implementation of the embryonic stem cell test (EST). The report and recommendations of an ECVAM/ReProTect Workshop. Altern Lab Anim. 2009;37:313–328. doi: 10.1177/026119290903700314. [DOI] [PubMed] [Google Scholar]
- Rao MS, Jacobson M. Dev Neurobiol. Kluwer Academic/Plenum Publishers: New York; 2005. [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology: transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Hartung T, Nicotera P. The dawning of a new age of toxicology. Altex. 2008;25:103–114. [PubMed] [Google Scholar]
- Irioka T, Watanabe K, Mizusawa H, Mizuseki K, Sasai Y. Distinct effects of caudalizing factors on regional specification of embryonic stem cell-derived neural precursors. Brain Res Dev Brain Res. 2005;154:63–70. doi: 10.1016/j.devbrainres.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, et al. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134:285–293. doi: 10.1242/dev.02727. [DOI] [PubMed] [Google Scholar]
- Hartung T, Hoffmann S. Food for thought … on insilico methods in toxicology. Altex. 2009;26:155–166. doi: 10.14573/altex.2009.3.155. [DOI] [PubMed] [Google Scholar]
- Lee S-H, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotech. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Lund S, Christensen KV, Hedtjarn M, Mortensen AL, Hagberg H, Falsig J, et al. The dynamics of the LPS triggered inflammatory response of murine microglia under different culture and in vivo conditions. J Neuroimmunol. 2006;180:71–87. doi: 10.1016/j.jneuroim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Mody M, Cao Y, Cui Z, Tay KY, Shyong A, Shimizu E, et al. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H, Kolde R, Adler P, Aksoy I, Anastassiadis K, Bader M, et al. The FunGenES database: a genomics resource for mouse embryonic stem cell differentiation. PLoS One. 2009;4:e6804. doi: 10.1371/journal.pone.0006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przyborski SA, Smith S, Wood A. Transcriptional profiling of neuronal differentiation by human embryonal carcinoma stem cells in vitro. Stem Cells. 2003;21:459–471. doi: 10.1634/stemcells.21-4-459. [DOI] [PubMed] [Google Scholar]
- van Dartel DA, Zeijen NJ, de la Fonteyne LJ, van Schooten FJ, Piersma AH. Disentangling cellular proliferation and differentiation in the embryonic stem cell test, and its impact on the experimental protocol. Reprod Toxicol. 2009;28:254–261. doi: 10.1016/j.reprotox.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Suter DM, Tirefort D, Julien S, Krause KH. A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells. 2009;27:49–58. doi: 10.1634/stemcells.2008-0319. [DOI] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler – a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucl Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.