Abstract
14-3-3s are binding proteins with survival functions in cells by interaction with proteins involved in the regulation of cell fate. The role of 14-3-3 during autophagy was investigated, thus, a forced expression of 14-3-3ζ reduces C2-ceramide-induced autophagy, whereas depletion of 14-3-3ζ promotes autophagy. The 14-3-3 role in autophagyc-related proteins was also investigated. The human vacuolar protein sorting 34 (hVps34), the class III phosphatidylinositol-3-kinase mediates multiple vesicle-trafficking processes such as endocytosis and autophagy, its activation being a requirement for autophagy initiation. Using chromatography techniques, hVps34 were eluted from a 14-3-3 affinity column, showing also a direct interaction with 14-3-3 proteins under physiological condition. Further analysis suggests that hVps34/14-3-3 association is a phorbol-12-myristate-13-acetate-dependent phosphorylated mechanism promoting a strong inhibition of the hVps34 lipid kinase activity, proteins kinase C being the likely kinase involved in phosphorylation and 14-3-3 binding of hVps34 under physiological conditions. Meanwhile, stimulation of autophagy leads to the dissociation of the 14-3-3/hVps34 complex enhancing hVps34 lipid kinase activity. Forced expression of 14-3-3ζ reduces hVps34 kinase activity and depletion of 14-3-3ζ promotes upregulation of this activity. In this study, 14-3-3ζ proteins are shown as a negative regulator of autophagy through regulation of a key component of early stages of the autophagy pathway, such as hVps34.
Keywords: 14-3-3, autophagy, starvation, C2-ceramide, hVps34
Macroautophagy (herein referred to as autophagy) is an evolutionarily conserved and regulated pathway characterized by an increase in the number of autophagosomes, vacuoles surrounded by a double membrane that sequesters cytoplasm or organelles. Subsequently, autophagosomes fuse with lysosomes to form autolysosomes, in which sequestered material is digested by lysosomal hydrolases.1 Focusing on an autophagy pathway component, one of the earliest characterized mammalian autophagy proteins is Beclin-1 (encoded by Becn1, the orthologue of yeast autophagy-related gene 6 (Atg6)/Vps30).2 This protein is essential for the initiation of autophagy, perhaps via its interaction with the phosphatidylinositol-3-kinase class III (PI3KC3)/human vacuolar protein sorting 34 (hVps34; mammalian homolog of yeast Vps34) forming a complex that participates in autophagosome formation.3 hVps34 specifically produces the lipid phosphatidylinositol 3-phosphate,4 this activity being essential for autophagy. Thus, an inhibitor of hVps34 activity, 3-methyladenine (3-MA), is a known inhibitor of autophagy.5 Similar to yeast, a regulatory mechanisms directing Atg6/hVps34 specificity have been identified in mammals, thus newly identified proteins, Atg14L (yeast Atg14-like) and Rubicon (RUN domain and cysteine-rich domain containing, Beclin-1-interacting protein) were identified in a complex with Beclin-1/hVps34. Characterization of these proteins revealed that Atg14L enhances hVps34 lipid kinase activity and upregulates autophagy, whereas Rubicon reduces hVps34 activity and downregulates autophagy.6 According to these results it seems clear that autophagy is strictly controlled, maybe because of the multiplicity of functions that this process can promote in vivo. In this sense, it is still unclear if autophagy is a survival or death mechanism, or both. First, autophagy seems to have a prosurvival function at the cellular level. Thus, in the case of starvation or growth factor deprivation, autophagy genes are essential to maintain cellular ATP energy production, macromolecular synthesis and cell survival.7 In addition, the absence of autophagy genes promotes apoptosis of starved HeLa cells.8 Several other connections have been found between autophagy and cell death and in this regard it is worth mentioning that C2-ceramide-promoted autophagy may also represent a form of caspases-independent cell death.9 Antiapoptotic protein B-cell lymphoma 2 (Bcl-2) binds to Beclin-1, which disrupts autophagy and autophagy gene-dependent cell death.10 Recently, the FLICE-like inhibitor protein (cFLIP) that protect cells from apoptosis mediated by a death receptor, has been shown to regulate autophagosome biogenesis and to suppress caspases-independent cell death with autophagy.11
In this study, the aim was to obtain further understanding of the autophagy mechanism in context of survival or cell death by analyzing the role of 14-3-3 proteins on autophagy regulation. 14-3-3 is a binding protein with survival functions in cells by interaction with large numbers of proteins involved in the regulation of apoptosis, thereby modulating the function of the associated partners.12 In most cases, 14-3-3-mediated sequestration of proapoptotic client proteins mediates suppression of apoptosis.
In this study, mammalian cells were used to assess whether or not 14-3-3 proteins regulate autophagy. It was shown that overexpression of 14-3-3ζ protein blocks autophagy, whereas depletion of 14-3-3ζ promotes autophagy. Furthermore, 14-3-3 directly interacts with hVps34, allowing a negative regulation of hVps34 kinase activity under physiological conditions, proteins kinase C being the likely kinase involved in phosphorylation and 14-3-3 binding of hVps34. Meanwhile, starvation or C2-ceramide-induced autophagy promotes dissociation of the 14-3-3/hVps34 complex leading to hVps34 activation, which is required during autophagy. These data provide, to my understanding, the first evidence that 14-3-3ζ functions as a negative regulator of autophagy through inhibition of hVps34 kinase activity, which is essential for the autophagosome membrane assembly in the early stages of the autophagic process.
Results
Overexpression of 14-3-3ζ proteins inhibit C2-ceramide-induced autophagy
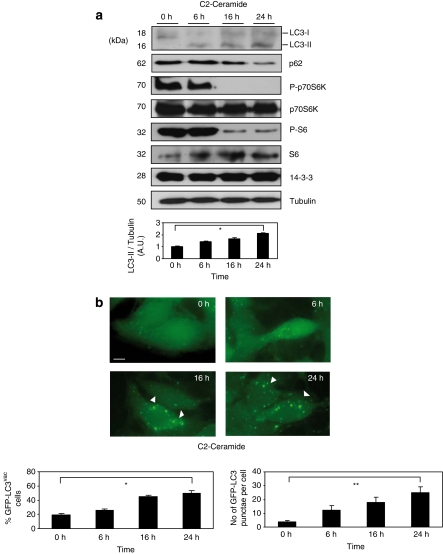
Ceramide is a bioactive sphingolipid associated with a large range of cell processes. Previous reports clearly show that C2-ceramide triggers autophagy by several mechanisms such as inhibition of the class I PI3K/AKT pathway and activation of protein phosphatases (PP2A and PP1) which promote inactivation of a number of progrowth cellular regulators (revised in Zheng et al.13). According to these data, Figure 1 clearly shows that 10 μM of C2-ceramide in a range of time triggers autophagy. Thus, C2-ceramide treatment induces the appearance of the structural component of autophagosomes microtubule-associated protein1 light chain 3 II (LC3-II; also known as Atg8), decreases the levels of a general autophagy marker such as p62/SQSTM1 and inhibits mammalian target of rapamycin (mTOR) activity (by analysis of P-p70S6K and P-S6; Figure 1a). C2-ceramide also increases the number of cells with autophagosome accumulation and the number of autophagosomes per cell seen by using microscopy analysis of autophagosome-associated protein LC3 (fused to green fluorescent protein (GFP)-LC3) distribution (Figure 1b). Autophagosome formation was significant at 16–24 h after the addition of 10 μM of C2-ceramide. Significantly, C2-ceramide within this concentration and time failed to induce death in HeLa cells (see Supplementary Figure S1). These data support C2-ceramide-induced autophagosome accumulation being dependent on the classic autophagy pathway through mTOR inactivation, reflecting an increase in functional autophagy as was previously reported.14
Figure 1.
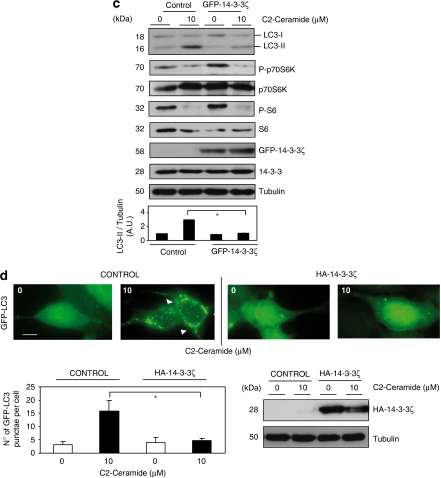
Overexpression of 14-3-3ζ proteins block C2-ceramide-induced autophagy. (a) Hela cells were incubated with 10 μM of C2-ceramide for indicated times. Cell extracts were harvested for immunoblot analysis of LC3-I processing into LC3-II, p62/SQSTM1, P-p70S6K, total p70S6K, P-S6, total 6S and 14-3-3 proteins. Tubulin was used as a protein loading control. Endogenous LC3-II/tubulin levels were quantified and the ratio presented as arbitrary units (N=3, *P=0.0006, Student t-test). (b) HEK293T cells transfected with GFP-LC3 were treated as in (a) and GFP-LC3 was analyzed by microscopy. Quantification of GFP-LC3vac cells number and number of GFP-LC3 punctae per cell were performed by counting in a blinded experiment. Columns, average of three different experiments, set up with duplicated samples and counting 6 × 100 cells per sample (quantification of GFP-LC3vac cells with more that five punctae per cell, is presented as a percentage of total cell numbers, N=3, *P=0.0001, **P=0.00087 Student t-test). (c) HEK293T cells transfected with GFP (control) or GFP-14-3-3ζ (GFP-14-3-3ζ) were left untreated or stimulated with C2-ceramide 10 μM for 24 h. Cell extracts were harvested for immunoblot analysis of LC3-I processing into LC3-II, P-p70S6K, total p70S6K, P-S6, total 6S, GFP-14-3-3ζ and total 14-3-3 proteins. Tubulin was used as a protein loading control. Endogenous LC3-II/tubulin levels were quantified and the ratio presented as arbitrary units. Columns, average of three different experiments (*P=0.00037, Student t-test). (d) HEK293T/GFP-LC3 cells were transfected with HA-control or HA-14-3-3ζ were left untreated or stimulated with C2-ceramide 10 μM for 24 h. GFP-LC3 was analyzed. Quantification of number of GFP-LC3 punctae per cell was performed by counting in a blinded experiment. Columns, average of three different experiments, set up with duplicated samples and counting 6 × 100 cells per sample, shown as percentage. Bars=10 μm (*P=0.00059, Student t-test). Western blot analysis shows levels of HA-14-3-3ζ. Tubulin was used as a loading control. A full-color version of this figure is available at Cell Death and Differentiation online
Ceramide clearly induces autophagy but whether ceramide-induced autophagy is a survival or cell death mechanism is under discussion and requires in-depth scrutiny. Thus, taking into consideration this controversy it becomes a matter of interest to study the function of survival 14-3-3 proteins during C2-ceramide-induced autophagy. Several reports suggest that 14-3-3ζ isoform has an important role in the cell survival regulation, in fact overexpression of this isoform leads the resistance to cell death (see Supplementary Figure S2). Thus, C2-ceramide-induced autophagy was evaluated in HEK293T cells transfected or not with GFP-14-3-3ζ. As was shown for HeLa cells, untransfected HEK293T cells responded to C2-ceramide by increasing autophagy (Figure 1c and d). It is noteworthy that overexpression of GFP-14-3-3ζ blocks autophagy induction (see ratio LC3-II/tubulin ratio in Figure 1c) even in the presence of low levels of mTOR activity checked by phosphorylation of its p70S6K substrate. This fact raises the possibility that the autophagy pathway could be directly regulated downstream of mTOR by 14-3-3 proteins. Interestingly, overexpression of HA-14-3-3ζ in C2-ceramide-treated HEK293T cells was able to considerably reduce the presence of the typical punctae structures of GFP-LC3 in autophagosomes formation during C2-ceramide treatment.
Downregulation of endogenous 14-3-3ζ sensitizes cells to autophagy
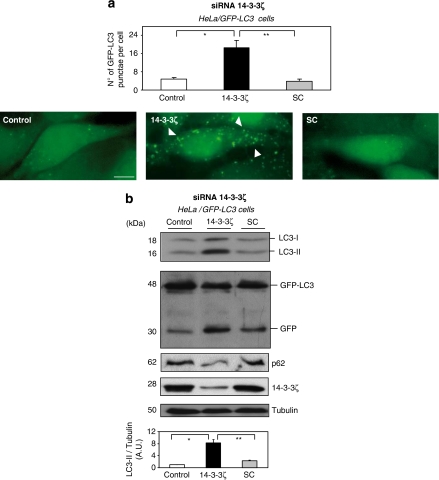
To further investigate the role of 14-3-3 in autophagy, 14-3-3ζ expression was knocked down in serum-grown cells by RNA interference (siRNA). Autophagosome formation was analyzed using fluorescence microscopy by checking increased punctae of GFP-LC3 in the transiently transfected HeLa cell. In control or scrambled siRNA-treated cells, few GFP-LC3 puncta were observed. In contrast, 14-3-3ζ siRNA transfection resulted in accumulation of a large GFP-LC3 puncta (Figure 2a). In addition, Hela cells, expressing GFP-LC3, 14-3-3ζ siRNA resulted in an increased LC3-II/tubulin ratio and in the processing of GFP-LC3 into GFP (caused by the autophagic-dependent degradation of LC3) in comparison with control or scramble siRNA (Figure 2b). Similar results were found using GFP-LC3 stable transfected MCF-7 breast carcinoma cell line (Supplementary Figure S3). It is worth mentioning that downregulation of endogenous 14-3-3ζ does not affect levels of expression of autophagy-related proteins under siRNA transfection conditions used in this study (Supplementary Figure S4). On the other hand, it has been well establish in the literature that 14-3-3 isoforms have the tendency to work redundantly in cell survival mechanisms, moreover, the 14-3-3σ isoform has an additional function of controlling cell cycle progression. The first analysis suggests that 14-3-3θ and 14-3-3ζ are best expressed in different cell types; 14-3-3σ was also identified in the MCF-7 cell line (Supplementary Figure S5A). Aimed at analyzing the impact of 14-3-3θ and 14-3-3σ isoforms on autophagy initiation, their expressions were knocked down in serum-grown cells by siRNA. Meanwhile this work was in revision, Wang et al.15 using U2OS cell lines stably expressing inducible or transient siRNA against 14-3-3τ (θ), using in house 19-nt target sequences for si14-3-3τ under the control of tetracycline operon in pSUPERIOR.puro vector, a significant decreases of the Beclin-1 protein expression was observed on depletion of 14-3-3τ, becoming a positive regulator of autophagy. Nevertheless, siRNA against either 14-3-3ζ, 14-3-3σ or 14-3-3θ from Santa Cruz Biotechnology (Santa Cruz, CA, USA) clearly depletes each 14-3-3 isoform expression specifically and induce autophagy, whereas it does not affect levels of expression of autophagy-related proteins, included Beclin-1 (Supplementary Figures S4 and S5).
Figure 2.
Downregulation of endogenous 14-3-3ζ sensitizes cells to autophagy. (a) HeLa cells expressing GFP-LC3 were transfected either with siRNA oligonucleotide targeting 14-3-3ζ or with a scrambled RNA oligonucleotide as described in Material and Methods. After 48 h, untransfected cells (control) or transfected either with siRNA 14-3-3ζ (14-3-3ζ) or scrambled siRNA (SC) were analyzed using GFP-LC3 by microscopy. Bars=10 μm. Quantification of number of GFP-LC3 punctae per cell was performed by counting in a blinded experiment. Columns, average of three different experiments, set up at duplicated samples and counting 6 × 100 cells per sample (N=3, *P=0.00034, **P=0.0003 Student t-test). (b) As in (a) extracts from untransfected cells (control) or transfected either with siRNA 14-3-3ζ (14-3-3ζ) or scrambled siRNA (SC) were harvested after 48 h for immunoblot analysis of LC3-I processing into LC3-II, GFP-LC3 processing into GFP, p62/SQSTM1 and endogenous 14-3-3ζ isoform. Tubulin was used as a protein loading control. Endogenous LC3-II/Tubulin levels were quantified and the ratio presented as arbitrary units (N=3, *P=0.0003, **P=0.0004, Student t-test). A full-color version of this figure is available at Cell Death and Differentiation online
All these data suggest that knocked down 14-3-3ζ protein clearly induces autophagy in cervix and breast cancer cell lines, whereas participation of the rest of the isoforms cannot be ruled out because at least another two additional isoforms (σ and θ) seem to work redundantly in autophagy regulation probably because of its tendency to form heterodimers.
The regulation of autophagy by 14-3-3 proteins raises the possibility that the autophagy pathway could be directly regulated by 14-3-3 proteins through binding to specific autophagy-related proteins.
14-3-3 Binding of proteins involved in autophagic pathway
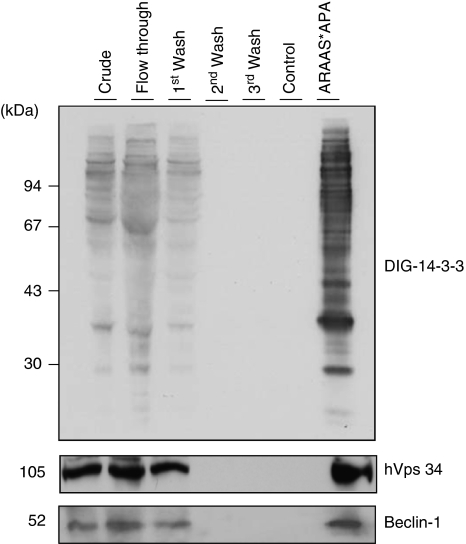
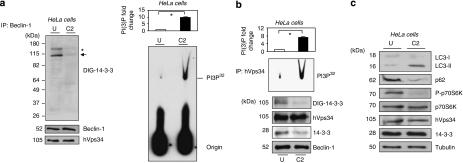
To approach this possibility, human 14-3-3-binding proteins were isolated using a 14-3-3 affinity chromatography. Using this method, among 200 14-3-3 affinity-purified proteins were identified in the past, a big proportion of them being studied in detail by their interaction to 14-3-3 proteins.16, 17 A crude extract of human HeLa cells continuously growing in serum were passed through a 14-3-3-Sepharose column. After extensive washes and ‘mock' elution with control peptides that do not bind to 14-3-3, proteins were eluted with a 14-3-3-binding phosphopeptide (ARAApSAPA, where pS is phosphorylated serine), with the aim of selectively purifying those proteins that were bound to the phosphopeptide-binding site on the immobilized 14-3-3s. The 14-3-3 overlay shows that the ARAApSAPA elution pool was highly enriched in 14-3-3-binding proteins compared with the crude extract, and that 14-3-3-binding proteins were not eluted with the control non-binding (phospho)peptide (Figure 3). Inmunoblotting analysis confirmed the presence of hVps34 and Beclin-1, with its appropriate molecular mass, in the ARAApSAPA elution pool. None of these proteins were eluted from the column by either extensive washing under high-salt conditions or mock elution. These results indicate that isolated proteins bind to 14-3-3 proteins in a phosphorylation-dependent manner, either directly or as components of protein complexes. Interestingly, both proteins involved in autophagosome formation during early and regulatory steps of autophagy have been reported to form a complex.3
Figure 3.
14-3-3 Affinity chromatography of human HeLa cell extracts. Clarified HeLa cell extract was chromatographed on 14-3-3-Sepharose, as described in Material and Methods. Column fractions were run on SDS/PAGE using 10% Tris-glycine gels, and transferred to nitrocellulose membranes. Amounts of protein run on SDS/PAGE were as follows: extract, flow through and beginning of salt wash (first wash), 40 μg of each; middle and end of salt wash (second wash and third wash, respectively), protein undetectable; control (phospho) peptide pool,<1 μg; and ARAApSAPA (ARAAS*APA) elution pool, 2 μg. Membranes were checked for binding to DIG-14-3-3 (top panel: Published in author's previous work.47 Copyright owners belong to author). Western blots were probed with antibodies against the indicated proteins related to autophagy (bottom panels)
These data suggest the possibility that 14-3-3 proteins can regulate the autophagy initiation process through association with essential proteins of this pathway. To test this hypothesis, 14-3-3 association with autophagy-related proteins was studied in detail.
hVps34 dissociates from 14-3-3 and is activated during C2-ceramide-induced autophagy
Autophagy-related proteins with essential regulatory roles in autophagy progression are Beclin-1 and hVps34. In this study, the amount of hVps34 associated with Beclin-1 complex was found to be similar under normal growth conditions or in C2-ceramide-induced autophagy in HeLa (Figure 4) and HEK293T cells (Supplementary Figure S6) using experiment of endogenous proteins co-immunoprecipitation. Analyzing digoxigenin (DIG)-14-3-3 overlay shows no sign of Beclin-1 size but a band that matched hVps34 size was found during serum-growth conditions, which become lost during C2-ceramide-induced autophagy in both cell lines (Figure 4, Supplementary Figure S6). An unknown 14-3-3-binding protein band (*) was observed in Beclin-1 immunoprecipitates, whose identity is now the focus of study. Considering that hVps34/14-3-3 association is a phosphorylated-dependent mechanism (see Figure 3) and that C2-ceramide may activate protein phosphatases (PP2A and PP1; revised in Zheng et al.13), 14-3-3/hVps34 dissociation may cause by protein phosphatase activation during C2-ceramide treatments. It is noteworthy also that hVps34/14-3-3 association/dissociation does not affect hVps34/Beclin-1 complex. On the other hand, 14-3-3 endogenous proteins co-immunoprecipitated with endogenous hVps34 under physiological conditions, this interaction being lost during autophagy (Figure 4b). An interesting observation is the fact that hVps34 lipid kinase activity, in both hVps34 and Beclin-1 immunoprecipitates, increases when it loses its association with 14-3-3 proteins during autophagy in HeLa and HEK293T cells. This raises the possibility that hVps34 kinase activity can be regulated by association with 14-3-3 proteins. To examine this hypothesis, the hVps34 kinase activity in mammalian cells overexpressing and lacking 14-3-3ζ proteins was measured.
Figure 4.
hVps34 dissociates from 14-3-3 and is activated during C2-ceramide-induced autophagy. HeLa cells continuously growing in serum were left untreated or stimulated with C2-ceramide (10 μM) for 24 h to promote autophagy. (a) Extracts (2 mg) from untreated (U) or C2-ceramide (C2) stimulated HeLa cells were incubated with 1 μg of Beclin-1 (Santa Cruz Biotecnology, Inc.) antibody bound to protein-G-Sepharose. The washed immunoprecipitates were resolved using SDS-PAGE, transferred to nitrocellulose and probed with DIG-14-3-3 (14-3-3 overlay), Beclin-1 (BD Biosciences) and hVps34 (Zymed) antibodies. The asterisk (*) shows an unknown protein band. The arrow corresponds to hVps34 size on DIG-14-3-3. The washed immunoprecipitates were also used for hVps34 in vitro kinase assay (right panel). Autoradiography shows levels of PI3P32. (Quantification of hVps34 in vitro kinase assay expressed as PI(3) fold change, N=3, *P=0.0007, Student t-test.) (b) Extracts (1 mg) from untreated (U) or C2-ceramide (C2) stimulated HeLa cells were incubated with 1 μg of hVps34 (Echelon Biosciences) antibody bound to protein-G-Sepharose. The washed immunoprecipitates were used for hVps34 in vitro kinase assay and also to analyze interaction with 14-3-3 (DIG-14-3-3) and presence of Beclin-1, hVps34 and 14-3-3 proteins in the immunoprecipitates (quantification of hVps34 in vitro kinase assay, N=3, *P=0.0007, Student t-test). Piece of the DIG-14-3-3 shown correspond to hVps34 size. (c) Extracts (30 μg) from HeLa cells continuously growing in serum or stimulated with C2-ceramide (10 μM) for 24 h to promote autophagy were run on SDS-PAGE, transferred to nitrocellulose membranes and probed with indicated antibodies
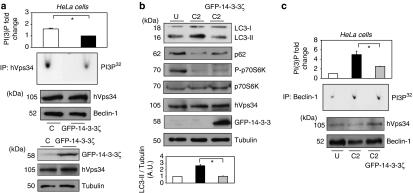
Increased expression of 14-3-3ζ decreases hVps34 activity, meanwhile blocking C2-ceramide-dependent autophagy
To examine the role of 14-3-3ζ proteins on hVps34 kinase activity and autophagy initiation, HeLa and HEK293T cells were transfected with GFP-14-3-3ζ. Those transfected with GFP-14-3-3ζ exhibit a significant decrease of hVps34 activity under physiological conditions suggesting that 14-3-3 proteins may directly inhibit hVps34 activity (Figure 5a, Supplementary Figure S7A). In addition, GFP-14-3-3ζ overexpression blocks C2-ceramide-dependent autophagy (Figures 1c, d and 5b, Supplementary Figure S7B) and significantly decreases hVps34 kinase activity in comparison with those levels found in untransfected cells treated with C2-ceramide (Figure 5c and Supplementary Figure S7C). The hVps34 function on autophagy progression being essential, the role of 14-3-3 proteins decreasing hVps34 kinase activity may contribute to 14-3-3-dependent inhibition of autophagy.
Figure 5.
Overexpression of 14-3-3ζ decreases hVps34 activity meanwhile blocks C2-ceramide-dependent autophagy. HeLa cells were transfected with GFP-control (C) or GFP-14-3-3ζ. (a) Continuously growing in serum HeLa cells extracts (1 mg) were incubated with 1 μg of hVps34 (Echelon Biosciences) antibody bound to protein-G-Sepharose. The washed immunoprecipitates were used for hVps34 in vitro kinase assay and also resolved to detect Beclin-1 and hVps34 proteins (top panel: quantification of hVps34 in vitro kinase assay expressed as PI(3) fold change, N=3, *P=0.0013, Student t-test). Cell extracts (30 μg) from both samples were run on SDS-PAGE, transferred to nitrocellulose membranes and probed with indicated antibodies (bottom panel). (b) HeLa cell extracts left untreated (U) or C2-ceramide stimulated (C2; 10 μM for 24 h) were run on SDS-PAGE, transferred to nitrocellulose membranes and probed with indicated antibodies. Endogenous LC3-II/Tubulin levels were quantified and the ratio presented as arbitrary units (N=3, *P=0.0029, Student t-test). (c) HeLa cell extracts (2 mg) from left untreated (U) or C2-ceramide stimulated (C2) (10 μM for 24 h) cells were incubated with 1 μg of Belin-1 (Santa Cruz Biotechnology, Inc.) antibody bound to Protein-G-Sepharose. The washed immunoprecipitates were used for hVps34 in vitro kinase assay and to analyze the presence of Beclin-1 and hVps34 proteins in immonoprecipitates. (On the top: quantifications of hVps34 in vitro kinase assays, N=3, *P=0.012, Student t-test.)
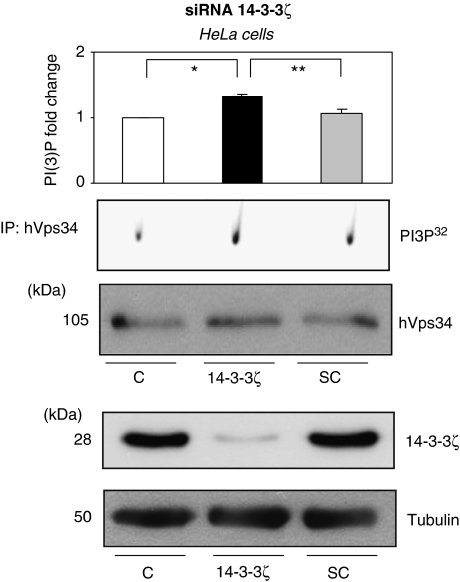
Downregulation of endogenous 14-3-3ζ increases hVps34 activity
Several assays were performed to determine the role of 14-3-3 in autophagy. Previously, it was shown that 14-3-3ζ overexpression promotes decrease of hVps34 activity and inhibition of autophagy; secondly, 14-3-3ζ expression was knocked down in serum-grown cells by RNA interference (siRNA) and autophagy was induced. When, hVps34 kinase activity was analyzed, a significant increase of activity was observed during the 14-3-3ζ knocked down in HeLa and MEF-7 in comparison with control cells (Figure 6 and Supplementary Figure S8). These data suggest that knocked down of 14-3-3ζ proteins clearly induces autophagy in cervix and breast cancer cell lines and the autophagy induction is coordinated with an increase in hVps34 lipid kinase activity.
Figure 6.
Downregulation of endogenous 14-3-3ζ increase hVps34 activity. HeLa cells were transfected either with siRNA oligonucleotide targeting 14-3-3ζ or with a scrambled RNA oligonucleotide as described in Material ad Methods. After 48 h, extract of untransfected (C) or transfected either with siRNA 14-3-3ζ (14-3-3ζ) or scrambled siRNA (SC) HeLa cells were incubated with 1 μg of hVps34 (Echelon Biosciences) antibody bound to Protein-G-Sepharose. The washed immunoprecipitates were used for hVps34 in vitro kinase assay and to analyze the presence of hVps34 protein in immonoprecipitates (top panel: quantification of hVps34 in vitro kinase assay, N=3, *P=0.038, **P=0.017, Student t-test). Extracts from untransfected cells (C) or transfected either with siRNA 14-3-3ζ (14-3-3ζ) or scrambled siRNA (SC) were harvested for immunoblot analysis of 14-3-3ζ isoform. Tubulin was used as a protein loading control (bottom panel)
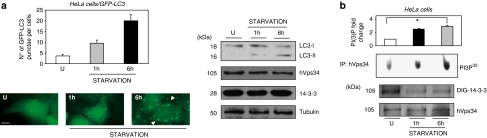
hVps34 dissociates from 14-3-3 and is activated during starvation-induced autophagy
The phosphatidylinositol-3-kinase class III, hVps34 is required for the induction of autophagy during nutrient deprivation.18 To further analyze the regulation of 14-3-3 proteins on hVps34 activity during the initial step of autophagy, HeLa and MCF-7 cells were left untreated or starved for several hours to induce autophagy. It was observed that hVps34 lipid kinase activity increases when it loses its association with 14-3-3 proteins during starvation-induced autophagy in HeLa and MCF-7 cells (Figure 7, Supplementary Figure S9). On the other hand, rapamycin and etoposide promote hVps34/14-3-3 dissociation as well while inducing autophagy, 3-MA blocks rapamycin- and etoposide-induced autophagy, whereas hVps34 remains associated with 14-3-3 (Supplementary Figure S10). Taking into consideration that hVps34/14-3-3 binding seems to be a phosphorylated-dependent mechanism (see Figure 3), several reports suggesting that rapamycin and etoposide promote phosphatase activities19 may explain the dephosphorylation of hVps34 leading its dissociation with 14-3-3 proteins.
Figure 7.
hVps34 dissociates from 14-3-3 and is activated during starvation-induced autophagy. (a) HeLa cells expressing GFP-LC3 left untreated or starved for indicated times were analyzed using GFP-LC3 by microscopy. Bars=10 μm. Quantification of number of GFP-LC3 punctae per cell was performed by counting in a blinded experiment. Columns, average of three different experiments, set up at duplicated samples and counting 6 × 100 cells per sample (N=3, *P=0.0047, Student t-test; left panel). Untreated and starved HeLa cells were harvested for immunoblot analysis of indicated proteins. Tubulin was used as a protein loading control (right panel). (b) HeLa cells were treated as in (a) and extracts from untreated and starved cells were incubated with 1 μg of hVps34 (Echelon Biosciences) antibody bound to Protein-G-Sepharose. The washed immunoprecipitates were used for hVps34 in vitro kinase assay and test presence of hVps34 protein in immunoprecipitates and 14-3-3 binding (DIG-14-3-3) (top panel: quantification of hVps34 in vitro kinase assay, N=3, *P=0.0001, Student t-test). A full-color version of this figure is available at Cell Death and Differentiation online
These data suggest that hVps34 binds 14-3-3 proteins under physiological conditions, this association being lost during autophagy initiation by starvation, C2-ceramide, rapamycin or etoposide.
Phorbol-12-myristate-13-acetate (PMA)-dependent 14-3-3 binding of hVps34 promotes a decreased hVps34 lipid kinase activity
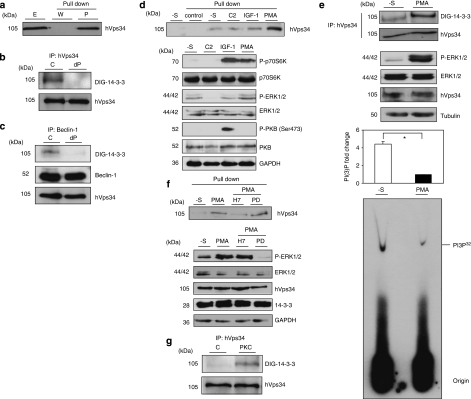
Aimed at further analyzing 14-3-3/hVps34 binding, hVps34 was also found to be present in 14-3-3-Sepharose precipitates from continuously serum-growing HeLa cells and lung mouse tissue extracts (Figure 8a, Supplementary Figure S11).
Figure 8.
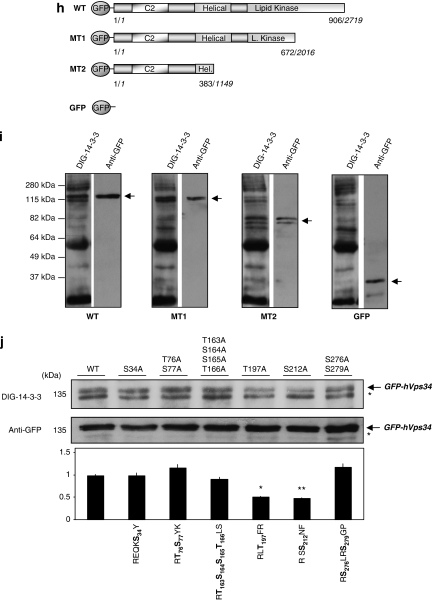
hVps34 binds 14-3-3 proteins by a phosphorylation-dependent mechanism. (a) HeLa cells continuously grown in the presence of serum were harvested as indicated and cell extracts (5 mg of protein) were incubated for 2 h with 50 μl of 14-3-3-Sepharose. Extract (E; 30 μg) and pellets, washed (W) and extracted with SDS-sample buffer (P), were run on SDS/PAGE and probed with indicated antibody. Serum-grown HeLa cell extract (1 mg) was incubated with 1 μg of hVps34 (Echelon Biosciences) (b) or Beclin-1 (Santa Cruz Biotechnology, Inc.) (c) antibodies bound to protein-G-Sepharose. The washed immunoprecipitates were used for dephosphorylation assay in the absence (C) or presence (dP) of 50 mU/ml PP2B and 15 mU/ml PP1 for 30 min at 30°C. Dephosphorylation was stopped using sample buffer and immunoprecipitates were resolved in SDS-PAGE, transferred to nitrocellulose and probed with DIG-14-3-3 (14-3-3 overlay), Beclin-1 (BD Biosciences) and hVps34 (Zymed) antibodies. (d) HeLa cells were treated with C2-ceramide (10 μM) for 24 h or serum-starved for 16 h (-S) and stimulated with IGF-1 (100 ng/ml, 20 min) or PMA (400 ng/ml, 15 min) as indicated. Cell extracts (5 mg of protein) were incubated for 2 h with 50 μl of 14-3-3-Sepharose according to Material and Methods and washed pellets (pull down) were extracted with SDS sample buffer, run on SDS/PAGE and probed with indicated antibody. Cell extract (30 μg) were run on SDS/PAGE transferred to nitrocellulose membranes and probed with indicated antibodies (bottom panel). (e) HeLa cells were serum starved for 16 h and left untreated (-S) or stimulated with PMA (400 ng/ml, 15 min). Cell extracts (1 mg) from both samples were incubated with 1 μg of hVps34 (Echelon Biosciences) antibody bound to protein-G-Sepharose. The washed immunoprecipitates were used for hVps34 in vitro kinase assay and also resolved to detect hVps34 protein and interaction with 14-3-3 analyzing with DIG-14-3-3. Cell extracts (30 μg) from both samples were run on SDS-PAGE, transferred to nitrocellulose membranes and probed with indicated antibodies (middle panel; quantification of hVps34 in vitro kinase assay expressed as PI(3) fold change, N=3, *P=0.0021, Student t-test). (f) HeLa cells grown in medium containing serum were serum-starved for 16 h (-S). Where indicated, cells were incubated with H-7 (100 μM), PD 184352 (2 μM) or no inhibitor for 1 h before stimulation with PMA (400 ng/ml) for a further 15 min. Cell extracts (5 mg) were incubated for 2 h with 50 μl of 14-3-3-Sepharose and washed pellets were extracted with SDS sample buffer, run on SDS/PAGE and probed with indicated antibody. Cell extract (30 μg) were run on SDS/PAGE transferred to nitrocellulose membranes and probed with indicated antibodies (bottom panel). (g) hVps34 was immunoprecipitated from HeLa cells that had been serum-starved for 16 h. The hVps34 immunoprecipitates were incubated in the presence of catalytic subunit of PKC (1 U/ml rat brain PKC) with MgATP for 30 min at 30°C, and analyzed by DIG-14-3-3 overlays. The lane marked C is a control with no PKC. (h) Schematic representation of hVps34 protein structure. N-terminal region of ∼50 amino acids, followed by C2 domain probably related to acidic phospholipids interactions, a possible regulatory helical domain and kinase domains with the extreme C-terminal 11 residues required for lipid kinase activity (26, 27). (i) GFP-tagged fragments of hVps34 were constructed: WT contains GFP-tagged full-length hVps34 (906 aa, 2719 pb), GFP-tagged fragment MT1 lost the residues required for hVps34 kinase activity and GFP-tagged fragment MT2 just contain the N-terminal part of the protein. GFP-tagged fragment of hVps34 were transiently expressed in HEK293T cells stimulated with PMA. GFP-tagged sequences were immunoprecipitate using anti-sheep GFP antibodies and carried out an in vitro binding reactions using DIG-14-3-3 overlay assay. Arrows show the correspondence GFP-tagged fragment reveal by anti-rabbit GFP antibody. (j) HEK293T cells were transfected with plasmid expressing GFP-hVps34 WT and several GFP-hVps34 mutants as indicated. After 24 h, cells were serum-starved for a further 16 h, then stimulated for 15 min with PMA (400 ng/ml). Cell extracts (0.5 mg) were incubated with 10 μg of anti-sheep GFP antibody bound to protein-G-Sepharose. The washed immunoprecipitates were used to detect GFP-hVps34 protein and its interaction with 14-3-3 proteins was analyzed by DIG-14-3-3 (bottom panel: quantification of DIG-14-3-3 signals correspond to GFP-hVps34 immunoprecipitates expressed as arbitrary unit, N=3, *P=0.0003, **P=0.0008, Student t-test)
In most cases, it was essential for the 14-3-3 binding that 14-3-3-interactor proteins were previously phosphorylated in vivo, thus, 14-3-3 proteins usually bind to specific Ser(P) and Thr(P) motifs within target proteins and two optimal 14-3-3 phosphopeptide ligands were reported with the consensus sequences: RSX(pS/T)XP (mode 1) and RX(Y/F)X(pS/T)XP (mode 2) (where pS/T represents phosphoserine or phosphothreonine and X is any amino acid).20 Some of the 14-3-3s bind to phosphorylated motifs different to the establish consensus sites.21 Data shown in Figure 3 suggests that hVps34 binds to 14-3-3s in a phosphorylation-dependent manner. Moreover, hVps34, isolated from continuous serum-growing HeLa cells, was dephosphorylated, using protein serine/threonine phosphatase 2B (PP2B) and PP1, and its binding to DIG-14-3-3 was abolished (Figure 8b and c). These data support that a phosphorylation-dependent mechanism is required for hVps34/14-3-3 association.
The HeLa cells used to identify hVps34/14-3-3 binding were cultured in medium with serum that contained a mixture of cell effectors. To identify the cellular stimuli that were responsible for promoting the phosphorylation of hVps34 at sites that facilitate binding to 14-3-3 proteins, cells were deprived of serum and stimulated in different ways. Among the tested stimuli, an increased amount of hVps34 protein bound to 14-3-3 from cells treated with the phorbol ester PMA was found (Figure 8d), whereas stimulation with IGF-1 or sphingolipid C2-ceramide does not promote hVps34/14-3-3 binding.
Moreover, PMA stimulation promoted 14-3-3/hVps34 association and as a result there was decreased hVps34 kinase activity (Figure 8e). It is interesting to note that hVps34 kinase activity increased when cells were exposed to a serum-free medium (in which autophagy has been induced, Supplementary Figure S12). Under this circumstance, as was mentioned above, a fast PMA-dependent phosphorylation of hVps34 promotes 14-3-3 binding and as a result a decreased kinase activity of the protein.
In addition to a well-characterized ability to activate protein kinase C (PKC) isoforms,22 PMA was found to stimulate the phosphorylation of p42/p44 MAPK (mitogen-activated protein kinase; extracellular-signal-regulated kinase 1 (ERK1)/ERK2) in HeLa cells (Figure 8d). Lack of SAPK2a activation (stress-activated protein kinase 2a/p38α) was found under stimulation with PMA, unlike C2-ceramide that activates the SAPK2a/p38a pathway (Chen et al.23; data not shown). Aimed at analyzing in detail kinase responsible for hVps34/14-3-3 interaction, data suggest that 14-3-3/hVps34 association in response to PMA was unaffected by the drug PD 184352, which inhibited the phosphorylation of the p42 and p44 MAPKs (Figure 8f).24 Contrarily, the response of hVps34 to PMA was blocked by H-7, which inhibits a number of other protein kinases, including the PMA-activated PKC25 in Hela (Figure 8f) and HEK293T cells (Supplementary Figure S13).
Together, these findings suggested that PKC isoforms, or a kinase activated downstream of PKC, were responsible for phosphorylation of at least one 14-3-3-binding site on hVps34, although the involvement of other protein kinases cannot be ruled out. Nevertheless, phosphorylation in vitro with PKC (catalytic subunit from rat brain) of hVps34 immunoprecipitates from serum-free stimulated HeLa cells enhanced hVps34/14-3-3 binding (Figure 8g).
Recently, the crystal structure of Vps34 was determined, its composition possessing strong homology with the catalytic subunits of others PI3Ks.26, 27 Aimed at analyzing the sequence domain of hVps34, which probably contains phosphorylated-binding sites required for hVps34/14-3-3 association, a series of deletion mutants (GFP-tagged fragments of hVps34) were constructed (Figure 8h and i). All GFP-tagged fragments of hVps34 are able to interact with 14-3-3 proteins by DIG-14-3-3 overlay. Even a recombinant GFP-tagged fragment (MT2) containing the N-terminal region binds 14-3-3s suggesting that one or several phophorylated sites included in GFP-tagged fragment (MT2) is responsible for the binding. In this context, the hVps34 sequence of GFP-tagged fragment MT2 (1–390 aa) was analyzed by mode 1 14-3-3-binding consensus sites and PKC phosphorylation consensus site, using the software Scansite.28 Mode 2 of 14-3-3-binding consensus sequence was also taken into consideration. Using low stringency in Scansite analysis, several possible 14-3-3-binding sites emerge such as Ser165 (RTSS165TLS) as mode 1 14-3-3-binding site and Ser34 (REQKS34Y), Thr166 (RTSST165LS), Thr197 (RLT197FR) and Ser276 (RS276LRSGP) as possible PKC phosphorylated sites. Completely covering hVps34 sequence of GFP-tagged fragment MT2 (1–390 aa) other possible 14-3-3-binding sites emerge such as Ser77 (RTS77YK), Ser212 (RSS212NF) and Ser279 (RSLRS279GP). Taking these data into consideration, GFP-hVps34 wild type (WT) and several mutants with specific serire or threonine changed by alanine were generated. Although, other 14-3-3-binding sites cannot be ruled out, after PMA-stimulation GFP-hVps34 WT shows 14-3-3-binding as shown above, whereas this association is reduced in GFP-hVps34 Thr197A and GFP-hVps34 Ser212A mutants (Figure 8j). Interestingly, both sites seem to contribute to hVps34/14-3-3 association giving a high level of complexity to hVps34/14-3-3 interaction (see Supplementary Figure S14). Several hVps34 lipid kinases assays were performed analyzing the consequence of both mutations in the activity, unfortunately lipid kinase activity levels in GFP-hVps34 WT and mutant immunoprecipitates were extremely low and insignificant in comparison with the endogenous protein. This fact has been reported before when overexpressed hVps34 was only 5% active relative to the endogenous enzyme, presumably because of the lack of hVps15 (p150) and other binding partners.18 Thus, hVps34 kinase activity of overexpressed protein is likely to be non-physiologically relevant, but the overexpressed proteins were interesting in terms of phosphorylation and 14-3-3-binding site analysis. Nevertheless, these data suggest multiple phosphorylated sites responsible for hVps34/14-3-3 interaction.
Discussion
As shown in this study, well-known antiapoptotic 14-3-3 proteins are also controlling cellular growth by regulating autophagy in cancer cell lines. Several observations strongly support this idea: (a) the initiator of autophagy, hVps34 directly interacts with 14-3-3 proteins under physiological conditions, through a PKC-dependent phosphorylation mechanism; (b) 14-3-3/hVps34 association inhibits hVps34 kinase activity; (c) overexpression of 14-3-3 proteins block hVps34 activation and counteracts ceramide-stimulated autophagy; (d) depletion of 14-3-3 proteins induces hVps34 activity and promotes autophagy and (e) starvation-induced autophagy promote dissociation of 14-3-3/hVps34 complex and activation of hVps34 kinase activity, its activation being physiologically important in autophagy initiation.
These data that connect PKCs and hVps34, are interesting because both activities have been implicated in autophagy regulation. The role of hVps34 in autophagy has been well established,29 whereas inhibitors of PKC such as safingol (-threo-sphinganine) induces autophagy in tumor cells,30 which implies PKC as a negative regulator of autophagy.
Data obtained suggest that N-terminal region of the protein seems to be required for the hVps34/14-3-3 binding, whereas Thr197 (LDRLT197FR) and Ser212 (EKRSS212NF) are likely involved in the binding. Taking into account the typical 14-3-3-binding motives, it is interesting to mention the presence of the usual arginine preceding the Ser/Thr responsible for the 14-3-3 binding, whereas they lack the proline residue that is in the +2 position of the canonical 14-3-3-binding phosphopeptides.20 Nevertheless, these data are consistent with recent reports suggesting the +2 proline residue occurs in less than half of the 14-3-3-binding sites of target proteins and raising the possibility that the two short stretch tandem phosphorylated sites on hVps34 may bind to the 14-3-3 dimer conforming a change in N-terminal tail of hVps34 altering its activity, which is the case for other 14-3-3-binding enzymes such as tyrosine hydroxylase.16, 31 A recent report form Dr. Yuan's laboratory shows that Vps34 was phosphorylated on Thr159 by Cdk1 during mitosis being a key regulatory mechanism that controls the class III PtdIns3 kinase activity in cell-cycle progression.32 This phosphorylated residue has been analyzed by its 14-3-3 binding but Thr159 (EPTKT159PG) does not seem responsible for hVps34/14-3-3 interaction, probably because do not conform even remotely a typical 14-3-3-binding motif.
14-3-3 proteins have been shown to directly interact with insulin receptor substrate 1 (IRS-1), which promotes dissociation of IRS-1/PI3K class I complex during insulin desensitization.33 On the other hand, 14-3-3 may contribute to autophagy regulation by association with two regulators of mTOR activation such as tuberous sclerosis complex 2 (TSC2) and proline-rich Akt/PKB substrate 40 kDa (PRAS40) proteins. The 14-3-3 binding to these proteins promotes activation of mTOR.34 Nutrient or serum deprivation promotes inactivation of mTOR through dissociation of TSC2/14-3-3 and PRAS40/14-3-3 complexes. It is interesting to note, that overexpression of 14-3-3ζ impaired C2-ceramide-induced autophagy even if mTOR is inhibited suggesting that interaction of 14-3-3 with proteins involved in autophagosomes formation during autophagy, such as hVps34, may suppose a later control of autophagy. These data give an idea of how finely regulated autophagy is and how important it is that the cell does not embark on a cellular degradation process if not strictly necessary. Beyond the regulation by 14-3-3 proteins of Vps34 kinase activity, certainly other 14-3-3-binding proteins involved in regulating autophagy possibly remain to be identified.
Although autophagy and apoptosis use fundamentally different molecular mechanisms to execute protective functions, they also seem to be highly interconnected, recently found in the case of Bcl-2 and cFLIP.35 14-3-3 proteins have been generally involved in survival mechanisms by regulation of essential proapoptotic proteins, such as Bcl-xL/Bcl-2-associated death promoter, that lead the blockade of apoptosis.36 Thus, the 14-3-3 proteins role show similarities to Bcl-210 and cFLIP11 functions, 14-3-3 proteins not only possessing anti-apoptotic activity but additionally serve as antiautophagy molecules through their interaction and inhibition of hVps34. Thus, the autophagy signal transduction pathway is under tight surveillance, for example, by the mTOR kinase in autophagy induction,37 by cFLIP in autophagosome elongation or by Bcl-2 and 14-3-3 proteins in autophagosome generation.
14-3-3 is also part of the hVsp34/Beclin-1 complex and joins the pool of proteins that mediate regulation of hVps34 kinase activity, such as hVps15,38 Atg 14L or Rubicon.6 Thus, 14-3-3 binds to hVps34 that leads the rapid inactivation of the enzyme under physiological condition. The dynamic change in protein composition between different functional Beclin-1/hVps34 complexes may have a central role in mediating the Beclin-1/hVps34 activity, which governs autophagy.
In summary, a new link between 14-3-3ζ isoform and autophagy is identified. These data shown in this study, contribute also to the understanding as to how regulators of autophagosome formation, such as hVps34, respond to survival signals in cells during physiological condition or antagonistic to growth and survival, such as ceramide or nutrients starvation, through association/dissociation with 14-3-3 proteins. The identification of this regulation will help to understand the molecular events that lead to activation of autophagy by anticancer drugs and may contribute to the design of new therapeutic strategies for inhibiting tumor growth.
Material and Methods
Cell lines
HeLa cells were maintained in DMEN media supplemented with 10% fetal bovine serum, at 37°C in a humidified 5% CO2, 95% air incubator. HeLa cells stably transfected with GFP-14-3-3ζ were maintained in DMEN media supplemented with 10% fetal bovine serum and 500 μg/ml geneticin sulfate (G-418). Human embryonic kidney 293 (HEK293T) cells were maintained in DMEN media supplemented with 10% fetal bovine serum and 10% glutamine. The MCF-7 breast epithelial cells expressing GFP-LC3 fusion protein described previously39 and generously provided by Dr M Jäättelä and Dr A Lopez-Rivas were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 10% glutamine and 500 μg/ml G-418 at 37°C in a humidified 5% CO2, 95% air incubator. Then appropriate cells where starved using Earle's balanced salt solution (starvation condition).
Reagents
Optimen, protease-inhibitor cocktail tablets and geneticin (G-418) were purchased from Invitrogen (Paisley, UK). Peptide (ARAApSAPA) was synthesized by Graham Bloomberg at the University of Bristol and generously provided by Dr. C MacKintosh. N-acetyl--sphingosine (C2-ceramide), phosphatidylinositol, E64d, 12-O-tetradecanoylphorbol 13-acetate (PMA), IGF-1, H-7 inhibitor, 3-MA, etoposide, rapamacyn and propidium iodide (PI) were obtained from Sigma Chemical Corp (St. Louis, MO, USA). PD 184352 inhibitor was from Axon Medchem (Groningen, The Netherlands). Frozen human HeLa cell pellets from 4c Biotech (Ghent, Belgium). All chromatographic columns were from Amersham-Pharmacia Biotech (Uppsala, Sweden). GFP-LC3 and TRAIL (TNF-related apoptosis-inducing ligand) was generously provided by Dr J Oliver and Dr A López-Rivas. HA-14-3-3ζ, PP1 and GFP-14-3-3ζ were a gift from Dr C MacKintosh. TLC coated silica gel 60 was purchased from Merk (Whitehouse Station, NJ, USA). The catalytic subunit of PKC from rat brain and PP2B were from Calbiochem (Nottingham, UK).
Antibodies
Anti-14-3-3 polyclonal, 14-3-3ζ, 14-3-3θ, 14-3-3σ, 14-3-3ɛ, 14-3-3γ, p150, GFP, p62/SQSTM1, GAPDH, P-p70S6K, p70S6K, Atg1, P-S6, S6 and Beclin-1 (D-18, used for immunoprecipitation) antibodies were purchased from Santa Cruz Biotechnology, Inc. Monoclonal antibody to α-tubulin was purchased from Sigma Chemical Corp (St Louis, MO, USA). LC3 from NanoTools (Teningen, Germany) and Novus Biologicals (Littleton, CO, USA) and Beclin-1 antibodies used by western blot was from BD Biosciences (San Jose, CA, USA). Atg16 and Atg12-Atg5 antibodies were from Novus Biologicals (Littleton, CO, USA). PIST, LAMP1 and LAMP2 were purchased from ProSci Incorporates (Poway, CA, USA). hVps34 antibody used for immunoprecipitation was purchased from Echelon Biosciences (Salt Lake City, UT, USA) and hVps34 antibody for western blot was from Zymed (San Francisco, CA, USA). P-ERK1/2, ERK1/2 and P-PKB (Ser473) were from Cell Signalling (Danvers, MA, USA). Total PKB was a gift from Division of Signal Transduction Therapy (DSTT; University of Dundee, UK). Appropriate peroxidase-conjugated secondary antibodies from DAKO (Glostrup, Denmark) and ECL immunoblotting reagents were from Millipore corporation (Billerica, MA, USA).
Immunoblotting and 14-3-3 overlay assay
Total cell lysate (30–50 μg) were heated at 90°C for 5 min in SDS sample buffer, and subjected to polyacrylamide gel electrophoresis and electrotransfer to nitrocellulose membrane. The membrane was blocked for 1 h in TBS-Tween buffer containing 5% (w/v) skimmed milk. The membrane was probed with 1 μg/ml of indicated antibodies in TBS-Tween, 5% (w/v) skimmed milk for 16 h at 4°C. Detection was performed using horseradish peroxidase-conjugated secondary antibodies and exposed using the ECL system (Amersham Biosciences, Uppsala, Sweden). For the immunoblot detection of LC3-I and LC3-II, the samples were treated with 10 μg/ml pepstatin A and 10 μg/ml E64d for the last 4 h before the lysis. Quantification of western blot signals was performed by densitometry with ImageQuant TL software after scanning the films on ImageScanner II (GE Healthcare, Buckinghamshire, UK). The relative expression of proteins was normalized to that of loading controls. The values are expressed as arbitrary units of the expression levels in control cells.
For 14-3-3 overlays, DIG-labelled 14-3-3s were used in place of a primary antibody. The membrane was incubated with 5 μg/ml total His-BMH1 and His-BMH2 (yeast 14-3-3 isoforms), in TBS-Tween containing 1 mg/ml BSA for 2 h at room temperature. After several washes, the membrane was incubated with an anti-DIG-horseradish peroxidise secondary antibody and exposed using the ECL system (Amersham Biosciences).
RNA interference
Cells were transfected with indicated siRNAs at 20 nM for 14-3-3ζ, 14-3-3σ and 14-3-3θ siRNAs using DharmaFECT transfection agent (Dharmacon Research, Lafayette, CO, USA) according to manufacturer's guidelines. After 48 h of transfection, cells were used for further analysis. siRNAs corresponding to control siRNA (5′-CGACCGAGACAAGCGCAAGtt-3′) from Dharmacon Research. siRNAs against 14-3-3ζ (sc-29583), siRNAs against 14-3-3σ (sc-29590) and siRNAs against 14-3-3θ (sc-29586) were purchased from Santa Cruz Biotechnology, Inc.
Affinity purification of 14-3-3-binding proteins
Clarified lysates of HeLa cells containing 5 mg total protein, were incubated at 4°C for 2 h on a vibrating platform with 50 μl of BMH1/2 CH-Sepharose (14-3-3-Sepharose). The beads were washed 10 times with 1 ml of lysis buffer containing 0.15 M NaCl, and 5 times with buffer A (50 mM Tris-HCl pH 7,4 plus 1 mM EGTA). The 14-3-3-binding proteins were treated with sample buffer and subjected to electrophoresis and immunoblotting.
14-3-3 Affinity chromatography of human HeLa cell extracts
HeLa cells were thawed and extracted in 20 ml lysis buffer (50 mM Tris-HCl pH 7.5, 1 mM EGTA, 1 mM EDTA, 0,2% (w/v) Nonidet P-40 (NP-40), 1 mM sodium orthovanadate, 10 mM sodium-β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 mM dithiothreitol (DTT) and a complete proteinase inhibitor cocktail). The broken cells were centrifuged at 15 000 g for 60 min, and the supernatant was collected, filtered and processed for affinity chromatography as previously described.16 Briefly, the clarified solution was mixed end-over-end for 1 h at 4 °C with 5 ml Sepharose linked to BMH1/BMH2 (the Saccharomyces cerevisiae 14-3-3 isoforms).40 The mixture was poured into a column, flow through collected, and the column washed with 600 column volumes of 50 mM Tris-HCl pH 7.5, 500 mM NaCl, 1 mM DTT (buffer A) and then 12 ml of a control synthetic phosphopeptide that does not bind 14-3-3s (1 mM RSRTRTDpSYSAGQSV in buffer A). Proteins that bind to the phosphopeptide-binding site of 14-3-3s were eluted with 12 ml of 1 mM ARAApSAPA phosphopeptide in buffer A. Samples (12 ml) from the beginning, middle and end of the 0.5 NaCl wash, the control peptide mock elution pool (12 ml) and the ARAApSAPA elution pool (12 ml) were each concentrated to ∼400 μl, desalted in vivaspin concentrators, and analyzed as indicated in Figure 2.
cDNA cloning
cDNA cloning and sequencing human hVps34 (catalytic phosphatidylinositol 3-kinase 3 (Homo sapiens) NP_002638.2) was cloned by PCR using EST source (IMAGE 5517878). Primers PI3KC3 33: (AAAAGAATTCGCGCTGGGGGAAGCAGAGAAGTTTCAC) and PI3KC3 34 (TTTTGGATCCTTTTCTCCAGTACTGGGCAAACTTGTGAATCTGTTC) were used to amplify the open reading frame and add a GFP tag at the 5′ terminus. The resulting 2719-bp cDNA fragment was digested with BamH1 and EcoRI and ligated into the same sites in pEGFP-C2 (Clontech, Mountain View, CA, USA) to produce pGFP-hVps34 WT. Fragment MT1 was amplify using primers PI3KC3 33: (AAAAGAATTCGCGCTGGGGGAAGCAGAGAAGTTTCAC) and PI3KC3 39: (GCGGATCCGCCATGTTTTGTACTGGTGGCTAACAC) and the MT2 fragment was amplify using primers PI3KC3 33: (AAAAGAATTCGCGCTGGGGGAAGCAGAGAAGTTTCAC) and PI3KC3 37: (GCGGATCCCTCATCATCGGCCTGTCGCAACCG). The resulting MT1 (2040-bp cDNA) and MT2 (1170-bp cDNA) fragments were digested with BamH1 and EcoRI and ligated into the same sites in pEGFP-C2 (Clontech) to produce pGFP-hVps34 MT1 and pGFP-hVps34 MT2.
The mutant at Ser34 (GFP-hVps34 Ser34A) was created by PCR (QuikChange II Site-Directed Mutagenesis Kits, Stratagene, La Jolla, CA, USA) using primers PI3KC3mut 41: (GGAAGAGAGAACAAAAGGCTTATAAAGCTGTCCTGG) and PI3KC3mut 42: (CCAGGACAGCTTTATAAGCCTTTTGTTCTCTCTTCG). The mutant at Thr163, Ser164, Ser165 and Thr166 (GFP-hVps34 Thr163A, Ser164A, Ser165A, Thr166A) was created using primers PI3KC3mut 45 (CAAAAACTCCTGGCAGAGCAGCTGCCGCTCTCTCAGAAGATCAG) and PI3KC3mut 46: (CTGATCTTCTGAGAGAGCGGCAGCTGCTCTGCCAGGAGTTTTTG). The mutant at Thr197 (GFP-hVps34 Thr197A) was created using primers PI3KC3mut 47: (GGCTGGATAGATTGGCA TTTAGAGAAATAGAAATG) and PI3KC3mut 48: (CATTTCTATTTCTCTAAATGCCAATCTATCCAGCC). The mutant at Ser212, (GFP-hVps34 Ser212A) was created using primers PI3KC3mut 49: (GAGAGTGAAAAACGAACTGCTAATTTC ATGTACCTG) and PI3KC3mut 50: (CAGGTACATGAAATTAGCAGTTCGTTTTTCACTCTC). The mutant at Ser276 and Ser279, (GFP-hVps34 Ser276A, Ser279A) was created using primers PI3KC3mut 51: (CACAAGCTTGCCCGGGCTTTAAGAGCTGGACCTTCTGACCAC) and PI3KC3mut 52: (GTGGTCAGAAGGTCCAGCTCTTAAAGCCCGGGCAAGCTTGTG). All constructs were confirmed by restriction analysis and sequencing and purified using Qiagen Plasmid Maxi (Qiagen, Germantown, MD, USA) according to the manufacturer's protocol.
Transfection of HEK293T and stable HeLa/GFP-14-3-3ζ cell line
Clones (GFP-hVps34 constructs, GFP-LC3, GFP-14-3-3ζ and HA-14-3-3ζ) were used to transfect HEK293T cells, which were cultured on density of 2 × 106 per 10 cm dish in DMEM containing 10% fetal calf serum for 24 h. Medium was changed into fresh medium and cells were transfected by a modified calcium phosphate method with 2 μg/ml DNA/plate then added 0,5 ml × 2 HBS (50 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid-HCl, pH 6.96, 0.28 M NaCl and 1.5 mM Na2HPO4) and 150 mM NaCl.41 The resulting mixture was vortexed for 1 min, allowed to stand at room temperature for 20 min and then added dropwise to a 10 cm dish of HEK293T cells. The cells were placed in an atmosphere of 5% CO2 for 16 h at 37°C, then the medium was aspirated and replaced with fresh DMEM containing 10% fetal calf serum.
To generate stable GFP-14-3-3ζ HeLa, the cells were cultured in 10 cm diameter dishes to 50–70% confluence and transfected with 3 μg pGFP-14-3-3ζ construct using Fugene 6 reagent (Roche, Tucson, AZ, USA) according to the manufacturer's instructions. After 24 h, G-418 was added to the medium to a final concentration of 3 μg/ml, and the medium was changed every 24 h maintaining G-418. After 14–20 days, individual surviving colonies expressing low levels of GFPfluorescence were selected and expanded. FACS analysis was also performed to ensure uniform expression of GFP in the selected cell lines and anti-14-3-3ζ immunoblotting analysis of lysed cells was performed to ensure a band at the expected apparent molecular mass. To generate transient expression of GFP-LC3 in HeLa cells 3 μg pGFP-LC3 and Fugene 6 reagent were used.
Immunoprecipitation assay
Cells were lysed in lysis buffer and the lysate was centrifuged at 4°C for 10 min at 13 000 g and an aliquot of the supernatant (0.5–2 mg protein) was incubated with 1 μg of Beclin-1 (Santa Cruz Biotechnology, Inc.), hVps34 (Echelon Biosciences) or 10 μg GFP antibodies (DSTT) coupling to 20 μl protein G-Sepharose for 3 h at 4°C. The suspension was centrifuged for 1 min at 13 000 g, the protein G-Sepharose/antibody/protein complex was washed twice with 1 ml lysis buffer containing 150 mM NaCl and twice with buffer A and the immunoprecipitate resolves in SDS-PAGE for DIG-14-3-3 or western blot analysis.
Autophagy detection
Several methods were used to assess autophagy such as monitoring phagophore and autophagosome formation using fluorescence microscopy by checking increase in punctuate of fluorescence highly specific autophagy marker GFP-LC3 (Atg8 in yeast) previously transfected in cells.42 In yeast Atg8, an ubiquitin-like protein, is present on autophagic membranes as a phosphatidylethanolamine (PE)-conjugated form (Atg8-PE) to expand the autophagic membranes.43 Lipid conjugation leads to the conversion of the soluble 18 kDa form of Atg8 (mammalian LC3-I) to an autophagic associated vesicle, fast migrating the 16 kDa form onto SDS-PAGE gels (mammalian LC3-II). Fluorescence analysis of GFP-LC3/Atg8 indicates a change from a difuse cytoplasmic or nuclear distribution to punctate dots that reflect autophagic vesicles. Thus, cells cultured on coverslips expressing GFP-LC3 were fixed in 3.7% formaldehyde and applied to a Leica CTR 6000 confocal microscope (Leica, Solms, Germany). Cells presenting a mostly diffuse distribution in the cytoplasm and nucleus were considered as non-autophagic, whereas cells with several intense LC3-GFP punctae were classified as autophagic. The percentage of cells with GFP-LC3 translocation or the number of GFP-LC3 punctae per cell were quantified (600 cells/sample on duplicated samples of three different experiments). Cross-sections with over five dots on LC3 were considered positive. Other methods of monitoring autophagosome formation were measuring turnover of LC3-II, GFP-LC3 processing, levels of a general marker of autophagy such as p62/SQSTM1 and mTOR activity (by analyzing P-p70S6K) by inmunoblot analysis. p62/SQSTM1 binds LC3 and it is selectively recognized and degraded by autophagy becoming a autophagy marker.44
hVps34 kinase assay
hVps34 kinase assay was performed as described previously.6 Hela and HEK293T cells were lysed in 1% NP-40 lysis buffer (20 mM Tris-pH 7.5, 137 mM NaCl, 1 mM MgC12, 1 mM CaC12, 100 mM NaF, 10 mM sodium pyrophosphate, 100 μM Na3VO4, 10% glycerol, 0.35 mg/ml PMSF, protease and phosphatase inhibitor cocktails). Immunoprecipitation was performed with 1 μg/IP of anti-hVps34 (Echelon Biosciences) or anti-Beclin (D-18; Santa Cruz Biotechnology, Inc.) affinity gel beads, according to the manufacturer's protocol. Beads (associated with purified proteins) were washed three times in lysis buffer, followed by three washes in washing buffer (100 mM Tris-HCl pH7.4 and 500 mM LiCl) and two washes in reaction buffer (10 mM Tri-HCl-pH7.4, 100 mM NaCl and 1 mM EDTA). Beads were resuspended in 60 μl of reaction buffer, MnCl2 (10 μl of 100 mM) and sonicated phosphatidylinositol (10 μl of 2 μg/μl) were added. The reaction was started by the addition of ATP (10 μl of 440 μM) containing γ-32P-ATP (10 μCi); beads were incubated for 20 min at room temperature. The reaction was terminated by adding HCl (20 μl of 8 M), and the organic phase was extracted with 160 μl chloroform:methanol (1 : 1). Extracted phospholipid products were resolved by TLC using TLC coated silica gel and a solvent composed of chloroform:methanol:H2O:ammonium acetate (v/v/v/v, 20 : 15 : 5 : 3), followed by visualization with Fujifilm FLA-5100 imaging system. Software used for activity quantification was Multi Gauge (Lifescience/Fujifilm, Valhalla, NY, USA).
Detection of cell death
The crystal violet assay as has been previously described was used to determine cell viability.45 Cells were seeded at approximately 10 000 cells per well in 48-well plates and allowed to adhere overnight. After treatment, cells were fixed in 100% methanol for 10 min, stained with 0.5% crystal violet solution in 25% methanol for 10 min, washed three times with 1 × PBS to remove excess dye and allowed to dry overnight. The following morning, crystal violet containing cells was solubilized in 1 ml 1% SDS and an absorbance reading was taken at 560 nm. Hypodiploid death cells (sub-G1) were detected using PI by flow cytometry as described previously.46 Flow cytometry was performed on a FACScan cytometer using Cell Quest software (Becton Dickinson, Mountain View, CA, USA).
Statistical analysis
Independent experiments were pooled when the coefficient of variance could be assumed identical. Statistical significance was evaluated by using t-test Student (N=number of independent experiments). P-values of under 0.05 were considered significant.
Acknowledgments
This study was supported by the ‘Ministerio de Educación y Ciencia' grant BFU2006-01088/BMC and ‘Programa Ramón y Cajal' contract (BOE 17/02/2004 ORDEN CTE/351/2004) to Mercedes Pozuelo Rubio. I thank J Oliver, A López-Rivas and M Jäättelä for tools provided and G Herrero and C Fumarola for helpful discussion. Thanks to C MacKintosh for antibodies, constructs and other helpful tools, P Chacón for mouse lung tissue extraction and P Domínguez, MJ Quintero, A Vela, N Leslie and JC Ostos for helpful assistance. I also thanks Diane Haun for spell checking.
Glossary
- 3-MA
3-methyladenine
- Atg6
autophagy-related gene 6 (orthologue of mammalian Beclin-1)
- Bcl-2
B-cell lymphoma 2
- BMH1/2
yeast 14-3-3 homolog
- cFLIP
FLICE-like inhibitor protein
- DIG
digoxigenin
- G-418
geneticin
- hVps34/(PI3KC3)
human vacuolar protein sorting 34 (phosphatidylinositol-3-kinase class III)
- LC3/Atg8
microtubule-associated protein1 light chain 3/autophagy-related gene 8
- mTOR
mammalian target of rapamycin
- NP-40
Nonidet P-40 (octyl phenoxylpolyethoxylethanol)
- PKC
protein kinase C
- PP1
protein phosphatase 1
- PP2B
protein serine/threonine phosphatase 2B
- PMA
phorbol-12-myristate-13-acetate
The author declares no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by E Baehrecke
Supplementary Material
References
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendil KB, Lauridsen AM, Seglen PO. Both endocytic and endogenous protein degradation in fibroblasts is stimulated by serum/amino acid deprivation and inhibited by 3-methyladenine. Biochem J. 1990;272:577–581. doi: 10.1042/bj2720577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14 L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter GW, Khuri FR, Fu H. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16:193–202. doi: 10.1016/j.semcancer.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, et al. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- Wang B, Ling S, Lin WC. 14-3-3Tau regulates Beclin 1 and is required for autophagy. PloS One. 2010;5:e10409. doi: 10.1371/journal.pone.0010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Peggie M, Wong BH, Morrice N, MacKintosh C. 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2, 6-bisphosphate kinase/phosphatase. EMBO J. 2003;22:3514–3523. doi: 10.1093/emboj/cdg363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, et al. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM. The regulation and function of class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Chen CL, Lin CF, Chiang CW, Jan MS, Lin YS. Lithium inhibits ceramide- and etoposide-induced protein phosphatase 2A methylation, Bcl-2 dephosphorylation, caspase-2 activation, and apoptosis. Mol Pharmacol. 2006;70:510–517. doi: 10.1124/mol.106.024059. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Aitken A. Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol Biol. 2002;50:993–1010. doi: 10.1023/a:1021261931561. [DOI] [PubMed] [Google Scholar]
- Parker PJ, Murray-Rust J. PKC at a glance. J Cell Sci. 2004;117:131–132. doi: 10.1242/jcs.00982. [DOI] [PubMed] [Google Scholar]
- Chen CL, Lin CF, Chang WT, Huang WC, Teng CF, Lin YS. Ceramide induces p38 MAPK and JNK activation through a mechanism involving a thioredoxin-interacting protein-mediated pathway. Blood. 2008;111:4365–4374. doi: 10.1182/blood-2007-08-106336. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Coward J, Ambrosini G, Musi E, Truman JP, Haimovitz-Friedman A, Allegood JC., et al., , Schwartz GK. Safingol (L-threo-sphinganine) induces autophagy in solid tumor cells through inhibition of PKC and the PI3-kinase pathway. Autophagy. 2009;5:184–193. doi: 10.4161/auto.5.2.7361. [DOI] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T., et al., , Yuan J. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Yuan M, Song Y, Ruderman N, Wen R, Luo Z. 14-3-3 facilitates insulin-stimulated intracellular trafficking of insulin receptor substrate 1. Mol Endocrinol. 2002;16:552–562. doi: 10.1210/mend.16.3.0790. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J Biol Chem. 1997;272:2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Mathiasen IS, Elling F, Jaattela M. Vitamin D analog EB1089 triggers dramatic lysosomal changes and Beclin 1-mediated autophagic cell death. Cell Death Differ. 2005;12:1297–1309. doi: 10.1038/sj.cdd.4401651. [DOI] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr Biol. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- Chen CA, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Wosikowski K, Kung W, Hasmann M, Loser R, Eppenberger U. Inhibition of growth-factor-activated proliferation by anti-estrogens and effects on early gene expression of MCF-7 cells. Int J Cancer. 1993;53:290–297. doi: 10.1002/ijc.2910530220. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ruiz C, Lopez-Rivas A. Mitochondria-dependent and -independent mechanisms in tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis are both regulated by interferon-gamma in human breast tumour cells. Biochem J. 2002;365:825–832. doi: 10.1042/BJ20020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Campbell DG, Morrice NA, Mackintosh C. Phosphodiesterase 3A binds to 14-3-3 proteins in response to PMA-induced phosphorylation of Ser428. Biochem J. 2005;392:163–172. doi: 10.1042/BJ20051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.