Abstract
Introduction
The Toll-like receptor 7 (TLR7) gene, encoded on human chromosome Xp22.3, is crucial for type I interferon production. A recent multicenter study in East Asian populations, comprising Chinese, Korean and Japanese participants, identified an association of a TLR7 single-nucleotide polymorphism (SNP) located in the 3' untranslated region (3' UTR), rs3853839, with systemic lupus erythematosus (SLE), especially in males, although some difference was observed among the tested populations. To test whether additional polymorphisms contribute to SLE in Japanese, we systematically analyzed the association of TLR7 with SLE in a Japanese female population.
Methods
A case-control association study was conducted on eight tag SNPs in the TLR7 region, including rs3853839, in 344 Japanese females with SLE and 274 healthy female controls.
Results
In addition to rs3853839, two SNPs in intron 2, rs179019 and rs179010, which were in moderate linkage disequilibrium with each other (r2 = 0.53), showed an association with SLE (rs179019: P = 0.016, odds ratio (OR) 2.02, 95% confidence interval (95% CI) 1.15 to 3.54; rs179010: P = 0.018, OR 1.75, 95% CI 1.10 to 2.80 (both under the recessive model)). Conditional logistic regression analysis revealed that the association of the intronic SNPs and the 3' UTR SNP remained significant after we adjusted them for each other. When only the patients and controls carrying the risk genotypes at the 3' UTR SNPpositionwere analyzed, the risk of SLE was significantly increased when the individuals also carried the risk genotypes at both of the intronic SNPs (P = 0.0043, OR 2.45, 95% CI 1.31 to 4.60). Furthermore, the haplotype containing the intronic risk alleles in addition to the 3' UTR risk allele was associated with SLE under the recessive model (P = 0.016, OR 2.37, 95% CI 1.17 to 4.80), but other haplotypes were not associated with SLE.
Conclusions
The TLR7 intronic SNPs rs179019 and rs179010 are associated with SLE independently of the 3' UTR SNP rs3853839 in Japanese women. Our findings support a role of TLR7 in predisposition for SLE in Asian populations.
Introduction
Toll-like receptors (TLRs) play a central role in detecting microbial pathogens. TLRs initiate innate immune responses and also induce adaptive immune responses [1]. Recently, TLRs have been strongly implicated in autoimmune diseases [2]. The TLR7 and TLR9 genes, which are expressed intracellularly in plasmacytoid dendritic cells (pDCs) and B cells, recognize single-stranded RNA and DNA containing cytidine-phosphate-guanosine motifs, respectively. Activation of pDCs by TLR7 and TLR9 induces a large amount of type I interferon (IFN). It has become evident that RNA- and DNA-containing immune complexes, which often exist in sera of patients with systemic lupus erythematosus (SLE), can activate TLR7 and TLR9 signaling [2].
Several lines of evidence support a role of TLR7 in SLE pathogenesis [2]. Male BXSB mice bearing the Y chromosome-linked autoimmune accelerator (Yaa) gene develop severe SLE. It has been revealed that Yaa mutation is caused by a translocation of a portion of the X chromosome containing TLR7 onto the Y chromosome [3,4]. Yaa-bearing mice have been demonstrated to have twofold overexpression of TLR7 protein and mRNA [3,4]. In contrast, lupus-prone MRL/Mplpr/lpr mice lacking TLR7 showed impaired production of antibodies to RNA-containing antigens, such as anti-Smith (anti-Sm) antibodies, and developed less severe disease [5]. Furthermore, upregulated expression of TLR7 mRNA in peripheral blood mononuclear cells (PBMNCs) was observed in human SLE [6].
Recently, a multicenter collaborative study including our group reported an association of TLR7, located in Xp22.3, with SLE in combined East Asian populations [7]. In a discovery panel consisting mainly of Chinese and Korean populations, the association of 27 single-nucleotide polymorphisms (SNPs) in the TLR7-TLR8 region with SLE was examined, and a significant association of the TLR7 3' untranslated region (3' UTR) SNP, rs3853839, was identified. Subsequently, the association of rs3853839 was replicated in two independent Chinese and Japanese case-control sets. The association was prominent in males with SLE. In addition, rs3853839 was associated with elevated expression of TLR7. The study also revealed some differences in the association of rs3853839 and other SNPs among Chinese, Korean and Japanese populations [7], indicating that systematic SNP screening should be performed in each population.
In this study, we examined the association of eight TLR7 tag SNPs with SLE in Japanese women and discovered a newly identified association of two intronic SNPs, rs179019 and rs179010, with SLE. These SNPs and the 3'UTR rs3853839 were found to independently contribute to the genetic risk for SLE.
Materials and methods
Patients and controls
Three hundred forty-four Japanese female patients with SLE (mean age ± SD, 42.9 ± 13.8 years) and 274 healthy female controls (mean age ± SD, 31.3 ± 8.9 years) were recruited at University of Tsukuba, Juntendo University, Sagamihara National Hospital, and at the University of Tokyo. Among them, 296 SLE patients and 250 healthy controls were also examined in a previous study to replicate the association of rs3853839 with SLE in Japanese, but other SNPs were not investigated in that study [7]. All patients and healthy individuals were native Japanese living in the central part of Japan. All patients with SLE fulfilled the American College of Rheumatology criteria for SLE [8].
This study was carried out in compliance with the Declaration of Helsinki. The study was reviewed and approved by the research ethics committees of University of Tsukuba, Sagamihara National Hospital, the University of Tokyo and Juntendo University. Informed consent was obtained from all study participants.
Genotyping
Eight tag SNPs in the TLR7 region were selected on the basis of the HapMap Phase II JPT (Japanese in Tokyo) data obtained from the HapMap database [9] with the criteria of minor allele frequency >0.1 and an r2 threshold of 0.9. Genotyping of the tag SNPs was carried out using the TaqMan genotyping assay on the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. Thermal cycling conditions consisted of initial denaturation at 95°C for 10 minutes, followed by 50 cycles at 95°C for 15 seconds each and at 60°C for one minute each. TaqMan probes used in this study were as follows: Assay ID: C__15757400_10 (rs2302267), C___2259585_10 (rs179019), C___7625717_10 (rs1634322), C___2259582_10 (rs179016), C___2259578_10 (rs179012), C___2259576_10 (rs179010), C___2259575_10 (rs179009), and C___2259573_10 (rs3853839).
Expression analysis by real-time quantitative reverse transcription polymerase chain reaction assay
Total RNA was extracted from PBMNCs of 18 females with SLE using the RNeasy Mini Kit (QIAGEN, Hilden, Germany), reverse transcribed into cDNA and used for real-time quantitative reverse transcription polymerase chain reaction (RT-PCR) assay. Expression of TLR7 was analyzed using the TaqMan Gene Expression Assay (Applied Biosystems), Hs00152971_m1. Amplification of cDNA was conducted using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems) under the following conditions: 50°C for 2 minutes and 95°C for 10 minutes, and 50 cycles at 95°C for 15 seconds and at 60°C for 1 minute, and then the cycle threshold (CT) value for each sample was obtained using Applied Biosystems 7300 System SDS version 1.4 software (Applied Biosystems). Relative quantitative levels were calculated on the basis of the CT value by a standard curve method and were normalized to β-actin (ACTB) expression (Hs99999903_m1). The experiments were done in triplicate for each sample.
Statistical analysis
Differences in allele and genotype frequencies between SLE patients and healthy controls were analyzed by using a χ2 test with 2 × 2 contingency tables. When one or more of the variables in the contingency tables was 20 or less, Fisher's exact test was employed. Linkage disequilibrium (LD) was analyzed using HaploView version 4.0 software (Broad Institute, Cambridge, MA, USA). Pairwise r2 values were calculated on the basis of the genotypes of 274 healthy controls. Estimation of haplotype frequencies and association tests were performed using HaploView version 4.0 software.
To examine whether each SNP independently contributes to susceptibility to SLE, conditional logistic regression analysis was employed. Dominant, codominant and recessive models were tested for each SNP, and the model that provided the lowest P value was selected as the best fit model. As a result, the following were used as independent variables: rs3853839, C/C = 0, G/C = 1 and G/G = 2 under the codominant model for the G allele; rs179019, C/C = 0, C/A = 0, A/A = 1 under the recessive model for the A allele; rs179010, C/C = 0, C/T = 0, and T/T = 1 under the recessive model for the T allele.
The association of TLR7 SNPs with TLR7 mRNA expression was assessed by using the Kruskal-Wallis test.
Results
Association of TLR7 SNPs with SLE in a Japanese female population
To systematically examine association of TLR7 SNPs with SLE in Japanese, eight tag SNPs in the TLR7 gene, including rs3853839 in the 3'UTR, which was recently shown to be associated with SLE in East Asian populations [7], were analyzed in 344 Japanese females with SLE and 274 healthy female controls. Because TLR7 is located on an X chromosome, male and female individuals needed to be analyzed for the association separately. However, because of the female predominance of SLE (9:1 female to male ratio), the sample size of male SLE patients was too small to be analyzed statistically. Therefore, male patients and controls were excluded from this study. No deviation from the Hardy-Weinberg equilibrium was observed in the controls (P > 0.05).
In addition to the association of rs3853839 reported separately [7], the association of two SNPs in intron 2, rs179019 and rs179010, was newly detected (Figure 1 and Table 1). Significant association of rs179019 and rs179010 was observed under the recessive model for the A and T alleles, respectively (rs179019: P = 0.016, odds ratio (OR) 2.02, 95% confidence interval (95% CI) 1.15 to 3.54; rs179010: P = 0.018, OR 1.75, 95% CI 1.10 to 2.80). LD was present between rs179019 and rs179010 (r2 = 0.53), while LD between rs3853839 and each of the intronic SNPs was modest (r2 = 0.02 and 0.04) (Figure 1).
Figure 1.
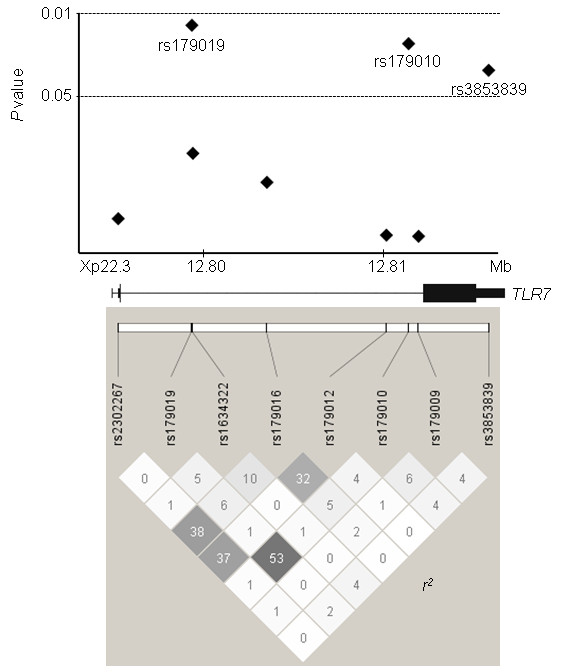
Association of tag single-nucleotide polymorphisms in the Toll-like receptor 7gene with systemic lupus erythematosus. Top: P values under the recessive model for minor alleles are indicated. Association was tested by χ2 analysis using 2 × 2 contingency tables. Bottom: r2 values based on data from 274 healthy Japanese women are shown.
Table 1.
Association of TLR7 SNPs with SLE in a Japanese populationa
| Allelic association | Dominant model | Recessive model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study population | Genotype, n (%) | Risk allele, n (%) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | ||
| rs3853839 | G/G | G/C | C/C | G | ||||||
| SLE | 197(57.3) | 125 (36.3) | 22 (6.4) | 519 (75.4) | 0.017 | 1.36 (1.06 to 1.75) | 0.030 | 1.87 (1.05 to 3.31) | 0.072 | 1.34 (0.97 to 1.84) |
| Controls | 137(50.0) | 106 (38.7) | 31 (11.3) | 380 (69.3) | ||||||
| rs179019 | A/A | C/A | C/C | A | ||||||
| SLE | 45 (13.1) | 131 (38.1) | 168 (48.8) | 221 (32.1) | 0.17 | 1.19 (0.93 to 1.52) | 0.77 | 1.05 (0.76 to 1.44) | 0.016b | 2.02 (1.15 to 3.54) |
| Controls | 19 (6.9) | 118 (43.1) | 137 (50.0) | 156 (28.5) | ||||||
| rs179010 | T/T | C/T | C/C | T | ||||||
| SLE | 61 (17.7) | 156 (45.3) | 127 (36.9) | 278 (40.4) | 0.062 | 1.25 (0.99 to 1.57) | 0.36 | 1.16 (0.84 to 1.61) | 0.018 | 1.75 (1.10 to 2.80) |
| Controls | 30 (10.9) | 133 (48.5) | 111 (40.5) | 193 (35.2) | ||||||
aTLR7, Toll-like receptor 7 gene; SNP, single-nucleotide polymorphism; 95% CI, confidence interval; OR, odds ratio; SLE, systemic lupus erythematosus. Genotype and allele frequencies are shown in parentheses (%). Association was tested by χ2 analysis or Fisher's exact test using 2 × 2 contingency tables under the indicated models for rs3853839G, rs179019A and rs179010 T alleles. bFisher's exact test was used.
To examine the contribution of each SNP to susceptibility to SLE, conditional logistic regression analysis was conducted. As shown in Table 2, the association of rs3853839 remained significant after adjustment for the intronic SNP genotypes. Adjusted P values (Padjusted) for rs3853839 under the codominant model were 0.040 and 0.047 after adjustment for rs179019 and rs179010, respectively. The association of rs179019 and rs179010 also remained significant after adjustment for rs3853839 (rs179019: Padjusted = 0.026; rs179010: Padjusted = 0.042). These results suggest that rs3853839 and the intronic SNPs are independently associated with SLE. In contrast, the association of rs179019 and rs179010 was eliminated when they were adjusted for each other as expected on the basis of LD between the two (Table 2 and Figure 1).
Table 2.
Conditional logistic regression analysis of TLR7 SNPsa
| P adjusted b | ||||||
|---|---|---|---|---|---|---|
| SNP | Risk allele | Model | P c | rs3853839 | rs179019 | rs179010 |
| rs3853839 | G | Codominant | 0.021 | NA | 0.040 | 0.047 |
| rs179019 | A | Recessive | 0.014 | 0.026 | NA | 0.24 |
| rs179010 | T | Recessive | 0.019 | 0.042 | 0.42 | NA |
aTLR7, Toll-like receptor 7 gene; SNP, single-nucleotide polymorphism; NA, not applicable; bP value adjusted for each SNP by conditional logistic regression analysis using the indicated model; cP value for each SNP calculated by logistic regression analysis. The indicated model showed the lowest P value for each SNP.
In agreement with these findings, when only the patients and controls carrying the risk genotypes of the 3' UTR SNP were analyzed, possession of both of the intronic SNP risk genotypes was significantly associated with SLE (P = 0.0043, OR 2.45, 95% CI 1.31 to 4.60) (Table 3).
Table 3.
Independent effect of intron 2 SNPs in the carriers of the 3' UTR risk genotypesa
| Risk genotype | Study group, n (%) | ||||||
|---|---|---|---|---|---|---|---|
| rs3853839 | rs179019 | rs179010 | SLE (N = 322) | Controls (N = 243) | P | OR | 95% CI |
| G/G or G/C | A/A | T/T | 42 (13.0) | 14 (5.8) | |||
| + | + | + | 280 (87.0) | 229 (94.2) | 0.0043 | 2.45 | 1.31 to 4.60 |
| + | Others | Reference | |||||
aSNP, single-nucleotide polymorphism; 3' UTR, 3' untranslated region; OR, odds ratio; 95% CI, 95% confidence interval. Genotype frequencies are shown in parentheses (%). P value was calculated using Fisher's exact test.
SLE-associated SNPs rs179019, rs179010 and rs3853839 were estimated to form five major haplotypes (Table 4). When haplotype frequencies were compared between female SLE patients and healthy controls, tendencies for an increase of haplotype 3 containing all of the SLE risk alleles and a decrease of haplotype 2 containing none of them were observed, although the differences did not reach statistical significance (permutation P, haplotype 3 = 0.081; permutation P, haplotype 2 = 0.068). We next examined the haplotype association under the recessive model. Individuals homozygous for all three SNPs were considered to be homozygous for the haplotype. A significant association of haplotype 3 was detected under the recessive model (haplotype 3/3 versus others: P = 0.016, OR 2.37, 95% CI 1.17 to 4.80), but haplotype 1 (P = 0.21, OR 1.32, 95% CI 0.86 to 2.05) and haplotype 4 (P = 1.0, OR 0.80, 95% CI 0.11 to 5.68), which also contained the 3'UTR risk allele but not both of the intronic SNPs, were not associated. These results suggest that the combination of the intronic and 3'UTR risk alleles may be associated with higher SLE risk.
Table 4.
Estimated haplotype frequencies in SLE and controlsa
| Haplotype | rs179019 | rs179010 | rs3853839 | SLE | Controls | Permutation P value |
|---|---|---|---|---|---|---|
| 1 | C | C | G | 40.6% | 38.0% | 0.94 |
| 2 | C | C | C | 18.2% | 24.1% | 0.068 |
| 3b | A | T | G | 26.1% | 20.3% | 0.081 |
| 4 | C | T | G | 8.5% | 8.8% | 1.0 |
| 5 | A | T | C | 5.2% | 5.6% | 1.0 |
aSLE, systemic lupus erythematosus; P values were calculated by permutation test (100,000 permutations) using HaploView version 4.0 software; beach haplotype was also tested for association under the recessive model. Individuals homozygous at all three SNPs were considered homozygous for the haplotype. Only haplotype 3 was significantly associated with SLE under the recessive model (SLE, 31 (9.0%) of 344; control, 11 (4.0%) of 274; P = 0.016 by Fisher's exact test; odds ratio 2.37, 95% confidence interval 1.17 to 4.80).
Association of TLR7 SNPs with clinical subsets of SLE
We examined whether TLR7 SNPs were associated with clinical phenotypes such as the presence of anti-Sm antibodies, anti-double-stranded DNA antibodies and renal disorder. Association was tested between SLE patients with each phenotype and healthy controls. The OR of rs179019 was slightly higher in the subset with renal disorder (P = 0.011, OR 2.25, 95% CI 1.21 to 4.18) than in all SLE patients (P = 0.016, OR 2.02, 95% CI 1.15 to 3.54) (Table 5), although no statistically significant association was observed in case-only analysis (SLE patients with renal disorder versus those without). The association of rs179019 with renal disorder remained significant after adjustment for rs3853839 on the basis of logistic regression analysis (Padjusted = 0.019, OR 2.10, 95% CI 1.13 to 3.93 under the recessive model).
Table 5.
Association study of TLR7 SNPs with clinical characteristics of SLEa
| SLE total | Anti-Sm antibodies | Anti-dsDNA antibodies | Renal disorder | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Model | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) |
| rs3853839 | Allele | 0.017 | 1.36 (1.06 to 1.75) | 0.032 | 1.65 (1.04 to 2.62) | 0.014 | 1.40 (1.07 to 1.84) | 0.025 | 1.40 (1.04 to 1.89) |
| rs179019 | Recessive | 0.016b | 2.02 (1.15 to 3.54) | 1.0b | 0.89 (0.29 to 2.73) | 0.029b | 1.93 (1.07 to 3.48) | 0.011b | 2.25 (1.21 to 4.18) |
| rs179010 | Recessive | 0.018 | 1.75 (1.10 to 2.80) | 0.67b | 1.16 (0.51 to 2.67) | 0.030 | 1.72 (1.05 to 2.83) | 0.042 | 1.73 (1.02 to 2.95) |
aTLR7, Toll-like receptor 7 gene; SNP, single-nucleotide polymorphism; SLE, systemic lupus erythematosus; anti-Sm, anti-Smith; dsDNA, double-stranded DNA; OR, odds ratio, 95% CI, confidence interval; bFisher's exact test was used. Association was tested by χ2 analysis or Fisher's exact test using 2 × 2 contingency tables under the indicated model for rs3853839G, rs179019A and rs179010 T allele. All SLE as well as each SLE subset were compared with healthy controls.
Analysis of association between TLR7 SNPs andTLR7 mRNA levels
To investigate the functional significance of the TLR7 SNPs, we analyzed the association betweenTLR7 SNPs and TLR7 mRNA levels (Figure 2). The TLR7 mRNA levels in PBMNCs from Japanese female SLE patients were measured using RT-PCR assay and were compared among individuals carrying each genotype. Although not statistically significant because of the limited sample size, a tendency toward an association of rs3853839G with elevated TLR7 mRNA levels was observed (P = 0.20 by Kruskal-Wallis test). This tendency was consistent with the observations in the Chinese population [7], which demonstrated increased TLR7 transcripts in individuals carrying rs3853839G. On the other hand, evidence for an association of the intronic SNPs with mRNA levels was not observed.
Figure 2.
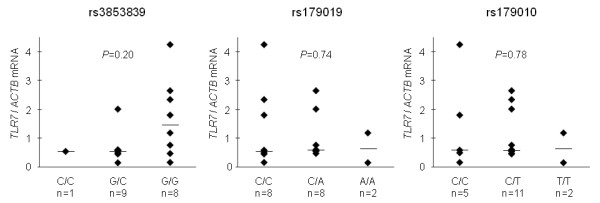
Association analysis of Toll-like receptor 7 genotypes with mRNA expression in peripheral blood mononuclear cells. Association between Toll-like receptor 7 (TLR7) single-nucleotide polymorphisms (SNPs) and TLR7 mRNA levels was examined by using the Kruskal-Wallis test. Relative quantitative levels of TLR7 mRNA were normalized to β-actin (ACTB) mRNA levels. Bars indicate median values in each group. The experiments were performed in triplicate.
Discussion
In the recently reported multicenter study, an association of rs3853839 was originally found by screening the TLR7-TLR8 region in Chinese and Korean populations and was subsequently replicated in Chinese and Japanese populations [7]. In the process of the study, some population difference was noted for rs3853839 and other SNPs, even among these East Asian populations. Because association between TLR7 and SLE had not been examined in a systematic manner in a Japanese population, we thought that TLR7 SNPs other than rs3853839 might also contribute to SLE.
To explore such a possibility, we analyzed the association of eight tag SNPs in TLR7 and the newly detected association of two SNPs in intron 2, rs179019 and rs179010. Conditional logistic regression analysis indicated that the association of the intronic SNPs cannot be explained by LD with rs3853839. In agreement with these results, the association of the intronic SNPs remained significant after excluding the effect of the 3'UTR SNP by testing the association only among individuals carrying the 3'UTR risk allele. Furthermore, haplotype analysis showed significant association of the haplotype containing all of the three SLE risk alleles, but not of the other haplotypes. All of these results support the possibility that the possession of both the 3'UTR and intronic risk alleles may confer further risk for SLE.
Although rs179019 and rs179010 were also investigated in the Discovery Panel in the previous study, the majority of whom were Chinese and Korean participants, no significant association was detected [7]. The Japanese patients and controls analyzed in this study were not included in the Discovery Panel. Population difference was also observed for rs3853839 between the Chinese and Korean populations, as this SNP was strongly associated with SLE in Chinese, but not in Koreans [7], suggesting that the genetic background with respect to TLR7 association with SLE might be somewhat different, even among the closely related East Asian populations. Minor allele frequencies of rs179019 and rs179010 in the HapMap CHB (Han Chinese in Beijing) samples (rs179019: 30.9%, rs179010: 37.3%) available in the International HapMap database [9] are similar to those in the Japanese observed in this study (rs179019: 28.5%, rs179010: 35.2%). Thus, the difference in the association cannot be explained by differences in the minor allele frequencies. We cannot rule out the possibility that another SNP tagged by rs179019 and rs179010 in Japanese, but not in Chinese or Koreans because of difference in the LD status, might play a causative role. Such a possibility would be addressed by resequencing the entire TLR7 region.
There is growing evidence to support involvement of type I IFN in the development of SLE. TLR7 is crucial for the production of type I IFN. Thus, the most plausible role of TLR7 SNPs in SLE pathogenesis is likely to be explained by elevated type I IFN production. The sera of SLE patients displayed elevated levels of type I IFN, and expression of IFN-inducible genes in PBMNCs was also upregulated in SLE [10]. Occasional occurrence of SLE symptoms following treatment with IFNα in patients with cancer or hepatitis underscored the relevance of type I IFN [10]. Type I IFN is thought to be a potential therapeutic target for SLE, and clinical trials of anti-IFNα antibodies in SLE are currently underway [11].
Recent genetic studies have identified an association of type I IFN pathway-related genes, IFN regulatory factor 5 (IRF5) and STAT4, with SLE in various populations [10,12-16]. An IRF5 SLE risk haplotype has been shown to be associated with high serum IFNα activity in SLE patients [17], whereas the STAT4 SLE risk variant was associated with increased sensitivity to IFNα in vivo [18]. These observations, as well as the previous study on TLR7 showing upregulation of TLR7 in the risk genotype [7], suggest that SLE-associated alleles in the type I IFN pathway are gain-of-function alleles in nature.
Another potential role of TLR7 polymorphisms may be related to the induction of proinflammatory cytokines. IRF5 is activated by TLR7 signaling and regulates the expression of many genes, including type I IFN and proinflammatory cytokines [19]. STAT4 is activated by type I IFN as well as interleukin 12 and plays a role in Th1 differentiation [20]. In view of these observations, the association between TLR7 SNPs and SLE might also be explained by overproduction of proinflammatory cytokines in addition to type I IFN.
There are conflicting reports about copy number variation (CNV) of TLR7. Initially, the existence of CNV was reported by Kelley et al. [21]. They showed that, although common CNV was observed in Caucasians and African-Americans, no association with SLE was detected [21]. Recently, García-Ortiz et al. [22] reported an association of CNV with childhood-onset SLE in a Mexican population. In contrast to these observations, Shen et al. [7] did not find common TLR7 CNV in multiple populations, including Asians. The latter observation is consistent with the fact that no CNV was registered in the Database of Genomic Variants [23], which includes results derived from the HapMap JPT (Japanese in Tokyo) samples.
Although our observation in the expression analysis supported the previous report that indicated the association between the risk allele of the 3'UTR SNP and elevated expression of TLR7 [7], evidence for the association of the intronic SNPs with levels of TLR7 mRNA was not observed, and therefore the molecular mechanism of the intronic SNPs requires further study. TLR7 is mainly expressed in pDCs and B cells. pDCs represent the major source of type I IFN, but constitute less than 1% of PBMNCs. If the intronic SNPs have a regulatory role in a cell type-specific fashion and influence the expression level of TLR7 in pDCs but not in other white blood cells, such an effect may not have been detected in the analysis of total PBMNCs. In addition, the sample size of this study may not have been large enough for us to conclude that the intronic SNPs have no effect on the expression of TLR7.
Because we focused only on the Japanese population, the sample size of this study was limited and the observed statistical association was modest. Therefore, the association of the intronic SNPs should be confirmed in future independent studies.
Conclusions
TLR7 intronic SNPs rs179019 and rs179010 are associated with SLE independently of 3'UTR SNP rs3853839 in Japanese women. Our findings support the genetic role of TLR7 SNPs in Asian populations with SLE.
Abbreviations
95% CI: 95% confidence interval; CNV: copy number variation; CpG: cytidine-phosphate-guanosine; IFN: interferon; LD: linkage disequilibrium; OR: odds ratio; PBMNCs: peripheral blood mononuclear cells; pDCs: plasmacytoid dendritic cells; RT-PCR: reverse transcription polymerase chain reaction; SLE: systemic lupus erythematosus; SNP: single-nucleotide polymorphism; ssRNA: single-stranded RNA; TLR: Toll-like receptor; UTR: untranslated region; Yaa: Y chromosome-linked autoimmune accelerator.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AK participated in the study design; carried out all genotyping, expression analysis and statistical analyses; and wrote the manuscript. HF, YK, SI, TH, MK, IM, ST, YT, HH and TS recruited the patients and controls and collected clinical information. NT designed and coordinated the study and helped in the manuscript preparation. All authors read and approved the final manuscript.
Contributor Information
Aya Kawasaki, Email: a-kawasaki@umin.net.
Hiroshi Furukawa, Email: h-furukawa@sagamihara-hosp.gr.jp.
Yuya Kondo, Email: c0830419@md.tsukuba.ac.jp.
Satoshi Ito, Email: s-ito@ra-center.com.
Taichi Hayashi, Email: t.hayashi@md.tsukuba.ac.jp.
Makio Kusaoi, Email: makio.k@med.juntendo.ac.jp.
Isao Matsumoto, Email: ismatsu@md.tsukuba.ac.jp.
Shigeto Tohma, Email: s-touma@sagamihara-hosp.gr.jp.
Yoshinari Takasaki, Email: tyoshi@med.juntendo.ac.jp.
Hiroshi Hashimoto, Email: hhashi@mountain.ocn.ne.jp.
Takayuki Sumida, Email: tsumida@md.tsukuba.ac.jp.
Naoyuki Tsuchiya, Email: tsuchiya-tky@umin.ac.jp.
Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research (B) (22390199) and Grant-in-Aid for Young Scientists (B) (21790935) from the Japan Society for the Promotion of Science (JSPS), Health and Labour Science Research Grants for the Research on intractable diseases from the Ministry of Health, Labour and Welfare of Japan, Japan Rheumatism Foundation, and Takeda Science Foundation.
References
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Tus K, Li QZ, Wang A, Tian XH, Zhou J, Liang C, Bartov G, McDaniel LD, Zhou XJ, Schultz RA, Wakeland EK. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci USA. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Komatsuda A, Wakui H, Iwamoto K, Ozawa M, Togashi M, Masai R, Maki N, Hatakeyama T, Sawada K. Up-regulated expression of Toll-like receptors mRNAs in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin Exp Immunol. 2008;152:482–487. doi: 10.1111/j.1365-2249.2008.03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Fu Q, Deng Y, Qian X, Zhao J, Kaufman KM, Wu YL, Yu CY, Tang Y, Chen JY, Yang W, Wong M, Kawasaki A, Tsuchiya N, Sumida T, Kawaguchi Y, Howe HS, Mok MY, Bang SY, Liu FL, Chang DM, Takasaki Y, Hashimoto H, Harley JB, Guthridge JM, Grossman JM, Cantor RM, Song YW, Bae SC, Chen S. et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci USA. 2010;107:15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- International HapMap Project. http://hapmap.ncbi.nlm.nih.gov/index.html.en
- Kyogoku C, Tsuchiya N. A compass that points to lupus: genetic studies on type I interferon pathway. Genes Immun. 2007;8:445–455. doi: 10.1038/sj.gene.6364409. [DOI] [PubMed] [Google Scholar]
- Rönnblom L, Elkon KB. Cytokines as therapeutic targets in SLE. Nat Rev Rheumatol. 2010;6:339–347. doi: 10.1038/nrrheum.2010.64. [DOI] [PubMed] [Google Scholar]
- Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, Bauer JW, Gillett C, Burtt N, Cunninghame Graham DS, Onofrio R, Petri M, Gunnarsson I, Svenungsson E, Rönnblom L, Nordmark G, Gregersen PK, Moser K, Gaffney PM, Criswell LA, Vyse TJ, Syvänen AC, Bohjanen PR, Daly MJ, Behrens TW. et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN); Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Xu JH, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- Kawasaki A, Kyogoku C, Ohashi J, Miyashita R, Hikami K, Kusaoi M, Tokunaga K, Takasaki Y, Hashimoto H, Behrens TW, Tsuchiya N. Association of IRF5 polymorphisms with systemic lupus erythematosus in a Japanese population: support for a crucial role of intron 1 polymorphisms. Arthritis Rheum. 2008;58:826–834. doi: 10.1002/art.23216. [DOI] [PubMed] [Google Scholar]
- Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, Matsumoto I, Ito S, Tsutsumi A, Koga M, Arinami T, Graham RR, Hom G, Takasaki Y, Hashimoto H, Behrens TW, Sumida T, Tsuchiya N. Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region. Arthritis Res Ther. 2008;10:R113. doi: 10.1186/ar2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-α activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-α in lupus patients in vivo. J Immunol. 2009;182:34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O'Shea JJ. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–156. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- Kelley J, Johnson MR, Alarcón GS, Kimberly RP, Edberg JC. Variation in the relative copy number of the TLR7 gene in patients with systemic lupus erythematosus and healthy control subjects. Arthritis Rheum. 2007;56:3375–3378. doi: 10.1002/art.22916. [DOI] [PubMed] [Google Scholar]
- García-Ortiz H, Velázquez-Cruz R, Espinosa-Rosales F, Jiménez-Morales S, Baca V, Orozco L. Association of TLR7 copy number variation with susceptibility to childhood-onset systemic lupus erythematosus in Mexican population. Ann Rheum Dis. 2010;69:1861–1865. doi: 10.1136/ard.2009.124313. [DOI] [PubMed] [Google Scholar]
- The Database of Genomic Variants. http://projects.tcag.ca/variation/


