Abstract
Introduction
The objective of this study is to determine if multiple systemic autoimmune diseases (SAID) share gene expression pathways that could provide insights into pathogenic mechanisms common to these disorders.
Methods
RNA microarray analyses (Agilent Human 1A(V2) 20K oligo arrays) were used to quantify gene expression in peripheral blood cells from 20 monozygotic (MZ) twin pairs discordant for SAID. Six affected probands with systemic lupus erythematosus (SLE), six with rheumatoid arthritis (RA), eight with idiopathic inflammatory myopathies (IIM), and their same-gendered unaffected twins, were enrolled. Comparisons were made between discordant twin pairs and these were also each compared to 40 unrelated control subjects (matched 2:1 to each twin by age, gender and ethnicity) using statistical and molecular pathway analyses. Relative quantitative PCR was used to verify independently measures of differential gene expression assessed by microarray analysis.
Results
Probands and unrelated, matched controls differed significantly in gene expression for 104 probes corresponding to 92 identifiable genes (multiple-comparison adjusted P values < 0.1). Differentially expressed genes involved several overlapping pathways including immune responses (16%), signaling pathways (24%), transcription/translation regulators (26%), and metabolic functions (15%). Interferon (IFN)-response genes (IFI27, OASF, PLSCR1, EIF2AK2, TNFAIP6, and TNFSF10) were up-regulated in probands compared to unrelated controls. Many of the abnormally expressed genes played regulatory roles in multiple cellular pathways. We did not detect any probes expressed differentially in comparisons among the three SAID phenotypes. Similarly, we found no significant differences in gene expression when comparing probands to unaffected twins or unaffected twins to unrelated controls. Gene expression levels for unaffected twins appeared intermediate between that of probands and unrelated controls for 6535 probes (32% of the total probes) as would be expected by chance. By contrast, in unaffected twins intermediate ordering was observed for 84 of the 104 probes (81%) whose expression differed significantly between probands and unrelated controls.
Conclusions
Alterations in expression of a limited number of genes may influence the dysregulation of numerous, integrated immune response, cell signaling and regulatory pathways that are common to a number of SAID. Gene expression profiles in peripheral blood suggest that for genes in these critical pathways, unaffected twins may be in a transitional or intermediate state of immune dysregulation between twins with SAID and unrelated controls, perhaps predisposing them to the development of SAID given the necessary and sufficient environmental exposures.
Introduction
Previous studies suggest that patterns of gene expression as measured by RNA microarray are correlated among healthy twins, siblings, and other first-degree family members [1-3]. These data support the importance of shared genetic and perhaps environmental influences on transcription and other controls of steady-state mRNA expression. Interestingly, similar profiles of global RNA expression are observed among first-degree family members discordant for autoimmune disease, suggesting that other healthy, first-degree relatives may have an underlying genetic predisposition to disease [2,4,5].
Shared patterns of global gene expression have also been observed among genetically unrelated individuals with systemic autoimmune diseases (SAIDs), including systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) [6-9]. These data support the hypothesis that different autoimmune diseases share certain clinical features and perhaps mechanisms of disease pathogenesis. Most notably, the type I IFN 'signature' associated with the transcriptional control of many IFN-regulated gene products has been consistently detected among patients with SLE, dermatomyositis (DM), or other SAIDs [8,10-16]. Conversely, disease-specific gene expression profiles are associated with particular autoimmune pathologies and may reflect target tissue specificity and other distinctions in genetic susceptibilities or environmental exposure histories or both [7,17-20].
To minimize the role of polymorphic gene effects, we enrolled 20 pairs of monozygotic (MZ) twins discordant for SAID (six probands with SLE, six with RA, and eight with idiopathic inflammatory myopathies, or IIMs) and used RNA microarrays (Agilent Human 1A(V2); 20K) to examine gene expression patterns in their peripheral blood cells. To determine whether unaffected twins might show preclinical evidence of enhanced susceptibility to SAID, we also compared probands and their unaffected twins with 40 unrelated, matched control subjects. One might expect etiologically relevant genes to show expression patterns in the unaffected twins that are intermediate between those in their affected twin counterparts and in unrelated, matched controls. Moreover, in an effort to identify shared molecular pathways involved in the pathogenesis of these SAIDs, we asked whether particular gene expression profiles were characteristic of affected probands regardless of specific disease diagnosis.
Materials and methods
Study subjects
Five adult (at least 18 years of age) and 15 juvenile MZ twin pairs discordant for SAID and 40 unrelated control subjects (two controls per twin pair) matched on age within 6 years, gender, and ethnicity were subjects in this study. These subjects were among those enrolled between 2001 and 2006 in the National Institutes of Health (NIH) investigational review board-approved Twins-Sib study assessing the pathogenesis of SAID. Ethical approval for this microarray study was also obtained from the NIH investigational review board, and all human subjects provided informed consent. Twin pairs enrolled within 4 years of probands' diagnoses included 19 non-Hispanic Caucasian twin pairs and a single Hispanic twin pair (with SLE). Probands fulfilled American College of Rheumatology criteria for adult or juvenile SLE (n = 4 and 2, respectively), RA or JRA (n = 1 and 5, respectively), juvenile dermatomyositis (JDM) (n = 7), or juvenile polymyositis (JPM) (n = 1); we excluded patients with inherited, metabolic, infectious, or other causes of disease. The juvenile probands ranged in age from 3 to 18 years (mean of 11.2 years), whereas adults ranged from 19 to 43 years (mean of 29.2 years). Twins included 14 female and 6 male pairs. Monozygosity was confirmed by short tandem repeat analysis of genomic DNAs (Proactive Genetics, Inc., Augusta, GA, USA). Unrelated, matched controls were free of infections, trauma, vaccines, and surgeries for 8 weeks and had no first-degree family members with SAID.
RNA was purified from peripheral whole blood samples collected in PAXgene RNA tubes (VWR Scientific, Radnor, PA, USA) by using a PAXgene RNA Isolation Kit (Qiagen, Inc., Valencia, CA, USA) in accordance with the recommendations of the manufacturer. Total RNA was quantified spectrophotometrically and then stored at -80°C until analysis. To minimize sources of variability, we collected whole blood samples in the morning, and immunosuppressive therapy was held at least 24 hours before collection.
Microarray analysis
Gene expression analysis was conducted by using Agilent Human 1A(V2) Oligo arrays with approximately 20,000 probes for known genes and expressed sequence tags (Agilent Technologies, Inc., Santa Clara, CA, USA). Each sample was hybridized against a human universal RNA control (Stratagene, La Jolla, CA, USA). Total RNA (500 ng) was amplified and labeled by using the Agilent Low RNA Input Fluorescent Linear Amplification Kit in accordance with the protocol of the manufacturer. For each two-color comparison, 750 ng of each Cy3- (universal control) and Cy5-labeled sample cRNA was mixed and fragmented by using the Agilent in situ Hybridization Kit. Hybridizations were performed for 17 hours in a rotating hybridization oven. Slides were washed and then scanned with an Agilent scanner. Data were obtained by using Agilent Feature Extraction software (version 7.5), with defaults for all parameters. The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise. The resulting data were processed by using the Rosetta Resolver system (version 7.2) (Rosetta Biosoftware, now part of Microsoft Corporation, Redmond, WA, USA). Resulting base 2 logarithms of expression ratios (subject over universal control) were exported from Rosetta Resolver for further analysis. The data reported in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) [21] and are accessible through GEO Series accession number [GEO:GSE24060].
Microarray data normalization and quality control
We used principal component analysis (PCA) to investigate the effect of known sample variables such as subject age, gender, race, disease status, disease activity, medications, and peripheral white blood cell counts and differentials as well as technical variables such as sample hybridizations performed at different times. We observed no obvious differences across the first three principal components due to age, gender, ethnicity, disease status, disease activity, or treatment. We noticed, however, that samples clustered according to their hybridization batch. To correct for this, we centered the ratios for each probe by using the median for their hybridization group. The PCA plots generated by using the corrected data showed no obvious groupings, so we used these data for further analyses.
Identification of differentially expressed probes
To identify probes that were differentially expressed in probands (P) as compared with their unaffected twin (U) and with unrelated, matched controls (C), we employed a mixed-effects model to calculate P values for each of the approximately 20,000 probes. Random effects included indicators for twin pairs, for P-versus-U comparisons, and for the strata defined by the matching variables for the P-versus-C and U-versus-C comparisons. With these adjustments, we performed three binary comparisons: (a) P versus C, in which all probands with SAID were compared with two unrelated, matched controls; (b) U versus C, in which all unaffected twins were compared with two unrelated, matched controls; (c) P versus U, in which all probands with SAID were compared with their unaffected twins in a paired analysis. We adjusted P values from each of the above three comparisons to control the false discovery rate (FDR) at 0.1 by using the Benjamini-Hochberg method [22].
Disease-specific gene expression analyses
The goal was to investigate whether blood cell gene expression profiles for probands with SAID - namely (J)RA, (J)SLE, and (J)IIM (that is, JDM + JPM) - show characteristic differences in gene expression. To answer this question, we used all probe expression data and employed three different types of statistical analyses: (a) Hierarchical clustering - we performed unsupervised hierarchical clustering by using all 20 proband samples to check whether probands cluster by their known SAID subtype. (b) Multiclass analysis of variance (ANOVA) - to assess whether the difference between the gene expression of a proband and their respective twin is SAID subtype-specific, we first obtained a measurement of the gene expression difference per twin pair for each probe by subtracting the log-transformed expression value of the unaffected twin from that of the respective affected twin. We next grouped twin pairs into the three major SAID diagnostic classes. Finally, we performed a three-category, one-way ANOVA by using the differences in log-transformed expression values for each oligo probe. The P values were then adjusted for multiple testing by using the Benjamini-Hochberg method with the FDR of 0.1 [22]. (c) Mixed-effects model - to assess whether the difference between the gene expression of an affected proband and their corresponding unrelated but matched controls is SAID subtype-specific, we employed a mixed-effects model allowing a random effect for each matched set and treating the disease subtype as a fixed effect. The P values were then adjusted for multiple testing by using the Benjamini-Hochberg method with the FDR of 0.1 [22].
Functional analysis of differentially expressed genes
We used Ingenuity Pathways Analysis (IPA) software (Ingenuity® Systems, Redwood City, CA, USA) to assess functional class membership for probes identified as differentially expressed between probands and unrelated, matched controls. Each gene of interest was mapped and overlaid onto a global molecular network developed from information contained in the IPA Knowledge Base. Networks of genes were then generated algorithmically on the basis of their connectivity as established in the published literature. Fisher's exact test was used to assess whether the number of probes mapped to each biologic function or pathway (or both) differs from the number expected due to chance alone.
Real-time polymerase chain reaction analysis
Relative quantitation (RQ) measurements of gene expression were by real-time polymerase chain reaction (RT-PCR) assays using cDNA prepared from Paxgene-purified whole peripheral blood RNA preparations collected from disease-discordant twins and unrelated, matched controls as described above. cDNA was prepared by using a High Capacity cDNA Reverse Transcription kit (1.0 μg of input RNA per subject) in accordance with the recommendations of the manufacturer (Applied Biosystems, Foster City, CA, USA). RT-PCRs were performed in triplicate by using commercially prepared cDNA primers and probes (TaqMan Gene Expression Assays; Applied Biosystems) for the following genes of interest: MAP2K6 (TaqMan Assay Hs00992389), IL1RN (Hs00893626), IFI27 (Hs00271467), FCER1A (Hs01090134), FYN (Hs00176628), LTK (Hs00914334), KRTCAP2 (Hs00744717), SYTL2 (Hs00262988), ANXA3 (Hs00971411), CEACAM6 (Hs00366002), DEFA4 (Hs00157252), TNFAIP6 (Hs01113602), TNFSF10 (Hs00234356), EIF2AK2 (Hs00169345), and LGR6 (Hs00663887). Genes of interest were selected from the microarray analysis by a combination of criteria, including degree of statistical significance, magnitude of differential gene expression, and possible biologic relevance to disease. Standard TaqMan assays (20 μL) were performed by using an Applied Biosystems Universal Assay Buffer with 10 ng of input cDNA per reaction and amplified for 40 cycles in an ABI Prism 7900HT Sequence Detection System (SDS software version 2.3). Co-amplication of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an endogenous assay and standardization control. RQ of gene expression was calculated from log-phase mean threshold cycle values normalized to GAPDH expression by using Applied Biosystems RQ Manager software (version 1.2).
Results
Microarray differential expression analysis
Significant differences (FDR-adjusted P values of less than 0.1) were observed for 104 oligo probes in analyses of SAID probands compared with the unrelated, matched controls (Table 1). Upon probe annotation, 92 genes representing several non-mutually exclusive categories, including immune function (16%), signaling pathways (24%), transcription/translation regulators (26%), and metabolic functions (15%), were identified (Table 2). Of these 92 genes, 73.9% were underexpressed (mean fold change of 0.71 and range of 0.91 to 0.56) and 26.1% were overexpressed (mean fold change of 2.2 and range of 1.1 to 7.2) in probands relative to unrelated, matched controls.
Table 1.
Genes that were differentially expressed between probands (n = 20) with systemic autoimmune disease and unrelated, matched controls (n = 40)
| Accession | Annotation | Fold changea | P valueb | FDR | Function/Category |
|---|---|---|---|---|---|
| Increased expression in probands | |||||
| TNFAIP6 | Tumor necrosis factor-alpha-induced protein 6 | 2.2 | 1.5 × 10-5 | 0.051 | Co-factor, immune |
| HP | Haptoglobin | 2.2 | 2.0 × 10-5 | 0.051 | Carrier, transport |
| HPR | Haptoglobin-related protein | 2.3 | 3.5 × 10-5 | 0.059 | Carrier, transport |
| ANXA3 | Annexin A3 | 2.8 | 5.1 × 10-5 | 0.074 | Regulation, signaling |
| APOBEC3A | Apolipoprotein B mRNA editing enzyme 3A | 1.4 | 5.8 × 10-5 | 0.074 | Regulation, translation |
| EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | 2.4 | 7.1 × 10-5 | 0.074 | Regulation, translation |
| BMX | Non-receptor tyrosine kinase | 2.2 | 8.6 × 10-5 | 0.074 | Kinase, signaling |
| CEACAM6 | Carcinoembryonic Ag-related cell adhesion molecule 6 | 4.7 | 8.7 × 10-5 | 0.074 | Receptor, targeting |
| PLSCR1 | Phospholipid scramblase 1 | 2.3 | 1.0 × 10-4 | 0.074 | Regulation, immune |
| LTF | Lactotransferrin | 2.3 | 1.1 × 10-4 | 0.074 | Regulation, immune |
| TNFSF10 | Tumor necrosis factor ligand 10 | 1.6 | 1.4 × 10-4 | 0.074 | Cytokine, immune |
| OASL | 2'-5'-Oligoadenylate synthetase-like | 2.2 | 1.7 × 10-4 | 0.074 | Binding, thyroid receptor |
| MAP2K6 | Mitogen-activated protein kinase kinase 6 | 1.4 | 2.2 × 10-4 | 0.080 | Kinase, signaling |
| ZCCHC2 | Zinc finger protein | 1.5 | 2.4 × 10-4 | 0.082 | Unknown, signaling |
| INHBB | Inhibin, beta B | 2.3 | 2.7 × 10-4 | 0.085 | Ligand, signaling |
| DEFA4 | Defensin, alpha 4, corticostatin | 2.3 | 2.9 × 10-4 | 0.088 | Ligand, signaling |
| IFI27 | Interferon, alpha-inducible protein 27 | 7.2 | 3.0 × 10-4 | 0.088 | Unknown, immune |
| SLC22A4 | Solute carrier family 22, member 4 | 1.7 | 3.3 × 10-4 | 0.088 | Transport |
| IL1RN | Interleukin 1 receptor antagonist | 1.7 | 3.4 × 10-4 | 0.088 | Ligand, immune |
| RRAGD | Ras-related GTP-binding protein D | 1.3 | 3.4 × 10-4 | 0.088 | Ligand, signaling |
| DEGS1 | Degenerative spermatocyte homolog 1 | 1.1 | 3.9 × 10-4 | 0.093 | Enzyme, metabolism |
| PGAP1 | GPI deacylase | 1.3 | 3.9 × 10-4 | 0.093 | Enzyme, signaling |
| F5 | Coagulation factor V | 1.4 | 4.2 × 10-4 | 0.094 | Ligand, coagulation |
| CDS2 | CDP-diacylglycerol synthase | 1.2 | 4.6 × 10-4 | 0.097 | Enzyme, metabolism |
| HIST1H2AB | Histone 1, H2ab | 1.3 | 4.7 × 10-4 | 0.097 | Chromatin, structural |
| Decreased expression in probands | |||||
| PACSIN1 | Protein kinase C substrate 1 in neurons | -1.4 | 3.3 × 10-6 | 0.042 | Kinase, signaling |
| KRTCAP2 | Keratinocyte-associated protein 2 | -1.2 | 5.9 × 10-6 | 0.042 | Enzyme, structural |
| FCER1A | Fc fragment of high affinity IgE receptor | -1.9 | 9.2 × 10-6 | 0.042 | Receptor, immune |
| FYN | Oncogene | -1.3 | 9.8 × 10-6 | 0.042 | Kinase, signaling |
| SYTL2 | Synaptotagmin-like 2 | -1.5 | 1.0 × 10-5 | 0.042 | Unknown, immune |
| CD99 | MIC2, single-chain type-1 glycoprotein | -1.2 | 1.8 × 10-5 | 0.051 | Receptor, immune |
| PECI | Peroxisomal D3,D2-enoyl-CoA isomerase | -1.3 | 3.3 × 10-5 | 0.059 | Enzyme, metabolism |
| CD81 | Target of antiproliferative antibody 1 | -1.2 | 3.4 × 10-5 | 0.059 | Receptor, immune |
| PBX4 | Pre-B-cell leukemia transcription factor 4 | -1.4 | 7.7 × 10-5 | 0.074 | Regulation, transcription |
| DYRK2 | Tyrosine-phosphorylation regulated kinase 2 | -1.2 | 8.1 × 10-5 | 0.074 | Kinase, signaling |
| ZBTB5 | Zinc finger and BTB domain containing 5 | -1.2 | 9.4 × 10-5 | 0.074 | Regulation, transcription |
| PABPC3 | Poly(A)-binding protein 3 | -1.3 | 9.8 × 10-5 | 0.074 | Regulation, translation |
| NELL2 | NEL-like 2 | -1.3 | 1.0 × 10-4 | 0.074 | Mitogen, cell division |
| LOC100133315 | Unknown cation channel/receptor | -1.2 | 1.0 × 10-4 | 0.074 | Receptor, signaling |
| DNAJA3 | DnaJ (Hsp40) homolog A3 | -1.3 | 1.2 × 10-4 | 0.074 | Chaperone, transport |
| EIF3S6IP | Eukaryotic translation initiation factor 3, subunit 6 | -1.3 | 1.2 × 10-4 | 0.074 | Regulation, translation |
| EIF3S8 | Eukaryotic translation initiation factor 3, subunit 8 | -1.2 | 1.2 × 10-4 | 0.074 | Regulation, translation |
| RBMX | RNA-binding motif protein | -1.1 | 1.2 × 10-4 | 0.074 | Regulation, transcription |
| GZMK | Granzyme K | -1.4 | 1.3 × 10-4 | 0.074 | Enzyme, immune |
| DNMT1 | DNA (cytosine 5)-methyltransferase 1 | -1.3 | 1.3 × 10-4 | 0.074 | Enzyme, metabolism |
| ZNF219 | Zinc finger protein 219 | -1.2 | 1.3 × 10-4 | 0.074 | Regulation, transcription |
| DVL1 | Dishevelled homologue (Drosophila) | -1.2 | 1.4 × 10-4 | 0.074 | Mitosis, signaling |
| IMP3 | U3 small ribonucleoprotein homologue (yeast) | -1.2 | 1.5 × 10-4 | 0.074 | Structural, translation |
| ANAPC5 | Anaphase promoting complex subunit 5 | -1.2 | 1.5 × 10-4 | 0.074 | Enzyme, cell cycle |
| AOF2 | Amine oxidase (flavin containing) domain 2 | -1.2 | 1.6 × 10-4 | 0.074 | Regulation, transcription |
| GPRASP1 | G protein-coupled receptor | -1.4 | 1.6 × 10-4 | 0.074 | Receptor, signaling |
| LTK | Leukocyte tyrosine kinase | -1.3 | 1.6 × 10-4 | 0.074 | Kinase, immune |
| MTA1 | Metastasis-associated 1 | -1.2 | 1.6 × 10-4 | 0.074 | Regulation, transcription |
| UXT | Ubiquitously expressed transcript | -1.2 | 1.6 × 10-4 | 0.074 | Regulation, transcription |
| KLRB1 | Killer cell lectin-like receptor B1 | -1.6 | 1.7 × 10-4 | 0.074 | Receptor, immune |
| SEMA4C | Semaphorin 4C | -1.3 | 1.7 × 10-4 | 0.074 | Neural development |
| STMN3 | Stathmin-like 3 | -1.4 | 2.0 × 10-4 | 0.079 | Cytoskeletal, signaling |
| ATP1A1 | ATPase Na+/K+ transporter | -1.2 | 2.0 × 10-4 | 0.079 | Transporter, signaling |
| PSBP | Prostatic steroid-binding protein | -1.3 | 2.2 × 10-4 | 0.080 | Receptor, signaling |
| RPL18 | Ribosomal protein L18 | -1.3 | 2.2 × 10-4 | 0.080 | Structural, translation |
| RNF220 | Ring finger protein | -1.1 | 2.2 × 10-4 | 0.080 | Unknown |
| SPOCK2 | Sparc/osteonectin/proteoglycan (testican) 2 | -1.2 | 2.3 × 10-4 | 0.081 | Extracellular, structural |
| DDX28 | DEAD box polypeptide | -1.2 | 2.3 × 10-4 | 0.081 | RNA helicase, transcription |
| SRP46 | Splicing factor, arginine/serine-rich, 46 kDa | -1.3 | 2.4 × 10-4 | 0.082 | Regulation, transcription |
| EIF3S7 | Eukaryotic translation initiation factor 3S7 | -1.2 | 2.5 × 10-4 | 0.084 | Regulation, translation |
| TNFRSF25 | Tumor necrosis factor receptor 25 | -1.3 | 2.6 × 10-4 | 0.085 | Receptor, immune |
| MYBL1 | v-Myb myeloblastosis viral oncogene homolog | -1.4 | 2.7 × 10-4 | 0.085 | Regulation, transcription |
| UBE2I | Ubiquitin-conjugating enzyme E2I | -1.1 | 2.8 × 10-4 | 0.087 | Enzyme, regulation |
| PABPC1 | Poly(A)-binding protein | -1.3 | 3.0 × 10-4 | 0.088 | Regulation, translation |
| LDOC1L | Leucine zipper, downregulated in cancer 1-like | -1.4 | 3.1 × 10-4 | 0.088 | Unknown |
| LGR6 | Leucine-rich repeat-containing G protein receptor 6 | -1.6 | 3.1 × 10-4 | 0.088 | Receptor, signaling |
| AES | Amino-terminal enhancer of split | -1.1 | 3.2 × 10-4 | 0.088 | Regulation, transcription |
| CXCR3 | Chemokine receptor 3 | -1.3 | 3.2 × 10-4 | 0.088 | Receptor, immune |
| ATP5G2 | ATP synthase, mitochondrial F0 complex | -1.2 | 3.3 × 10-4 | 0.088 | Enzyme, metabolism |
| RPLP2 | Ribosomal protein P2 | -1.3 | 3.3 × 10-4 | 0.088 | Structural, translation |
| TC2N | Membrane targeting C2 domain | -1.4 | 3.6 × 10-4 | 0.090 | Unknown, signaling |
| ERGIC3 | Breast cancer antigen 84 | -1.2 | 3.8 × 10-4 | 0.092 | Unknown, cell growth |
| GPR183 | Epstein-Barr virus induced gene 2 | -1.5 | 3.9 × 10-4 | 0.093 | Receptor, signaling |
| CSNK1E | Casein kinase 1, epsilon | -1.2 | 4.1 × 10-4 | 0.094 | Kinase, regulation |
| CRIP1 | Cysteine-rich protein 1 | -1.3 | 4.1 × 10-4 | 0.094 | Zn-binding, transport |
| ARIH2 | Ariadne homolog 2 | -1.1 | 4.2 × 10-4 | 0.094 | Ubiquitin ligase, regulation |
| POLS | DNA polymerase sigma | -1.2 | 4.2 × 10-4 | 0.094 | Enzyme, metabolism |
| PUF60 | Ro-RNP-binding protein | -1.2 | 4.3 × 10-4 | 0.096 | RNP-binding, regulation |
| MFNG | Fucose-specific β-1,3-N-acetylglucosaminyltransferase | -1.2 | 4.7 × 10-4 | 0.097 | Enzyme, development |
| PRPSAP2 | Phosphoribosyl pyrophosphate synthetase protein 2 | -1.2 | 4.7 × 10-4 | 0.097 | Enzyme, metabolism |
| STAG3 | Stromal antigen 3 | -1.3 | 4.7 × 10-4 | 0.097 | Cohesion subunit, meiosis |
| FBL | Fibrillarin | -1.2 | 4.9 × 10-4 | 0.098 | snRNP-binding, pre-rRNA |
| RPS19 | Ribosomal protein S19 | -1.3 | 4.9 × 10-4 | 0.098 | Structural, translation |
| DDB1 | Damage-specific DNA-binding protein 1 | -1.2 | 5.0 × 10-4 | 0.098 | DNA repair |
| FBXO21 | F-box protein 21 | -1.2 | 5.0 × 10-4 | 0.098 | Ubiquitin ligase, regulation |
| PTDSS1 | Phosphatidylserine synthase 1 | -1.2 | 5.1 × 10-4 | 0.100 | Enzyme, metabolism |
| AKR1B1 | Aldo-keto reductase family 1, member B1 | -1.2 | 5.2 × 10-4 | 0.100 | Enzyme, metabolism |
aFold change values indicate increased (positive value) or decreased (negative value) levels of gene expression in probands relative to unrelated, matched controls. Oligo probes for hypothetical or unidentified genes were omitted. bUncorrected. FDR, false discovery rate.
Table 2.
Functional categorization of genes differentially expressed between SAID probands and unrelated, matched controls
| Immune | Signaling | Gene expression | Metabolism | Transport | Structural | Cell cycle | Other/Unknown |
|---|---|---|---|---|---|---|---|
| TNFAIP6 | ANXA3 | EIF2AK2 | DEGS1 | HP | HIST1H2AB | NELL2 | ZCCHC2 |
| PLSCR1 | APOBEC3A | PBX4 | CDS2 | HPR | KRTCAP2 | DVL1 | RNF220 |
| LTF | BMX | ZBTB5 | PECI | SLC22A4 | SPOCK2 | ANAPC5 | LDOC1L |
| TNFSF10 | CEACAM6 | PABPC3 | DNMT1 | DNAJA3 | SEMA4C | DDB1 | |
| IFI27 | OASL | EIF3S6IP | ATP5G2 | CRIP1 | ERGIC3 | ||
| IL1RN | MAP2K6 | EIF3S8 | POLS | STAG3 | |||
| FCER1A | INHBB | RBMX | MFNG | ||||
| SYTL2 | DEFA4 | ZNF219 | PRPSAP2 | ||||
| CD99 | RRAGD | IMP3 | PTDSS1 | ||||
| CD81 | PGAP1 | AOF2 | AKR1B1 | ||||
| GZMK | PACSIN1 | MTA1 | |||||
| LTK | FYN | UXT | |||||
| KLRB1 | DYRK2 | RPL18 | |||||
| TNFRSF25 | LOC100133315 | DDX28 | |||||
| CXCR3 | GPRASP1 | SRP46 | |||||
| STMN3 | EIF3S7 | ||||||
| ATP1A1 | MYBL1 | ||||||
| PSBP | PABPC1 | ||||||
| UBE2I | AES | ||||||
| LGR6 | RPLP2 | ||||||
| TC2N | FBL | ||||||
| GPR183 | RPS19 | ||||||
| CSNK1E | |||||||
| ARIH2 | |||||||
| PUF60 | |||||||
| FBXO21 | |||||||
| F5 |
SAID, systemic autoimmune disease.
Comparisons between probands with SAID and their unaffected twins produced no significant differences in gene expression values (that is, the smallest adjusted P values were greater than 0.1), although several of the top-ranking genes were shared with those identified as significant in comparisons of probands and unrelated, matched controls. Comparisons of unaffected twins and unrelated, matched controls likewise revealed no significant differences in gene expression.
We conducted an unsupervised, hierarchical cluster analysis for all 80 subjects (20 probands, 20 unaffected twins, and 40 unrelated, matched controls) by using the expression data for the 104 probes with significant differential expression on the basis of the comparison of probands versus unrelated controls. The resulting heat map (Figure 1) demonstrated a clear segregation of the probands with SAID and the unrelated, matched controls. In contrast, the unaffected twins were interspersed among and between the probands and unrelated controls. The first two major branches of the dendrogram divided the heat map into approximately equal numbers of study subjects (that is, 40 in the top half and 40 in the bottom half of the heat map). While all probands segregate into the top half of the map (that is, Figure 1, branch 1), the unaffected twins were equally distributed between the two major branches. The unrelated, matched controls were distributed between the two major branches (approximately 1/3 in branch 1 and 2/3 in branch 2). We also observed that several probands mapped outside the primary proband cluster (branch 1). Further analyses of proband outliers did not reveal any significant differences in age, medications, disease duration, physical exam findings, patient- and physician-assessed levels of disease activity, white blood or platelet counts, or hematocrit when compared with probands in the primary cluster (data not shown).
Figure 1.
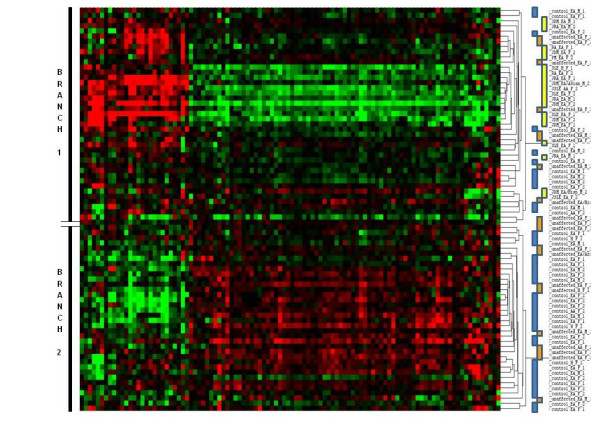
Heat map representing an unsupervised, hierarchical cluster analysis of the three study groups. The groups consisted of 20 probands with a systemic autoimmune disease, their 20 unaffected twins, and 40 unrelated, matched controls, respectively. The heat map uses 104 oligo probes that exhibited statistically significant differential gene expression (multiple comparison-adjusted P values (false discovery rate) of less than 0.1) between probands and unrelated, matched controls. Color codes to the immediate right of the dendrogram correspond to probands (yellow), unaffected twins (orange), and unrelated, matched controls (blue). The first major partition in the dendrogram (branches 1 and 2) is marked to the left of the heat map.
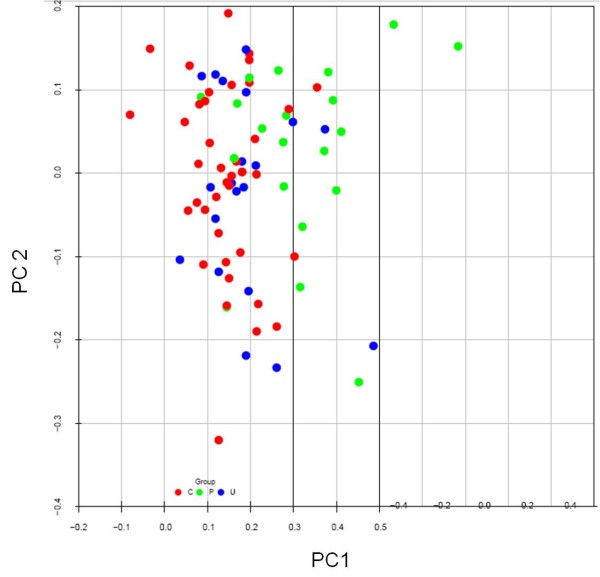
A PCA of the 80 study subjects by using the same 104 differentially expressed probes again revealed segregation of probands from unaffected twins and unrelated, matched controls (Figure 2). No differences due to age, gender, ethnicity, disease status, or treatment were observed across the first three principal components.
Figure 2.
Principal component analysis of the three study groups. The groups consisted of 20 probands with a systemic autoimmune disease (green), their 20 unaffected twins (blue), or 40 unrelated, matched controls (red), respectively. The analysis uses 104 oligo probes that exhibited statistically significant differential gene expression (multiple comparison-adjusted P values (false discovery rate) of less than 0.1) between probands and unrelated, matched controls. C, unrelated, matched control; P, proband; PC, principal component; U, unaffected twin.
While the distinction between unaffected twins and unrelated, matched controls was less obvious in this PCA, our inability to detect any significant differences in gene expression between unaffected twins and either probands or unrelated, matched controls suggested that the gene expression profile of unaffected twins may signify an intermediate or transitional state between the healthy and disease phenotypes. We observed further that the average gene expression for unaffected twins was intermediate between that of twins with SAID and unrelated, matched controls for 6,535 (32%) of a total of approximately 20,000 probes on the chip (about one third of all probes might be predicted to show this pattern by chance alone). By contrast, the same intermediate ordering was observed for 84 out of 104 probes (81%) that were significantly different between probands and controls.
Disease subtypes
By performing an unsupervised hierarchical clustering of all proband samples, we further examined whether probands might also segregate by disease subtype (that is, SLE, RA, and IIM). The clustering did not segregate probands into groups dominated by SAID subtypes (data not shown). Second, to identify probes that were significantly different among the disease groups, for each probe and each twin pair we calculated differences in log-transformed gene expression levels between unaffected and affected twins and compared these differences among disease groups with ANOVA. After adjustments for multiple testing, we did not detect any probes with significant P values among twin pairs whose probands had different diagnoses (data not shown).
To further assess these findings, we employed a mixed-effects model comparing probe expression values from probands and unrelated, matched controls, stratified by including a random effect for the matched sets, and comparing across disease phenotypes. Again, after adjustments for multiple testing, no significant differences in the change in gene expression (that is, the fixed-effects interaction between disease status and phenotype category) were observed. Together, these data suggest that either our disease-specific sample sizes were too small to detect significant differences or patients with different SAIDs possess similar gene expression patterns.
Pathway analysis
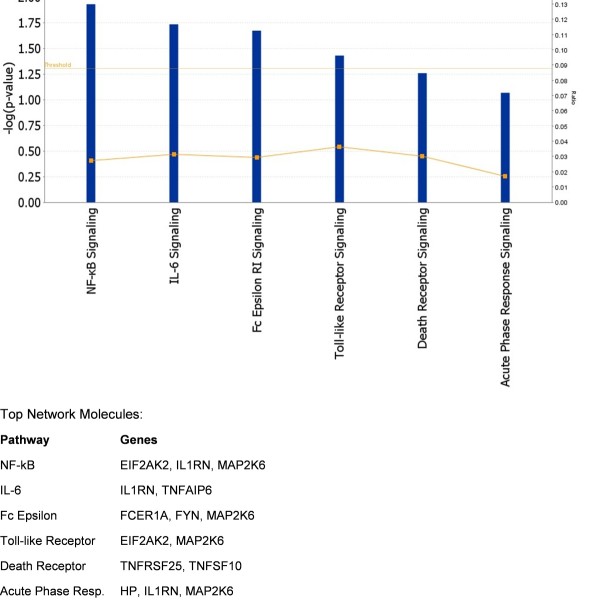
To examine the possible functional clustering of genes that were differentially expressed in SAID, we performed IPAs to assess whether differentially expressed genes might be linked by common biologic pathways. As shown in Figure 3, those findings having the greatest significance (that is, lowest P values) included several canonical biologic pathways involved in immune response and inflammation (signaling molecules for NF-κB, IL-6, FcεRI, Toll-like receptor signaling, apoptotic, and acute-phase responses). In some instances, genes expressed differentially in SAID were found to play a regulatory role in multiple immune response and inflammatory pathways. For example, mitogen-activated protein kinase kinase 6 (MAP2K6) plays a regulatory role in NF-κB, IL-6, FcεRI, Toll-like receptor, and acute-phase response pathways. Similarly, the interleukin 1 receptor antagonist (IL1RN) influences the NF-κB, IL-6, and acute-phase response pathways, and eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) is likewise involved in NF-κB and Toll-like receptor signaling. These results suggest that alterations in expression of a limited number of genes may, in turn, influence the dysregulation of numerous integrated immune response and inflammatory pathways that are common to several SAIDs.
Figure 3.
Ingenuity pathways analysis of 104 probes with false discovery rate-adjusted P values of less than 0.1. The analysis compares 20 twins affected with systemic autoimmune disease and 40 unrelated, matched controls. P values (blue bars) describing the confidence of the association of the data set with a given pathway are shown on the left vertical axis (-log(P value)). The cutoff threshold value (defined as P = 0.05) is also shown (horizontal yellow line). The ratio of the number of genes from the data set that map to a given pathway divided by the total number of molecules that comprise the pathway (orange line connecting blue bars) is shown on the right vertical axis. IL-6, interleukin-6; NF-κB, nuclear factor-kappa-B.
RQ-PCR validation
RQ of differential gene expression among disease-discordant twin pairs and unrelated, matched controls was independently evaluated by RT-PCR analyses for several genes of interest (KRTCAP2, MAP2K6, IL1RN, IFI27, SYTL2, FYN, LTK, EIF2AK2, FCERA1, LGR6, DEFA4, ANXA3, TNFAIP6, CEACAM6, and TNFSF10) as detailed in Materials and methods. As summarized in Table 3, 13 of the 15 genes evaluated by RQ-PCR showed the same trend (that is, increased or decreased levels of gene expression) as those observed from the microarray analysis in comparisons of twin probands and unrelated, matched controls. Moreover, the observed magnitude of differential expression for these genes was generally similar for these independent microarray and RQ-PCR assays. An opposite trend of differential expression was observed for the FYN gene by these assays, although the magnitudes of differential gene expression were small (approximately 1.2-fold). For the LTK gene, RQ-PCR did not detect a difference in gene expression between probands and unrelated, matched controls whereas microarray analysis estimated a 2.3-fold increase in gene expression. Similar comparative analyses of unaffected twins demonstrated intermediate levels of differential gene expression versus unrelated, matched controls for the majority of genes surveyed.
Table 3.
Comparison of differential gene expression values determined by relative quantitative-polymerase chain reaction and microarray analyses
| Gene | RQ-PCRa | Microarraya | ||
|---|---|---|---|---|
| Affected | Unaffected | Affected | Unaffected | |
| TNFAIP6 | 3.04 | 2.17 | 2.24 | 2.15 |
| TNFSF10 | 1.50 | 1.37 | 1.58 | 1.33 |
| MAP2K6 | 1.20 | 1.20 | 1.38 | 1.21 |
| IL1RN | 1.37 | 1.21 | 1.67 | 1.17 |
| IFI27 | 1.20 | 2.01 | 7.24 | 6.21 |
| ANXA3 | 3.18 | 1.82 | 2.79 | 2.43 |
| CEACAM6 | 5.70 | 1.15 | 4.70 | 1.10 |
| DEFA4 | 7.01 | 1.47 | 2.32 | 1.18 |
| EIF2AK2 | 2.20 | 1.12 | 2.36 | 1.70 |
| FCERA1 | -1.82 | -1.48 | -1.76 | -1.22 |
| SYTL2 | -1.93 | -1.61 | -1.49 | -1.10 |
| LGR6 | -1.25 | -1.13 | -1.64 | -1.11 |
| KRTCAP2 | -1.20 | -1.20 | -1.16 | -1.07 |
| LTK | 1.02 | 1.21 | 2.32 | 1.06 |
| FYN | 1.23 | 1.19 | -1.26 | -1.01 |
aFold change values indicating increased (positive value) or decreased (negative value) levels of gene expression in systemic autoimmune disease-affected or -unaffected twins compared with unrelated, matched controls. RQ-PCR, relative quantitative-polymerase chain reaction.
Discussion
Pathology in human autoimmune diseases is thought to result from a complex interplay of multiple, often polymorphic, genetic loci and accumulating environmental exposures. The importance of genetic predisposition is evidenced by high disease concordance rates and genetic linkage analyses among family members [23,24]. Further evidence for the role of genetics in SAID comes from the identification of candidate susceptibility loci, especially genes encoding polymorphic variants of HLA class II antigen-presenting molecules [25]. These findings are supported by single-nucleotide polymorphism-based genome-wide association studies (GWASs) identifying large numbers of putative susceptibility markers positioned along multiple chromosomes [26]. Many of the confirmed GWAS disease markers map in close proximity to or within genes controlling immune response signaling pathways (PTPN22, TNFAIP3, TRAF1, CD40, and REL), transcriptional activation (STAT4 and IRF5), and cytokine production (IL-12 and IL-23) [26,27]. Interestingly, the majority of these disease susceptibility loci individually confer only modest disease risks (relative risk values of 1.2 to 1.5) [28]. Moreover, many genetic risk factors are shared among multiple autoimmune diseases (for example, SLE, RA, type 1 diabetes, and multiple sclerosis) and across different ethnic backgrounds, suggesting that the dysregulation of fundamental immune regulatory pathways underlies shared pathogenetic mechanisms [2,4,29].
We used RNA microarrays to measure differential gene expression among MZ twins discordant for SAID and unrelated, healthy controls matched on age, gender, and ethnicity. Gene expression levels may be influenced by subjects' genetic background, age, gender, and environmental exposures as well as by many experimental variables relating to clinical sampling, processing, and data analysis. Despite these potential confounders, an array-based study of peripheral blood cells from MZ twins demonstrated significantly less variation in the proportion of differentially expressed genes among twins affected with an SAID (<2%) compared with unrelated controls (>14%) [1]. Interestingly, MZ twin-derived lymphoblastoid cell lines exhibit near identical patterns of monoallelic expression from otherwise heterozygous, polymorphic loci, suggesting, at least in part, a role for shared genetic controls of transcription or genetic imprinting or both [30]. Unexpectedly, we did not detect any significant differences in gene expression among disease-discordant MZ twins in our study after correction for multiple comparisons. Similar profiles of gene expression were described for array analyses examining eight pairs of MZ twins discordant for multiple sclerosis, in which investigators found only a single gene, IFN alpha-inducible protein GIP3, differentially expressed in over half of the eight twin pairs [5]. In contrast, larger numbers of differentially expressed genes were identified from lymphoblastoid cell lines derived from 11 MZ twin pairs discordant for RA, although this analysis was not adjusted for multiple comparisons [31]. Curiously, similar patterns of differential gene expression have been observed among SAID patients and their healthy, first-degree relatives [2,23]. Collectively, these data support a role for both genetic and other non-genetic variables, including perhaps environmental exposures influencing disease development. Also, small changes in levels of differential gene expression (generally 1.2- to 2.5-fold variations) may not be statistically observable in studies of smaller numbers of twin pairs, including our present study.
For 84 out of 104 probes (81%) with statistically significant differential expression between twins with an SAID and unrelated, matched controls, we observed that gene expression values of unaffected twins are positioned between affected twins and unrelated controls, suggesting that unaffected twins may represent an intermediate or transitional state of disease development. Together, our results and previous studies of MZ twins argue for the importance of shared genetic determinants in regulating gene expression profiles. However, additional factors, including epigenetic modifications that accumulate in an age-dependent fashion (that is, epigenetic drift) as well as accumulating environmental exposures, may also influence asymmetric patterns of disease development among MZ twins [32,33]. Differential patterns of gene expression between the affected probands and unrelated controls could reflect either acquired modifications that were present before the onset of disease and could (to some extent) have conferred susceptibility to developing an SAID or phenotypic features of the syndrome itself, having less to do with disease etiology.
Heterogeneous phenotypes characterize the spectrum of clinical entities comprising SAID (for example, SLE, RA, and IIM). However, as a group, these diseases share many clinical, laboratory, genetic, autoantibody, and possibly pathogenetic features [34]. In fact, many patients present clinically with overlap syndromes sharing diagnostic criteria for multiple autoimmune diseases. Previous microarray studies of peripheral blood cells isolated from patients with four different autoimmune diagnoses (SLE, RA, multiple sclerosis, and insulin-dependent diabetes mellitus) were found to have similar profiles of differential gene expression relative to unrelated, healthy controls [4,6]. Moreover, a meta-analysis of gene expression array profiles demonstrated high concordance between SLE and RA, further emphasizing disease similarities [7]. Although our power to detect differences was limited, our data indirectly support these conclusions in that we were unable to detect significant differences in gene expression among SAID subtypes (six with (J)SLE, six with (J)RA, and eight with JIIM) by using three independent statistical analyses (ANOVA, mixed-effects models, and hierarchical clustering). Together, these data suggest that shared alterations in gene expression may underlie similar pathogenic mechanisms among different SAIDs.
Our findings have corroborated previous studies identifying upregulation of type I IFN response genes observed predominantly in SLE but also in RA and IIM [7,10,12,14,16,19,35,36]. In fact, altered expression of IFN-inducible genes has been correlated with disease activity in SLE and IIM (DM and polymyositis) patients undergoing therapy [13,17,36]. We likewise observed upregulation of several SLE-associated IFN response genes, including OASL (2.2-fold), PLSCR1 (2.3-fold), EIF2AK2 (2.4-fold), and IFI27, the last of which exhibited the highest degree of increased gene expression (7.2-fold) in our study. Moreover, our findings are consistent with a report of IFN response-associated gene expression in DM whereby factors TNFAIP6, TNFSF10, OASF, PLSCR1, EIF2AK2, and IFI27 were elevated in peripheral blood [16].
Our study has several limitations, including small sample sizes and limited statistical power, resulting from the challenges of identifying and recruiting qualified MZ twins discordant for SAID. Studies of MZ twins, however, help to mitigate confounding factors associated with genetic polymorphisms in studies of unrelated human subjects. Variations in peripheral blood cell composition, disease activity, and effects of immunosuppressive therapies may also present difficulties; however, statistical evaluations of these variables did not reveal evidence of effects on gene expression patterns. Methodological and other technical variables were evaluated in part by the use of independent RQ-PCR assays, which corroborated our microarray data for the majority of genes studied. Other limitations included the phenotypic heterogeneity of the SAIDs studied and variations in environmental exposure histories. Despite these variables, many of the differentially expressed genes and candidate molecular pathways identified in our study are consistent with findings reported in other studies of SAID [7,12-14,16,36]. Functional analysis of genes found differentially expressed in our SAID probands identified multiple immunoregulatory (NF-κB, IL-6, and apoptosis) and proinflammatory (acute-phase response and Toll-like receptor) pathways. Moreover, several of these genes (for example, MAP2K6, EIF2AK2, and IL1RN) have coordinated functions in the integration of multiple, non-mutually exclusive regulatory pathways that together may contribute to common features of the SAID phenotype. The functional implications of each of these pathways in autoimmune disease were reviewed recently [37].
Conclusions
Gene expression profiling of biofluids and tissues from SAID patients and matched controls has provided a wealth of information into possible mechanisms of disease development, chronicity, and therapeutic response. Despite the variability of data anticipated with human clinical studies, some meaningful consensus has emerged [4,8]. As expected, we observed different gene expression profiles between disease probands and unrelated controls. Our identification of an SAID-associated profile of genes influencing multiple molecular pathways, including the immune response, cellular signaling, inflammation, transcriptional/translational, and other regulatory controls, is consistent with GWASs demonstrating large numbers of genetic susceptibility loci, each of which contributes only a modest degree of overall risk [26,28,29]. Moreover, the lack of statistically significant differences in gene expression detected between affected and unaffected twins emphasizes the importance of genetic susceptibility to disease whereby unaffected twins may represent a transitional state between health and disease. Our report of shared profiles of altered gene expression among SAID patients with different clinical phenotypes is consistent with the hypothesis that many SAIDs have common features of disease pathogenesis. The identification of relevant genes whose products regulate and integrate multiple physiologic pathways might permit the development of targeted therapeutics benefiting a broader spectrum of patients with multiple SAID phenotypes.
Abbreviations
ANOVA: analysis of variance; C: unrelated, matched control; DM: dermatomyositis; FDR: false discovery rate; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GEO: Gene Expression Omnibus; GWAS: genome-wide association study; IFN: interferon; IIM: idiopathic inflammatory myopathy; IL-6: interleukin-6; IPA: ingenuity pathways analysis; JDM: juvenile dermatomyositis; JPM: juvenile polymyositis; JRA: juvenile rheumatoid arthritis; MZ: monozygotic; NF-κB: nuclear factor-kappa-B; NIH: National Institutes of Health; P: proband; PCA: principal component analysis; RA: rheumatoid arthritis; RQ: relative quantitation; RQ-PCR: relative quantitation polymerase chain reaction; RT-PCR: real-time polymerase chain reaction; SAID: systemic autoimmune disease; SLE: systemic lupus erythematosus; U: unaffected twin.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TPO'H contributed to sample preparation, RQ-PCR studies, data and bioinformatic analyses, and manuscript preparation. LGR contributed to patient recruitment, clinical assessments and data analyses, and manuscript editing. LG contributed to sample preparation, RQ-PCR studies, data analysis, and manuscript editing. RF contributed to microarray analyses and data processing, bioinformatic analyses, and manuscript preparation and editing. RSP contributed to microarray analyses and data processing, bioinformatic analyses, and manuscript editing. DMU and CRW contributed to statistical analyses and manuscript editing. RRS contributed to data processing and analysis and to manuscript editing. DM contributed to data processing and analysis. MFG contributed to patient recruitment, clinical assessments, and manuscript editing. FWM contributed to study design, patient recruitment, clinical assessments, data analyses, and manuscript preparation and editing. All authors read and approved the final manuscript.
Contributor Information
Terrance P O'Hanlon, Email: ohanlont@niehs.nih.gov.
Lisa G Rider, Email: riderl@niehs.nih.gov.
Lu Gan, Email: ganl@niehs.nih.gov.
Rick Fannin, Email: fannin@niehs.nih.gov.
Richard S Paules, Email: paules@niehs.nih.gov.
David M Umbach, Email: umbach@niehs.nih.gov.
Clarice R Weinberg, Email: weinberg@niehs.nih.gov.
Ruchir R Shah, Email: shahr2@niehs.nih.gov.
Deepak Mav, Email: Deepak_mav@sra.com.
Mark F Gourley, Email: gourleym@niehs.nih.gov.
Frederick W Miller, Email: millerf@niehs.nih.gov.
Acknowledgements
The authors thank Robert Colbert and Nina Raben for their critical review of the manuscript. This work was supported by the intramural research program of the National Institute of Environmental Health Sciences. The funding sponsors had no role in the design, conduct, or interpretation of the study. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
References
- Sharma A, Sharma VK, Horn-Saban S, Lancet D, Ramachandran S, Brahmachari SK. Assessing natural variations in gene expression in humans by comparing with monozygotic twins using microarrays. Physiol Genomics. 2005;21:117–123. doi: 10.1152/physiolgenomics.00228.2003. [DOI] [PubMed] [Google Scholar]
- Maas K, Chen H, Shyr Y, Olsen NJ, Aune T. Shared gene expression profiles in individuals with autoimmune disease and unaffected first-degree relatives of individuals with autoimmune disease. Hum Mol Genet. 2005;14:1305–1314. doi: 10.1093/hmg/ddi141. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Li X, Sundquist K, Sundquist J. Shared familial aggregation of susceptibility to autoimmune diseases. Arthritis Rheum. 2009;60:2845–2847. doi: 10.1002/art.24749. [DOI] [PubMed] [Google Scholar]
- Olsen NJ, Moore JH, Aune TM. Gene expression signatures for autoimmune disease in peripheral blood mononuclear cells. Arthritis Res Ther. 2004;6:120–128. doi: 10.1186/ar1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkijarvi S, Kuusisto H, Paalavuo R, Levula M, Airla N, Lehtimaki T, Kaprio J, Koskenvuo M, Elovaara I. Gene expression profiles in Finnish twins with multiple sclerosis. BMC Med Genet. 2006;7:11. doi: 10.1186/1471-2350-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Maas K, Aune TM. Identification of gene expression signatures in autoimmune disease without the influence of familial resemblance. Hum Mol Genet. 2006;15:501–509. doi: 10.1093/hmg/ddi466. [DOI] [PubMed] [Google Scholar]
- Silva GL, Junta CM, Mello SS, Garcia PS, Rassi DM, Sakamoto-Hojo ET, Donadi EA, Passos GA. Profiling meta-analysis reveals primarily gene coexpression concordance between systemic lupus erythematosus and rheumatoid arthritis. Ann N Y Acad Sci. 2007;1110:33–46. doi: 10.1196/annals.1423.005. [DOI] [PubMed] [Google Scholar]
- van Baarsen LG, Bos CL, van der Pouw Kraan TC, Verweij CL. Transcription profiling of rheumatic diseases. Arthritis Res Ther. 2009;11:207. doi: 10.1186/ar2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junta CM, Sandrin-Garcia P, Fachin-Saltoratto AL, Mello SS, Oliveira RD, Rassi DM, Giuliatti S, Sakamoto-Hojo ET, Louzada-Junior P, Donadi EA, Passos GA. Differential gene expression of peripheral blood mononuclear cells from rheumatoid arthritis patients may discriminate immunogenetic, pathogenic and treatment features. Immunology. 2009;127:365–372. doi: 10.1111/j.1365-2567.2008.03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Onda H, Tanigawa A, Ohshima S, Fujiwara H, Mima T, Katada Y, Deguchi H, Suemura M, Miyake T, Miyatake K, Kawase I, Zhao H, Tomiyama Y, Saeki Y, Nojima H. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 2005;12:429–439. doi: 10.1093/dnares/dsi020. [DOI] [PubMed] [Google Scholar]
- Mandel M, Achiron A. Gene expression studies in systemic lupus erythematosus. Lupus. 2006;15:451–456. doi: 10.1191/0961203306lu2332oa. [DOI] [PubMed] [Google Scholar]
- Nzeusseu Toukap A, Galant C, Theate I, Maudoux AL, Lories RJ, Houssiau FA, Lauwerys BR. Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum. 2007;56:1579–1588. doi: 10.1002/art.22578. [DOI] [PubMed] [Google Scholar]
- Nikpour M, Dempsey AA, Urowitz MB, Gladman DD, Barnes DA. Association of a gene expression profile from whole blood with disease activity in systemic lupus erythaematosus. Ann Rheum Dis. 2008;67:1069–1075. doi: 10.1136/ard.2007.074765. [DOI] [PubMed] [Google Scholar]
- van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, Ibrahim SM, Fero M, Dijkmans BA, Tak PP, Verweij CL. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007;66:1008–1014. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pouw Kraan TC, van Baarsen LG, Wijbrandts CA, Voskuyl AE, Rustenburg F, Baggen JM, Dijkmans BA, Tak PP, Verweij CL. Expression of a pathogen-response program in peripheral blood cells defines a subgroup of rheumatoid arthritis patients. Genes Immun. 2008;9:16–22. doi: 10.1038/sj.gene.6364438. [DOI] [PubMed] [Google Scholar]
- Walsh RJ, Kong SW, Yao Y, Jallal B, Kiener PA, Pinkus JL, Beggs AH, Amato AA, Greenberg SA. Type I interferon-inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheum. 2007;56:3784–3792. doi: 10.1002/art.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, Bennett L, Allantaz F, Mejias A, Ardura M, Kaizer E, Monnet L, Allman W, Randall H, Johnson D, Lanier A, Punaro M, Wittkowski KM, White P, Fay J, Klintmalm G, Ramilo O, Palucka AK, Banchereau J, Pascual V. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Dalakas MC. Gene expression profile in the muscles of patients with inflammatory myopathies: effect of therapy with IVIg and biological validation of clinically relevant genes. Brain. 2005;128:1887–1896. doi: 10.1093/brain/awh518. [DOI] [PubMed] [Google Scholar]
- Tezak Z, Hoffman EP, Lutz JL, Fedczyna TO, Stephan D, Bremer EG, Krasnoselska-Riz I, Kumar A, Pachman LM. Gene expression profiling in DQA1*0501+ children with untreated dermatomyositis: a novel model of pathogenesis. J Immunol. 2002;168:4154–4163. doi: 10.4049/jimmunol.168.8.4154. [DOI] [PubMed] [Google Scholar]
- Huber R, Hummert C, Gausmann U, Pohlers D, Koczan D, Guthke R, Kinne RW. Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008;10:R98. doi: 10.1186/ar2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Prahalad S, Shear ES, Thompson SD, Giannini EH, Glass DN. Increased prevalence of familial autoimmunity in simplex and multiplex families with juvenile rheumatoid arthritis. Arthritis Rheum. 2002;46:1851–1856. doi: 10.1002/art.10370. [DOI] [PubMed] [Google Scholar]
- Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, Pons-Estel BA. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- O'Hanlon TP, Carrick DM, Targoff IN, Arnett FC, Reveille JD, Carrington M, Gao X, Oddis CV, Morel PA, Malley JD, Malley K, Shamim EA, Rider LG, Chanock SJ, Foster CB, Bunch T, Blackshear PJ, Plotz PH, Love LA, Miller FW. Immunogenetic risk and protective factors for the idiopathic inflammatory myopathies: distinct HLA-A, -B, -Cw, -DRB1, and -DQA1 allelic profiles distinguish European American patients with different myositis autoantibodies. Medicine (Baltimore) 2006;85:111–127. doi: 10.1097/01.md.0000217525.82287.eb. [DOI] [PubMed] [Google Scholar]
- Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116–121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Dermitzakis ET. Using gene expression to investigate the genetic basis of complex disorders. Hum Mol Genet. 2008;17:R129–134. doi: 10.1093/hmg/ddn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Hakonarson H. Microarray technology and applications in the arena of genome-wide association. Clin Chem. 2008;54:1116–1124. doi: 10.1373/clinchem.2008.105395. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Bruzel A, Burdick JT, Morley M, Devlin JL, Spielman RS. Monozygotic twins reveal germline contribution to allelic expression differences. Am J Hum Genet. 2008;82:1357–1360. doi: 10.1016/j.ajhg.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas CS, Creighton CJ, Pi X, Maine I, Koch AE, Haines GK, Ling S, Chinnaiyan AM, Holoshitz J. Identification of genes modulated in rheumatoid arthritis using complementary DNA microarray analysis of lymphoblastoid B cell lines from disease-discordant monozygotic twins. Arthritis Rheum. 2006;54:2047–2060. doi: 10.1002/art.21953. [DOI] [PubMed] [Google Scholar]
- Petronis A. Epigenetics and twins: three variations on the theme. Trends Genet. 2006;22:347–350. doi: 10.1016/j.tig.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Moreland LW, (Eds) Arthritis and Allied Conditions: A Textbook of Rheumatology. 15. Philadelphia, PA: Lippincott Williams & Wilkins; 2005. [Google Scholar]
- Zhou X, Dimachkie MM, Xiong M, Tan FK, Arnett FC. cDNA microarrays reveal distinct gene expression clusters in idiopathic inflammatory myopathies. Med Sci Monit. 2004;10:191–197. [PubMed] [Google Scholar]
- Greenberg SA. A gene expression approach to study perturbed pathways in myositis. Curr Opin Rheumatol. 2007;19:536–541. doi: 10.1097/BOR.0b013e3282efe261. [DOI] [PubMed] [Google Scholar]
- Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–571. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]