Abstract
The objective of this study was to determine if the within-herd prevalence of fecal Salmonella shedding is higher in dairy herds with clinical outbreaks of disease, as compared to herds with subclinical infections only. Data were collected prospectively from dairy herds throughout New York that had at least 150 lactating cows and that received clinical service from participating veterinarians. After enrollment, Salmonella surveillance consisted of both environmental screening and disease monitoring within the herd. Herds positive by either environmental or fecal culture were sampled during three visits to estimate the within-herd prevalence of Salmonella. We characterized isolates by serovar and antimicrobial resistance pattern. Among 57 enrolled herds, 44 (77%) yielded Salmonella-positive samples during the study period; 27 (61%) of the positive herds had Salmonella isolated from environmental samples only, and 17 (39%) had one or more laboratory-confirmed clinical cases. The within-herd prevalence of fecal Salmonella shedding ranged from 0 to 53%. Salmonella Cerro was the predominant serovar, accounting for 56% of all isolates. Antimicrobial resistance ranged from zero to nine drugs, and 14 (32%) of the positive farms generated multidrug-resistant isolates. Herds with laboratory-confirmed clinical cases had a higher prevalence of fecal Salmonella shedding than herds that only generated positive environmental samples, as estimated by a Poisson regression model (prevalence ratio, 2.7; p = 0.01). An association between dairy herd outbreaks of salmonellosis and a higher prevalence of asymptomatic shedding should help guide strategies for reducing the public health threat of Salmonella, as the ability to recognize high-risk herds by clinical laboratory submissions presents an obvious opportunity to maximize food safety at the preharvest level. This is in contrast with other foodborne zoonotic pathogens, such as Campylobacter jejuni and Escherichia coli O157:H7, which occur widely in adult cattle without accompanying clinical disease.
Introduction
Salmonella enterica is an important zoonotic pathogen that causes an estimated 1.4 million illnesses, 16,000 hospitalizations, and between 400 and 600 deaths annually in the United States alone (Mead et al., 1999; Voetsch et al., 2004). Although primarily a cause of self-limiting acute enteritis (diarrhea, abdominal pain, and fever, with a typical duration of 4 to 7 days), Salmonella can produce invasive infections that lead to sepsis and death. Young children, the elderly, and those with compromised immune systems are especially susceptible to severe disease. The prevalence of multidrug resistance among Salmonella strains has increased over the past two decades (Glynn et al., 1998; Dunne et al., 2000; Gupta et al., 2003; Davis et al., 2007), making treatment failures more common among those with serious disease. In addition, infections with resistant strains of Salmonella tend to be more severe and lead to higher rates of hospitalization than those caused by susceptible strains (Helms et al., 2002; Helms et al., 2004; Varma et al., 2005a, 2005b).
People generally acquire salmonellosis through foodborne exposure, although direct contact with infected animals is another possible route (Mead et al., 1999; L Plym and Wierup, 2006). Preliminary CDC FoodNet data from 2008 show that Salmonella accounted for 40% of all laboratory-confirmed cases of foodborne infections, based on surveillance in 10 states (CDC, 2009b). Dairy cattle are considered an important source of several Salmonella serovars that are a threat to human health, including multidrug-resistant (MDR) Newport and Typhimurium (Gupta et al., 2003; Dechet et al., 2006; Varma et al., 2006; Karon et al., 2007). Fecal contamination of beef carcasses at the time of slaughter is thought to represent the predominant source of transmission. According to the 1996 U.S. Department of Agriculture National Animal Health Monitoring System (NAHMS) Dairy report, 14.9% of culled dairy cows were shedding Salmonella at livestock markets, and 66.0% of markets had at least one cow shedding Salmonella (Wells et al., 2001). Contamination of crops, either by manure used as fertilizer or by irrigation water that has been contaminated by manure run-off, is another key source of transmission (Islam et al., 2004; Sivapalasingam et al., 2004; CDC, 2008a). Those who work or otherwise interact with livestock are also at risk of infection via direct exposure when cattle are shedding Salmonella.
Introduction of Salmonella onto a dairy farm can occur through a variety of routes, including purchased cattle, contaminated feed or water, wild animals such as rodents and birds, human traffic, and insects (Bender, 1994; Evans and Davies, 1996; Sanchez et al., 2002; Nielsen et al., 2007). Clinical signs of bovine salmonellosis may include diarrhea, fever, anorexia, dehydration, decreased milk production, abortion, and evidence of endotoxemia, although many infections remain asymptomatic (Divers and Peek, 2008). Infected cattle can shed the organism for variable periods and intermittently after either clinically apparent or subclinical infections. Widespread environmental contamination can result from Salmonella shedding, and the organism can survive for prolonged periods in suitable conditions outside a host (Wray and Wray, 2000; You et al., 2006). Fecal Salmonella shedding can also augment the risk of within-herd transmission and inadvertent spread to other herds. In addition to impacting the health and productivity of dairy cattle, these factors lead to an increased risk of zoonotic transmission. Although the prevalence of fecal Salmonella shedding among asymptomatic dairy cattle has been estimated in a number of studies (Wells et al., 2001; Huston et al., 2002; Fossler et al., 2004; Blau et al., 2005), the relationship between clinical outbreaks of salmonellosis and fecal shedding is not well understood.
Our hypothesis was that the within-herd prevalence of fecal Salmonella shedding is higher in herds with clinical outbreaks of disease, as compared to herds with subclinical infections only. Thus, the objective of this study was to determine the effect of clinical disease (salmonellosis) on the prevalence of asymptomatic fecal Salmonella shedding within dairy herds. The identification of such a link would provide a clear point of intervention to mitigate public health risk; herds posing the greatest danger to human health could therefore be recognized by clinical laboratory submissions, potentially reducing the need for surveillance among herds without clinical disease. In addition, we described the serovars and antimicrobial resistance patterns of the isolates to enhance our understanding of the epidemiology of Salmonella on dairy farms.
Materials and Methods
Study design
Data for this study were collected prospectively from a convenience sample of dairy herds throughout New York that had at least 150 lactating cows and that received clinical service from participating veterinarians. After enrollment, Salmonella surveillance consisted of both environmental screening and disease monitoring within the herd for at least 12 months. Environmental surveillance involved the repeated collection of samples from four locations per herd for Salmonella culture (cow housing, calf housing, manure storage area, and sick pen); the targeted interval between sample collections was monthly. In addition, veterinarians submitted fecal samples from suspected clinical cases for Salmonella culture. The diagnostic criteria provided to the veterinarians included diarrhea with blood, mucus, or a foul odor; fever of at least 103°F; depression; and decreased appetite, as well as sudden death in the absence of specific clinical signs or death after a course of diarrhea. To encourage the submission of samples from every clinical suspect animal, all shipping and laboratory costs were covered by the study. A positive culture result arising by either surveillance method prompted a series of three herd visits (at 4- to 8-week intervals) for cattle sampling by project personnel, with the goal of estimating the within-herd prevalence of Salmonella. The number of animals sampled at each visit ranged from 50 to 70, depending on herd size (<500 lactating cows: 50 animals sampled; 500–1000 lactating cows: 60; and >1000 lactating cows: 70). A subset of each sample was comprised of preweaned calves; that is, a total of 10, 15, and 20 calves made up the samples of 50, 60, and 70 animals, respectively. A conscious effort was made to sample cattle from each pen on the farm, and animals within a given pen were sampled systematically to the extent possible. No attempt was made to collect samples from the same cattle during the subsequent herd visits, though some animals may have been sampled again by chance.
Sample collection and processing
Environmental samples were collected using sterile 4 × 4-inch gauze pads saturated in double-strength skim milk, which had been placed beforehand into a sterile flip-top container. For each of the four sampling locations per farm, four different gauze pads were used to collect samples and were subsequently pooled into a single flip-top container. Locations sampled in the cow housing area included four sites on the floor within high-traffic sections of the barn. Calf housing samples consisted of either four swabs of the floor in group housing areas or four swabs of the bedding in individual hutches or pens. Manure storage areas were sampled by sticking an instrument deep into the lagoon or slurry pit and then swabbing it. Sick pen samples consisted of either four swabs of the floor in group pens or four swabs of the bedding in individual sick pens. All environmental samples were maintained at 4°C until processing; samples were shipped to the research laboratory for bacteriologic culture.
Fecal samples from suspected clinical cases were collected by veterinarians via rectal retrieval, with a new glove being used to collect each sample. Approximately 10 g of fecal matter was placed into a Para-Pak bottle (Meridian Bioscience, Cincinnati, OH) and sealed. These samples were shipped to the Animal Health Diagnostic Center (College of Veterinary Medicine, Cornell University, Ithaca, NY) for bacteriologic culture.
Fecal samples obtained by project personnel during the three monthly visits were collected via rectal retrieval, again with a new glove being used to collect each sample. Approximately 10 g of fecal matter was placed into a Para-Pak bottle and sealed. All of these samples were transported to the research laboratory for bacteriologic culture.
Standard culture methods were used to isolate Salmonella from feces. Individual fecal swabs from sample bottles were enriched in 10 mL of Tetrathionate broth (Difco, Detroit, MI) containing 0.2 mL of iodine solution; the mixture was incubated at 42°C for 18–24 hours. After incubation, the sample–broth mixture was streaked onto Brilliant Green agar with novobiocin (Becton Dickinson and Company, Franklin Lakes, NJ) and Xylose Lysine Tergitol 4 (XLT-4) selective media, and both plates were incubated at 37°C for 18–24 hours. Red colonies (lactose nonfermenting bacteria) on Brilliant Green agar with novobiocin and black colonies (H2S-producing bacteria) on XLT-4 were inoculated into Kligler Iron Agar slants and then incubated at 37°C for 18–24 hours. XLT-4 plates without suspected colonies were reincubated at 37°C for an additional 18–24 hours before checking again for characteristic black colonies. Colonies on Kligler Iron Agar slants that exhibited the biochemical properties of Salmonella were then serogrouped by slide agglutination using standard protocols. Those colonies that were positive by slide agglutination were then identified as Salmonella using the Sensititre Automated Microbiology System's A80 panel (TREK Diagnostic Systems, Cleveland, OH). Confirmed Salmonella isolates were sent to the National Veterinary Services Laboratories (Animal and Plant Health Inspection Service, U.S. Department of Agriculture) in Ames, Iowa, for serotyping using standard protocols.
Antimicrobial susceptibility testing
Antimicrobial susceptibility of Salmonella isolates was determined by use of the broth dilution method. Minimal inhibitory concentrations (MIC) were established for each isolate against a panel of up to 15 antimicrobial agents (Sensititre; TREK Diagnostic Systems). The panel used for Salmonella organisms isolated via environmental and follow-up sampling included 15 drugs (amikacin, amoxicillin/clavulanic acid, ampicillin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, tetracycline, and trimethoprim/sulfamethoxazole); the panel used for clinical isolates included 11 drugs (ampicillin, ceftiofur, chlortetracycline, enrofloxacin, florfenicol, gentamicin, neomycin, oxytetracycline, spectinomycin, sulfadimethoxine, and trimethoprim/sulfamethoxazole). Clinical and Laboratory Standards Institute (CLSI) guidelines were used to interpret MIC values when available (CLSI, 2008). Otherwise, MIC values were interpreted using National Antimicrobial Resistance Monitoring System breakpoints (CDC, 2009a). Isolates were classified as being resistant or susceptible to each agent; those isolates with intermediate susceptibility were categorized as being susceptible. Quality control was performed weekly using four strains of bacteria: Escherichia coli ATCC 25922, Staphylococcus aureus 29213, Enterococcus faecalis 29212, and Pseudomonas aeruginosa 27853. The MIC ranges for quality control recommended by the CLSI were used, and results were accepted if the MIC values were within expected ranges for these bacterial strains.
Data analysis
Study herds were considered Salmonella-positive either if Salmonella was isolated from one or more environmental samples or if there was at least one laboratory-confirmed clinical case. The median within-herd prevalence of Salmonella shedding was estimated for herds that only yielded Salmonella-positive environmental samples and for herds that had one or more clinical cases confirmed by bacteriologic culture. Descriptive analysis of serovar data and level of antimicrobial resistance was performed, stratified by positive herd type. The proportion of MDR isolates by serovar was also determined. In this study, multidrug resistance was defined as having in vitro resistance to five or more antimicrobial agents.
Repeated measures Poisson regression analysis was performed to study the relationship between the within-herd prevalence of Salmonella shedding and the dichotomous predictor variable for positive herd status (positive environmental samples only vs. laboratory-confirmed clinical cases). The response variable was the number of cattle positive for Salmonella upon follow-up sampling, and the offset variable was the logarithm of the number of animals tested via this method. A random effects regression model was used to account for the clustering of the three sequential prevalence estimates per herd. Time of sampling for the three within-herd prevalence measurements was added to the model as a fixed effect, thus allowing a comparison of the change in prevalence over time for the two positive herd types. Herd size was forced into the final model because it was considered an important potential confounding variable. This Poisson regression model was of the form
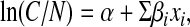 |
where C was the number of cattle positive for Salmonella upon follow-up sampling, N was the number of cattle tested via follow-up, α was the intercept term, xi were the model covariates, and βi were the regression coefficients. The generalized estimating equations method was used for this regression model.
Separate logistic regression models were utilized to determine whether herds with confirmed clinical cases were more likely to yield either serovars that are important human pathogens (Newport and Typhimurium) or MDR isolates, as compared to herds with positive environmental samples only. Logistic regression analysis was also used to investigate any associations between herd size and Salmonella status. All data analysis was performed in SAS (version 9.1; SAS Institute, Cary, NC), and p-values <0.05 were considered significant.
Results
Thirty-four veterinarians representing 11 veterinary practices participated in this study. A total of 62 dairy farms were enrolled, but 5 farms withdrew their involvement. Among the remaining 57 study herds, the median herd size was 875 female dairy cattle (range: 245–7412). Forty-four herds (77.2%) yielded positive samples over the course of the study period. Salmonella was isolated from 22.0% (409/1857) of environmental surveillance samples, 42.1% (120/285) of suspected clinical case samples, and 9.1% (674/7400) of follow-up samples for estimating within-herd prevalence. Of the positive clinical cases, 101 (84.2%) were adult cows and 19 (15.8%) were calves. The prevalence of fecal Salmonella shedding among clinical suspects was 49.3% (101/205) for adults and 23.8% (19/80) for calves. Of the positive follow-up samples, 588 (87.2%) were from adult cows and 86 (12.8%) were from calves. The prevalence of fecal Salmonella shedding among nonsuspect animals tested through follow-up was 10.1% (588/5797) for adults and 5.4% (86/1603) for calves.
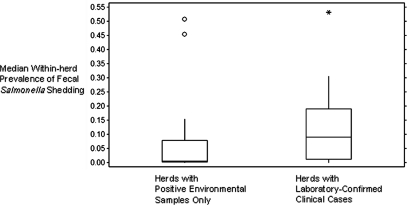
Twenty-seven (61.4%) of the positive herds had Salmonella isolated from environmental samples only, and 17 (38.6%) had one or more laboratory-confirmed clinical cases (median: 3 confirmed clinical cases; range: 1–48). The within-herd prevalence of fecal Salmonella shedding ranged from 0 to 53%. The median prevalence in herds that only generated Salmonella-positive environmental samples (0.5%) was significantly lower (Wilcoxon rank sum p-value = 0.01) than the median prevalence in herds that had at least one laboratory-confirmed clinical case (8.9%, Fig. 1).
FIG. 1.
Box-and-whiskers plot illustrating the median within-herd prevalence of fecal Salmonella shedding among herds in both categories of Salmonella-positive status. (Statistical outliers are noted.)
Serotyping was performed on 96.7% (1163/1203) of the Salmonella isolates, yielding 31 serovars during the study period (Table 1). The predominant serovar among herds with only positive environmental samples was Cerro, which accounted for 57.5% (253/440) of the isolates, followed by Kentucky (14.8%, 65/440), Anatum (including the 15+ variant; 11.1%, 49/440), and Meleagridis (2.7%, 12/440). Cerro was the most common environmental serovar (44.5%, 87/182) among these herds, as well as the one most frequently isolated during follow-up sampling (64.3%, 166/258). The most common serovar among herds that had at least one laboratory-confirmed clinical case was also Cerro, accounting for 55.6% (402/723) of the isolates, followed by Kentucky (14.0%, 101/723), Typhimurium (including the Copenhagen variant; 9.4%, 68/723), and Newport (5.9%, 43/723). Among these herds, Cerro was the leading serovar in all sampling categories (environment: 40.8%, 84/206; clinical cases: 59.2%, 71/120; follow-up sampling: 62.2%, 247/397). Of the predominant serovars in this study, Cerro and Kentucky were the most widespread among farms based on follow-up cattle sampling; Salmonella Cerro was isolated from 10 herds, of which 5 had clinical disease, whereas Salmonella Kentucky was isolated from 9 herds, of which 4 had clinical disease. Salmonella Newport (6 herds [4 clinical]) and Salmonella Typhimurium (5 herds [4 clinical]) were isolated from fewer farms but were more closely associated with clinical disease.
Table 1.
Salmonella Serovars Isolated from Dairy Cattle and the Environment of Farms in New York
| |
|
|
|
Total |
|
|---|---|---|---|---|---|
| Serovar | Number of environmental isolates | Number of clinical case isolates | Number of isolates from follow-up sampling | No. | % |
| 3,10:-:1,5 | 3 | — | 1 | 4 | 0.3 |
| 3,10:-:1,w | 1 | — | — | 1 | 0.1 |
| 3,10:e,h:- | 1 | 1 | — | 2 | 0.2 |
| 4,12:i:- | — | 2 | — | 2 | 0.2 |
| 4,5,12:i:- | — | — | 1 | 1 | 0.1 |
| 6,7:-:1,5 | 1 | — | — | 1 | 0.1 |
| 8,20:-:z6 | 1 | — | — | 1 | 0.1 |
| Agona | 4 | 1 | — | 5 | 0.4 |
| Anatum | 19 | 3 | 21 | 43 | 3.7 |
| Anatum var. 15+ | 4 | — | 13 | 17 | 1.5 |
| Cerro | 171 | 71 | 413 | 655 | 56.3 |
| Heidelberg | 1 | — | — | 1 | 0.1 |
| Infantis | 7 | 3 | — | 10 | 0.9 |
| Kentucky | 59 | 9 | 98 | 166 | 14.3 |
| Lexington | — | — | 1 | 1 | 0.1 |
| Mbandaka | 7 | — | — | 7 | 0.6 |
| Meleagridis | 26 | 8 | 20 | 54 | 4.6 |
| Minnesota | 3 | — | — | 3 | 0.3 |
| Montevideo | 7 | — | 1 | 8 | 0.7 |
| Muenster | 3 | 2 | 7 | 12 | 1.0 |
| Newport | 18 | 9 | 38 | 65 | 5.6 |
| Oranienburg | 5 | 1 | — | 6 | 0.5 |
| Orion var. 15+,34+ | 2 | — | — | 2 | 0.2 |
| Paratyphi B var. L-tartrate+ | 1 | — | — | 1 | 0.1 |
| Rubislaw | — | — | 1 | 1 | 0.1 |
| Senftenberg | — | — | 1 | 1 | 0.1 |
| Tennessee | 1 | — | — | 1 | 0.1 |
| Thompson | — | 1 | 1 | 2 | 0.2 |
| Typhimurium | 25 | 6 | 26 | 57 | 4.9 |
| Typhimurium var. Copenhagen | 9 | — | 5 | 14 | 1.2 |
| Rough isolates | — | 2 | — | 2 | 0.2 |
| Untypeable | 9 | 1 | 7 | 17 | 1.5 |
Forty isolates excluded from this table due to lack of serovar data.
Antimicrobial susceptibility testing was performed on 98.3% (1183/1203) of the Salmonella isolates. Antimicrobial resistance among isolates ranged from zero (pan-susceptible) to nine drugs. Herds with only positive environmental samples generated 26/449 (5.8%) MDR isolates and 404/449 (90.0%) that were pan-susceptible. Multidrug resistance was seen in 4.8% (9/186) of the environmental samples and 6.5% (17/263) of the follow-up samples from these herds. Among herds that had at least one laboratory-confirmed clinical case, 47/734 (6.4%) isolates were MDR and 580/734 (79.0%) were pan-susceptible. The MDR phenotype was observed in 8.1% (17/211) of the environmental samples, 6.7% (8/120) of the clinical case samples, and 5.5% (22/403) of the follow-up samples from these herds. Fourteen (31.8%) of the Salmonella-positive herds in our study yielded MDR isolates from cattle, the environment, or both.
There was considerable variation in antimicrobial resistance across the serovars most commonly isolated in this study. For instance, 86.2% (56/65) of the Newport isolates and 8.5% (6/71) of the Typhimurium isolates were MDR, whereas the proportion of isolates that were MDR among the Cerro, Kentucky, Anatum, and Meleagridis serovars ranged from 0% to 1.7%.
Using a multivariable Poisson regression model to account for the effects of other factors, we found that positive herd status (positive environmental samples only vs. laboratory-confirmed clinical cases) was a significant predictor of Salmonella prevalence within the herd (Table 2). Herds with confirmed clinical cases had a higher prevalence than herds with just positive environmental samples (prevalence ratio, 2.7; p = 0.01). Herd size was forced into the model but was not associated with within-herd Salmonella prevalence. There was not a significant interaction between positive herd status and time of sampling (p = 0.1). Among herds that had at least one laboratory-confirmed clinical case, however, there was a significant decrease in the median within-herd prevalence between the first sampling visit (12.9%) and the last (2.9%; Wilcoxon rank sum p-value = 0.04). The median prevalence for the first and last sampling visits did not differ significantly among herds that only generated Salmonella-positive environmental samples.
Table 2.
Association Between Within-Herd Salmonella Prevalence and Positive Herd Status Among Dairy Cattle in New York, When Forcing Herd Size into a Poisson Regression Model
| Variable | Prevalence ratio | 95% Confidence interval | p-Value |
|---|---|---|---|
| Positive herd status | |||
| Laboratory-confirmed clinical cases | 2.7 | (1.3, 5.9) | 0.01 |
| Positive environmental samples only | 1.0 | — | — |
| Herd size (female dairy cattle) | |||
| ≥1500 | 0.6 | (0.2, 1.5) | 0.3 |
| 1000–1499 | 0.5 | (0.2, 1.7) | 0.3 |
| 500–999 | 0.7 | (0.2, 2.4) | 0.6 |
| <500 | 1.0 | — | — |
Logistic regression analysis showed that herds with confirmed clinical cases were more likely (odds ratio [OR], 5.6; p = 0.03) to yield either Newport or Typhimurium on follow-up sampling than were herds with positive environmental samples only (Table 3). Clinical herds also tended to be more likely (OR, 4.4; p = 0.06) to yield MDR isolates on follow-up sampling than herds with just positive environmental samples.
Table 3.
Association Between Herd-Level Isolation of Salmonella Newport/Typhimurium and Positive Herd Status Among Dairy Cattle in New York, as Estimated by a Logistic Regression Model
| Variable | Odds ratio | 95% Confidence interval | p-Value |
|---|---|---|---|
| Positive herd status | |||
| Laboratory-confirmed clinical cases | 5.6 | (1.2, 26.1) | 0.03 |
| Positive environmental samples only | 1.0 | — | — |
Deviance = 41.9 (df = 42).
Herd size was not a significant predictor of whether or not Salmonella was isolated from a study herd. Among the positive herds, however, herd size was significantly associated with the presence of laboratory-confirmed clinical cases (Table 4). Larger herds with at least 1500 female dairy cattle were more likely to have clinical cases than smaller herds with fewer than 500 female dairy cattle (OR, 7.2; p = 0.04). The odds of having clinical cases among the three smaller herd size categories (1000–1499, 500–999, and <500 female dairy cattle) did not differ significantly.
Table 4.
Association Between the Presence of Laboratory-Confirmed Clinical Cases of Salmonellosis and Herd Size Among Salmonella-Positive Dairy Herds in New York, as Estimated by a Logistic Regression Model
| Variable | Odds ratio | 95% Confidence interval | p-Value |
|---|---|---|---|
| Herd size (female dairy cattle) | |||
| ≥1500 | 7.2 | (1.1, 48.0) | 0.04 |
| 1000–1499 | 3.0 | (0.3, 25.9) | 0.3 |
| 500–999 | 1.2 | (0.2, 9.0) | 0.9 |
| <500 | 1.0 | — | — |
Likelihood ratio chi-square = 6.8 (df = 3).
Discussion
A number of studies have described the prevalence of fecal Salmonella shedding among apparently healthy dairy cattle (Wells et al., 2001; Huston et al., 2002; Fossler et al., 2004; Blau et al., 2005), but there is little in the literature regarding the incidence of clinical disease associated with Salmonella infections in cattle (Cummings et al., 2009). To our knowledge, no studies have investigated the relationship between clinical outbreaks of bovine salmonellosis and the within-herd prevalence of fecal Salmonella shedding. This study had the advantage of utilizing both environmental surveillance and disease monitoring to assess the occurrence of Salmonella within each enrolled dairy herd. These herds were located throughout New York and were characterized by a wide range of sizes and management types representative of the dairy industry in this area of the country. Another strength of this study was the longitudinal sampling approach for estimating the within-herd prevalence of Salmonella when herds were identified as being positive. A random effects regression model accounted for the clustering of the three sequential prevalence estimates per herd, and the addition of sampling time to the model as a fixed effect allowed us to characterize the prevalence over time for the two positive herd types.
Environmental surveillance has been shown to be a relatively effective means of monitoring for the presence of Salmonella on dairy farms (Warnick et al., 2003). However, it is possible that the number of clinical cases in the present study was underestimated if clinically affected cattle went undetected by herd managers or veterinarians. Further, fecal culture does not have perfect sensitivity for detecting the presence of Salmonella, and we recognize that some positive cattle were presumably missed by culturing. Thus, the possibility exists that we could have missed Salmonella-positive herds altogether or misclassified herds with clinical cases as being positive by virtue of environmental samples only. Such nondifferential misclassification would bias our results toward the null.
It is also conceivable that participating veterinarians had a tendency to enroll client herds that had a history of diarrhea over herds without clinical disease. However, only 17 of the 57 enrolled herds had one or more laboratory-confirmed clinical cases over the duration of the study. Moreover, it was not our intention for study herds to be representative of herds throughout New York in terms of the herd-level prevalence of Salmonella infection. Our goal was to have a sufficient number of study herds positive by either method (positive environmental samples only vs. laboratory-confirmed clinical cases) to allow a comparison between the two.
Of the positive study herds, 39% had at least one confirmed case of salmonellosis, whereas the remaining 61% had Salmonella isolated from the environment only. The median within-herd prevalence of asymptomatic fecal Salmonella shedding was significantly higher in herds that had at least one confirmed clinical case, as compared to herds that only generated positive environmental samples. Positive herd status was also a significant predictor of within-herd Salmonella prevalence in a Poisson regression model controlling for herd as a random effect, after adjusting for the effect of herd size. Those herds with a high prevalence of asymptomatic fecal Salmonella shedding presumably represent a greater threat to public health than herds with few shedders. An association between dairy herd outbreaks of salmonellosis and higher within-herd Salmonella prevalence should help guide strategies for reducing this threat, as the ability to recognize high-risk herds by clinical laboratory submissions presents an obvious opportunity to maximize food safety at the preharvest level. This is in contrast with other foodborne zoonotic pathogens, such as Campylobacter jejuni and E. coli O157:H7, which occur widely in adult cattle without accompanying clinical disease (Dunn et al., 2004; Cho et al., 2006; Kwan et al., 2008; Ellis-Iversen et al., 2009; Huang et al., 2009). The occurrence of disease outbreaks among herds with a higher within-herd prevalence of Salmonella shedding also provides a tangible incentive for dairy producers to enhance their biosecurity and hygiene efforts on the farm. Salmonellosis can be a costly disease for producers on account of mortality, treatment expenses, reduced milk yield, and weight loss within the herd (Peters, 1985; Huston et al., 2002); the economic benefits of Salmonella control, coupled with the promotion of public health, should encourage the implementation of strategies for preventing the introduction and spread of this pathogen. Finally, the association between clinical disease and high-risk herds implies that steps to reduce the public health threat of Salmonella on dairy farms can yield improvements that are directly observed and quantified.
Among herds with at least one laboratory-confirmed clinical case, there was a significant decrease in the within-herd prevalence of fecal Salmonella shedding between the first and last sampling visits; such was not the case among herds with Salmonella-positive environmental samples only. The concentration of Salmonella within the manure of an infected cow ranges from 102 to 107 organisms per gram of feces (You et al., 2006). It seems likely that the presence of cattle with diarrhea due to salmonellosis will lead to an initial peak in the within-herd prevalence of fecal Salmonella shedding; the prevalence would be expected to decline as clinical signs of salmonellosis abate in the herd. In contrast, herds with Salmonella contamination of the environment but no clinical cases would presumably have consistent, repeated exposure over time. The within-herd prevalence of fecal Salmonella shedding is likely to persist at some level without peaking at any point.
Salmonella Cerro was the major serovar in this study, accounting for 56% (655/1163) of the isolates and leading all herd sampling categories. Cerro was the only serovar isolated from the four herds in this study with the highest within-herd prevalence of fecal Salmonella shedding (31%, 45%, 51%, and 53%). Two of these herds had at least one laboratory-confirmed case of salmonellosis, whereas the other two had Salmonella isolated from environmental samples only. The role of Salmonella Cerro in causing clinical disease in cattle is unclear (Cummings et al., in press). This serovar has been a rare isolate among people in the United States with laboratory-confirmed Salmonella infections, accounting for 0.1% of the cases in 2006 (CDC, 2008b).
Whereas Salmonella Cerro and Salmonella Kentucky were predominant among both positive herd types, Salmonella Newport and Salmonella Typhimurium (including the Copenhagen variant) were common only among those herds that had at least one laboratory-confirmed clinical case. Newport and Typhimurium were previously shown to be the two leading serovars in a large study on the incidence of clinical disease due to Salmonella infection among dairy herds in the northeastern United States (Cummings et al., 2009). CDC FoodNet data from 2008 showed that these serovars were also two of the three most common Salmonella serovars isolated from people with laboratory-confirmed foodborne infection, accounting for 26% of the human cases (CDC, 2009b). In contrast, the serovars most commonly isolated in studies of fecal Salmonella shedding among clinically healthy cattle differ from those that most frequently cause human disease. According to the 1996 NAHMS Dairy report, Salmonella Montevideo (21%) was the most prevalent serovar isolated from healthy lactating cows, and neither Newport nor Typhimurium was among the 10 most common serovars isolated (Wells et al., 2001). The 2002 NAHMS Dairy study found Salmonella Meleagridis (24%) to be the most prevalent serovar, whereas Newport and Typhimurium accounted for only 3% and 10% of all isolates, respectively (Blau et al., 2005). Similarly, Salmonella Meleagridis was the most common serovar isolated from the environment of dairy herds without clinical signs of salmonellosis; neither Newport nor Typhimurium was identified on these farms (Peek et al., 2004). In the present study, herds experiencing clinical disease were more likely to yield either Newport or Typhimurium on follow-up sampling than were herds with Salmonella-positive environmental samples only. Thus, herds with clinical outbreaks of salmonellosis may present a greater threat to public health, as there is a higher probability that the cattle are shedding serovars that are also important human pathogens.
With the exception of Salmonella Newport, there was a low frequency of antimicrobial resistance among isolates from any of the sampling categories in this study. Over 86% of the Newport isolates across all sample types displayed multidrug resistance, but virtually none of the isolates representing other common serovars showed an MDR phenotype. Studies of fecal Salmonella shedding among clinically healthy cattle across the United States have shown antimicrobial resistance to be uncommon (Wells et al., 2001; Blau et al., 2005; Ray et al., 2007). However, multidrug resistance was found to be highly prevalent among isolates from cattle with clinical signs of salmonellosis in the northeastern United States (Cummings et al., 2009), particularly among those serovars that were common in the present study such as Newport and Typhimurium. In this study, multidrug resistance was infrequent among both the broad population of cattle (follow-up sampling) as well as the cattle with salmonellosis, likely due to the distribution of serovars among the clinical isolates. Although 77.8% (7/9) of the clinical Newport isolates were MDR, Salmonella Newport was the isolated serovar in only 7.5% (9/120) of the clinical isolates. The low prevalence of MDR isolates in the dairy farm environment is consistent with another study on antimicrobial resistance among Salmonella on dairy farms, in which investigators found that 9.7% of environmental samples yielded isolates resistant to five or more antimicrobial agents (Ray et al., 2007). Despite the generally low levels of antimicrobial resistance detected in the present study, herds experiencing clinical disease tended to be more likely to yield MDR isolates on follow-up sampling than herds with Salmonella-positive environmental samples only. This further reinforces the notion that herds with clinical outbreaks of salmonellosis may pose a greater risk to public health.
Among the Salmonella-positive herds, herd size was significantly associated with the presence of laboratory-confirmed clinical cases. The largest herds in this study were more likely to have clinical cases than herds of the three smaller-size categories. Herd size was also found to be a significant predictor of the incidence of salmonellosis among dairy herds in the northeastern United States when included in a multivariable regression model (Cummings et al., 2009). Larger herds may have a greater likelihood of purchasing cattle from outside sources, with the accompanying risk of introducing Salmonella via a subclinical shedder that has been stressed by transport. High cattle density may also occur in larger herds and could promote Salmonella transmission and clinical disease; animal crowding enhances contact among cattle and may also encourage stressful group dynamics. Finally, larger herds may be characterized by management practices that somehow play a role in increasing the incidence of salmonellosis. Herd size is a risk factor that does not easily lend itself to practical intervention due to the management trends and economic constraints that prevail in the modern dairy industry. However, it is possible that certain attributes of larger herds that contribute to the incidence of salmonellosis could in fact be modified to reduce the occurrence of this disease.
Conclusions
In this study, dairy herds with laboratory-confirmed clinical cases of salmonellosis had a higher prevalence of fecal Salmonella shedding than herds that only generated positive environmental samples. Herds with confirmed clinical cases were also more likely to yield either Salmonella Newport or Salmonella Typhimurium on follow-up sampling, and they had a greater tendency to generate MDR isolates. Clinical laboratory submissions may thus aid in recognizing herds that represent an increased threat to public health. Salmonella Cerro was the predominant serovar isolated from clinical cases, asymptomatic cattle, and the farm environment.
Acknowledgments
The authors thank the veterinarians and dairy herd owners who participated in this study. This project was supported in part by the Cornell University Zoonosis Research Unit of the Food and Waterborne Diseases Integrated Research Network, funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract number N01-AI-30054.
Disclosure Statement
No competing financial interests exist.
References
- Bender JB. Reducing the risk of Salmonella spread and practical control measures in dairy herds. Bovine Pract. 1994;28:62–65. [Google Scholar]
- Blau DM. McCluskey BJ. Ladely SR, et al. Salmonella in dairy operations in the United States: prevalence and antimicrobial drug susceptibility. J Food Prot. 2005;68:696–702. doi: 10.4315/0362-028x-68.4.696. [DOI] [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Outbreak of Salmonella serotype Saintpaul infections associated with multiple raw produce items—United States, 2008. MMWR Morb Mortal Wkly Rep. 2008a;57:929–934. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Salmonella Surveillance: Annual Summary, 2006. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2008b. [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2006. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2009a. [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 States, 2008. MMWR Morb Mortal Wkly Rep. 2009b;58:333–337. [PubMed] [Google Scholar]
- Cho S. Bender JB. Diez-Gonzalez F, et al. Prevalence and characterization of Escherichia coli O157 isolates from Minnesota dairy farms and county fairs. J Food Prot. 2006;69:252–259. doi: 10.4315/0362-028x-69.2.252. [DOI] [PubMed] [Google Scholar]
- [CLSI] Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard—Third Edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. CLSI document M31-A3. [Google Scholar]
- Cummings KJ. Warnick LD. Alexander KA, et al. The incidence of salmonellosis among dairy herds in the northeastern United States. J Dairy Sci. 2009;92:3766–3774. doi: 10.3168/jds.2009-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KJ. Warnick LD. Elton M, et al. Salmonella enterica serotype Cerro among dairy cattle in New York: an emerging pathogen? Foodborne Pathog Dis. (in press). [DOI] [PMC free article] [PubMed]
- Davis MA. Besser TE. Eckmann K, et al. Multidrug-resistant Salmonella Typhimurium, Pacific Northwest, United States. Emerg Infect Dis. 2007;13:1583–1586. doi: 10.3201/eid1310.070536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechet AM. Scallan E. Gensheimer K, et al. Outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium Definitive Type 104 infection linked to commercial ground beef, northeastern United States, 2003–2004. Clin Infect Dis. 2006;42:747–752. doi: 10.1086/500320. [DOI] [PubMed] [Google Scholar]
- Divers TJ. Peek SF. Rebhun's Diseases of Dairy Cattle. St. Louis: Saunders Elsevier; 2008. [Google Scholar]
- Dunn JR. Keen JE. Thompson RA. Prevalence of Shiga-toxigenic Escherichia coli O157:H7 in adult dairy cattle. J Am Vet Med Assoc. 2004;224:1151–1158. doi: 10.2460/javma.2004.224.1151. [DOI] [PubMed] [Google Scholar]
- Dunne EF. Fey PD. Kludt P, et al. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA. 2000;284:3151–3156. doi: 10.1001/jama.284.24.3151. [DOI] [PubMed] [Google Scholar]
- Ellis-Iversen J. Cook AJ. Smith RP, et al. Temporal patterns and risk factors for Escherichia coli O157 and Campylobacter spp., in young cattle. J Food Prot. 2009;72:490–496. doi: 10.4315/0362-028x-72.3.490. [DOI] [PubMed] [Google Scholar]
- Evans S. Davies R. Case control study of multiple-resistant Salmonella Typhimurium DT104 infection of cattle in Great Britain. Vet Rec. 1996;139:557–558. [PubMed] [Google Scholar]
- Fossler CP. Wells SJ. Kaneene JB, et al. Prevalence of Salmonella spp. on conventional and organic dairy farms. J Am Vet Med Assoc. 2004;225:567–573. doi: 10.2460/javma.2004.225.567. [DOI] [PubMed] [Google Scholar]
- Glynn MK. Bopp C. Dewitt W, et al. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- Gupta A. Fontana J. Crowe C, et al. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J Infect Dis. 2003;188:1707–1716. doi: 10.1086/379668. [DOI] [PubMed] [Google Scholar]
- Helms M. Simonsen J. Molbak K. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J Infect Dis. 2004;190:1652–1654. doi: 10.1086/424570. [DOI] [PubMed] [Google Scholar]
- Helms M. Vastrup P. Gerner-Smidt P, et al. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg Infect Dis. 2002;8:490–495. doi: 10.3201/eid0805.010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL. Xu HY. Bao GY, et al. Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiol Infect. 2009;137:1111–1120. doi: 10.1017/S0950268809002039. [DOI] [PubMed] [Google Scholar]
- Huston CL. Wittum TE. Love BC, et al. Prevalence of fecal shedding of Salmonella spp. in dairy herds. J Am Vet Med Assoc. 2002;220:645–649. doi: 10.2460/javma.2002.220.645. [DOI] [PubMed] [Google Scholar]
- Islam M. Morgan J. Doyle MP, et al. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog Dis. 2004;1:27–35. doi: 10.1089/153531404772914437. [DOI] [PubMed] [Google Scholar]
- Karon AE. Archer JR. Sotir MJ, et al. Human multidrug-resistant Salmonella Newport infections, Wisconsin, 2003–2005. Emerg Infect Dis. 2007;13:1777–1780. doi: 10.3201/eid1311.061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan PS. Birtles A. Bolton FJ, et al. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl Environ Microbiol. 2008;74:3626–3633. doi: 10.1128/AEM.01669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L Plym F. Wierup M. Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech. 2006;25:541–554. [PubMed] [Google Scholar]
- Mead PS. Slutsker L. Dietz V, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LR. Warnick LD. Greiner M. Risk factors for changing test classification in the Danish surveillance program for Salmonella in dairy herds. J Dairy Sci. 2007;90:2815–2825. doi: 10.3168/jds.2006-314. [DOI] [PubMed] [Google Scholar]
- Peek SE. Hartmann FA. Thomas CB, et al. Isolation of Salmonella spp. from the environment of dairies without any history of clinical salmonellosis. J Am Vet Med Assoc. 2004;225:574–577. doi: 10.2460/javma.2004.225.574. [DOI] [PubMed] [Google Scholar]
- Peters AR. An estimation of the economic impact of an outbreak of Salmonella Dublin in a calf rearing unit. Vet Rec. 1985;117:667–668. doi: 10.1136/vr.117.25-26.667. [DOI] [PubMed] [Google Scholar]
- Ray KA. Warnick LD. Mitchell RM, et al. Prevalence of antimicrobial resistance among Salmonella on midwest and northeast USA dairy farms. Prev Vet Med. 2007;79:204–223. doi: 10.1016/j.prevetmed.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Sanchez S. Hofacre CL. Lee MD, et al. Animal sources of salmonellosis in humans. J Am Vet Med Assoc. 2002;221:492–497. doi: 10.2460/javma.2002.221.492. [DOI] [PubMed] [Google Scholar]
- Sivapalasingam S. Friedman CR. Cohen L, et al. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J Food Prot. 2004;67:2342–2353. doi: 10.4315/0362-028x-67.10.2342. [DOI] [PubMed] [Google Scholar]
- Varma JK. Greene KD. Ovitt J, et al. Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984–2002. Emerg Infect Dis. 2005a;11:943–946. doi: 10.3201/eid1106.041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma JK. Marcus R. Stenzel SA, et al. Highly resistant Salmonella Newport-MDRAmpC transmitted through the domestic US food supply: a FoodNet case-control study of sporadic Salmonella Newport infections, 2002–2003. J Infect Dis. 2006;194:222–230. doi: 10.1086/505084. [DOI] [PubMed] [Google Scholar]
- Varma JK. Molbak K. Barrett TJ, et al. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis. 2005b;191:554–561. doi: 10.1086/427263. [DOI] [PubMed] [Google Scholar]
- Voetsch AC. Van Gilder TJ. Angulo FJ, et al. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38(Suppl 3):S127–S134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- Warnick LD. Kaneene JB. Ruegg PL, et al. Evaluation of herd sampling for Salmonella isolation on midwest and northeast US dairy farms. Prev Vet Med. 2003;60:195–206. doi: 10.1016/s0167-5877(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Wells SJ. Fedorka-Cray PJ. Dargatz DA, et al. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J Food Prot. 2001;64:3–11. doi: 10.4315/0362-028x-64.1.3. [DOI] [PubMed] [Google Scholar]
- Wray C. Wray A. Salmonella in Domestic Animals. Oxford, New York: CABI Pub.; 2000. [Google Scholar]
- You Y. Rankin SC. Aceto HW, et al. Survival of Salmonella enterica serovar Newport in manure and manure-amended soils. Appl Environ Microbiol. 2006;72:5777–5783. doi: 10.1128/AEM.00791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]