Abstract
We have cloned, expressed and purified a hexameric human DNA helicase (hHcsA) from HeLa cells. Sequence analysis demonstrated that the hHcsA has strong sequence homology with DNA helicase genes from Saccharomyces cerevisiae and Caenorhabditis elegans, indicating that this gene appears to be well conserved from yeast to human. The hHcsA gene was cloned and expressed in Escherichia coli and purified to homogeneity. The expressed protein had a subunit molecular mass of 116 kDa and analysis of its native molecular mass by size exclusion chromatography suggested that hHcsA is a hexameric protein. The hHcsA protein had a strong DNA-dependent ATPase activity that was stimulated ≥5-fold by single-stranded DNA (ssDNA). Human hHcsA unwinds duplex DNA and analysis of the polarity of translocation demonstrated that the polarity of DNA unwinding was in a 5′→3′ direction. The helicase activity was stimulated by human and yeast replication protein A, but not significantly by E.coli ssDNA-binding protein. We have analyzed expression levels of the hHcsA gene in HeLa cells during various phases of the cell cycle using in situ hybridization analysis. Our results indicated that the expression of the hHcsA gene, as evidenced from the mRNA levels, is cell cycle-dependent. The maximal level of hHcsA expression was observed in late G1/early S phase, suggesting a possible role for this protein during S phase and in DNA synthesis.
INTRODUCTION
DNA helicases play important roles in a number of cellular processes. These enzymes utilize the energy derived from nucleoside triphosphate (NTP) hydrolysis to unwind the DNA double helix by disrupting hydrogen bonds and other non-covalent interactions between complementary base pairs. Multiple DNA helicases are found in prokaryotic and eukaryotic cells and can be grouped according to the type of nucleic acid substrates that they unwind. DNA helicases unwind duplex DNA (1) and function in processes such as DNA replication (2–6), recombination and repair (7–9), while RNA helicases function in processes such as transcription, pre-mRNA processing, regulation of RNA stability, ribosome assembly and protein translation (10–12). Helicases, however, are quite function-specific, and the enzyme that functions in DNA replication is distinct from that which operates in DNA repair or recombination. Consequently, it is not surprising that Escherichia coli contains at least eight different DNA helicases (13).
Many helicases have been described over the last few decades, from E.coli to human (1,14–16). Systematic studies of DNA unwinding enzymes present in human cells have led to the identification of several DNA helicases (17–23). It is now evident that there are several different DNA helicases, which may exist in the same cell, and that the specific functions remain to be elucidated.
Here we describe the cloning, expression, purification and characterization of a novel human DNA helicase (hHcsA). The biochemical and genomic properties of this DNA helicase have been investigated, as well as its expression in the various stages of the cell cycle.
MATERIALS AND METHODS
Nucleic acids, enzymes and other reagents
High fidelity thermostable Pfu DNA polymerase was obtained from Stratagene (La Jolla, CA). Yeast replication protein A (RPA) was purified to homogeneity from wild-type yeast as described by Brill and Stillman (24). Human RPA was received as a gift from Dr E.Baril (Worcester Foundation for Experimental Biology, Shrewsbury, MA). Terminal deoxynucleotidyl transferase was purchased from United States Biochemical Corp. (Cleveland, OH). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) and were of high purity (≥95%), as determined by autoradiography of the phosphorylated products. The oligonucleotides used in helicase assays were further purified by reversed phase HPLC. All chemicals used to prepare buffers and solutions were ACS reagent grade and were purchased from Fisher Chemical Company (Pittsburgh, PA). Vectashield mounting solution was obtained from Vector Laboratories (La Jolla, CA).
Buffers
Buffer A contained 25 mM Tris–HCl pH 7.9, 10% sucrose and 0.25 M NaCl. Buffer B contained 25 mM Tris–HCl pH 7.5, 10% (v/v) glycerol, 1 mM DTT and NaCl as indicated. Buffer C was analogous to Buffer B, except that it contained 25 mM HEPES pH 7.5 instead of Tris–HCl. Buffer D contained 25 mM Tris–HCl pH 7.5, 10% (v/v) glycerol, 0.1 mg/ml BSA and 5 mM DTT. Buffer E contained 20 mM Tris–HCl pH 8, 0.5 M NaCl, and imidazole at the indicated concentration.
PCR amplification, cloning and expression of the human helicase A gene
Amplification of the open reading frame (ORF) of the hHcsA gene was carried out using high fidelity Ultma DNA polymerase (Applied Biosystems, Foster City, CA) and primers specific to the 5′- (starting at the ATG initiator codon) and 3′-ends of helicase A, based on the nucleotide sequence (25). The primers were designed such that the amplified DNA would contain the restriction endonuclease sites for SalI and KpnI at its 5′- and 3′-ends, respectively, in order to facilitate subsequent subcloning. The primer sequences were as follows: left, 5′-CTC CTC GGT ACC CAT ATG GCC TCG GCA GCT GT-3′; right, 5′-CTC GTC GAC TTC CTC CAC ATT GG ACA TCG GAA. These primers were used to amplify the entire coding region of the hHcsA gene from HeLa cDNA (Clontech). PCR was carried out for 25 cycles with Ultma DNA polymerase according to the manufacturer’s instructions, and using high fidelity conditions such as low dNTP concentration and a limited number of cycles. The absence of fortuitous mutations introduced by PCR was verified by DNA sequencing carried out at the Core DNA sequencing facility of the Kimmel Cancer Center of Thomas Jefferson University.
Following amplification from HeLa cDNA, the PCR DNA was digested using the appropriate restriction endonucleases, purified and cloned into a T7 expression vector, pET29b (Novagen Inc., Madison, WI) following the manufacturer’s recommendations. Plasmid DNA was prepared following transformation of DH5αF cells. Several clones were selected, and the authenticity of the clones was verified initially by restriction endonuclease mapping based on sites predicted to be present within the sequence and finally by automated DNA sequencing. Escherichia coli strain BL21(DE3) was then transformed with pET29b-hHcsA plasmid for subsequent expression of the protein.
Assay for ATPase activity
ATPase assays were carried out based on methods previously described (26–28). The amount of enzyme used in the assays was selected such that the rate of hydrolysis would be linear in the time range examined.
Helicase assays
The helicase assays were based on the methods previously described by Biswas et al. (27).
Helicase substrates. The substrate used in the characterization of human helicase A was prepared as follows. A synthetic 60mer oligonucleotide complementary to a 50 bp sequence between nucleotides 6268 and 6317 of M13mp19 single-stranded (ss)DNA and labeled at its 5′-end using T4 polynucleotide kinase: the labeled oligonucleotide was hybridized to M13mp19 ssDNA as previously described (27). The oligomer was complementary to a 50 bp sequence between nucleotides 6268 and 6317 of M13mp19 ssDNA and contained 5′ nucleotide tails (non-homologous regions) on both 5′ and 3′ termini. Excess unhybridized labeled oligomer was removed by spin column purification (Promega Biotech, Madison, WI) The purified substrate was diluted to a final concentration of 17 fmol/µl (10 000–20 000 c.p.m./µl) with 10 mM Tris–HCl pH 7.5, 1 mM EDTA.
Directionality substrate. The substrate used to determine the polarity of translocation of DNA helicases was prepared as follows. A 75mer oligonucleotide was synthesized of which 65 bp (6–70 bp) was complementary to nucleotides 6232 and 6296 of M13mp19 ssDNA was hybridized to M13mp19 ssDNA. The oligonucleotide was 32P-labeled at its 5′- or 3′-end, as described (27). Excess unhybridized labeled 75mer was removed by spin column purification. The substrate was then digested with SmaI restriction endonuclease, generating a linear substrate with duplex ends consisting of a 30mer at the 5′ terminus and a 45mer at the 3′ terminus with 32P-labeling at the 5′- or 3′-ends. Translocation of the helicase in the 5′→3′ direction would liberate the 45mer oligonucleotide, while movement in a 3′→5′ direction would result in the 30mer oligonucleotide being liberated.
Assay conditions. Reaction mixtures were set up on ice as follows. A standard 20 µl reaction volume contained buffer D, 10 mM MgCl2, 3.4 mM ATP, 100 mg/ml RPA, 17 fmol (10 000–20 000 c.p.m./µl) of substrate and the indicated amount of enzyme. The reaction mixtures were incubated at 37°C for the times indicated and were terminated by the addition of 4 µl of 2.5% SDS, 60 mM EDTA and 1% bromophenol blue. A fraction (25%) of each reaction mixture was analyzed on 8% polyacrylamide gels in 1× TBE and 0.1% SDS. Electrophoresis was carried out in 1× TBE, 0.1% SDS for 1 h at 160 V. Following electrophoresis, the gels were dried and exposed to Fuji RX film at –80°C for 12 h. Helicase activity was quantitated by scintillation counting of excised substrate/product bands from the dried gels as described by Lebowitz and McMacken (2) and/or by scanning densitometry of the corresponding autoradiogram.
Fluorescence in situ hybridization (FISH) and digital imaging
The in situ hybridization and digital imaging were based on methods described by Nagele et al. (29). The hHcsA probe used for hybridization was complementary to a unique sequence within the hHcsA ORF and carried a digoxigenin (DIG) label. Its oligonucleotide sequence was as follows: 5′-CCT CTT GCT CAC AGT GAT GAA CCT C-3′. Semi-confluent HeLa cell cultures were harvested and samples were adhered to glass microslides by cytospinning (Shandon-Lipshaw Inc.). The cells were then fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. The fixed slides were washed with PBS, followed by three 10 min washes in 2× SSC at room temperature. The probe was denatured for 10 min at 95°C in 70% formamide/2× SSC. Cover slips were placed onto slides spotted with 12 µl of probe solution and sealed using rubber cement. Hybridization was allowed to proceed overnight in a humidified chamber at 37°C. Following hybridization, the slides were washed with 0.5× SSC at 72°C for 5 min and then with 4× SSC containing 0.1% Tween-20 at room temperature for 10 min.
Immunodetection of the hybridized DIG-labeled probe was then carried out as follows. The slides were first incubated in a solution of 4% BSA, 4× SSC, 0.1% Tween-20 to block non-specific binding of the antibodies. This was followed by incubation in blocking solution containing fluorescein isothiocyanate (FITC)-labeled sheep anti-DIG IgG (Boehringer Mannheim, Indianapolis, IN) at 37°C for 20 min. Unbound antibody was then removed by washing three times in 1× SSC containing 0.1% Tween-20. The slides were mounted using Vectashield mounting solution (Vector Laboratories) containing DAPI as DNA counter-stain. Negative controls for hybridization included (i) hybridization without the DIG-labeled probe and (ii) detection with solution containing no DIG antibody. The slides were then viewed under a Nikon FXA microscope equipped with epifluorescence optics and a Princeton Instruments CCD camera. The DAPI image was used to define the nuclear boundary. Photographs were obtained using a ×60 aperture objective and recorded on Fujichrome 400 color film. Separate digital images of the FITC signals and the DAPI nuclei were acquired, aligned and processed using ImagePro Plus (Phase 3 Imaging).
Cell cycle analysis
Confluent HeLa cell cultures were analyzed using the Cell Analysis System 200 (CAS 200). This system utilizes a stoichiometric Feulgen staining reaction in conjunction with microdensitometry to quantify the DNA content of individual nuclei (29). This was also used to generate a cell cycle profile for a given population of cells. As the procedure can be carried out on cells already affixed to slides, a given cell can be analyzed by FISH and for nuclear DNA content. The amount of DNA present in the nucleus of any given cell can be used to assign it to a specific phase in the cell cycle. The reliability of the standard curve measurements was tested by comparing measurements of 25 consecutive images of the same nucleus.
RESULTS
Identification of the ORF encoding human helicase A
We have identified a 2.98 kb human ORF that is homologous (>50%) to yHCS1 (yHcsA), which was purified and cloned previously (30–32). The deduced primary sequence of this protein contains 993 amino acids. Sequence alignment of yHcsA with hHcsA and a putative DNA helicase (F43G6.1 ORF) from Caenorhabditis elegans (Fig. 1) indicates evolutionary conservation of this gene. Approximately 40% homology was observed between hHcsA and the C-terminal 611 amino acids of the 1105 amino acid nematode protein. A total of 27 homologous domains were shared between yHcsA, hHcsA and nematode DNA helicase. Based on its sequence homology with yHcsA and hHcsA, we have designated the DNA helicase from C.elegans as nHcsA.
Figure 1.
Sequence alignment of hHcsA with yHcsA and C.elegans nHcsA. The polypeptide sequences were aligned by the matchbox server multi-align algorithm. Protein sequence alignments were carried out using the facilities at the Baylor College of Medicine (http://dot.imgen.bcm.tmc.edu:9331 and http://www.fundp.ac.be matchbox server). The matched domains shown were predicted as statistically significant. The residues that are identical between the helicases are highlighted in bold.
The peptide sequence of hHcsA revealed several important structural motifs: (i) a Walker type I (30) nucleotide-binding motif located at amino acid 214 and (ii) a bipartite nuclear localization signal, KKLSELSNQRTSRRKER, located at amino acids 975–991.
Cloning and heterologous expression of hHcsA
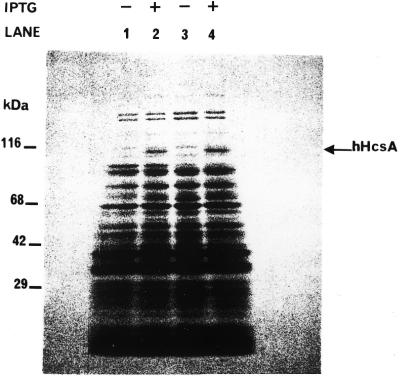
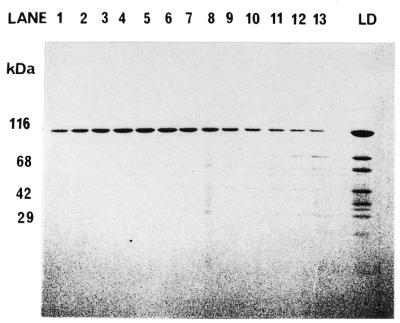
The gene encoding hHcsA was expressed using a T7 expression vector for subsequent production in E.coli. Reasonably good expression levels were obtained in the BL21(DE3) strain using 100 µM IPTG for 1 h at 37°C. Under these conditions, hHcsA contributed to ∼10% of the total cellular protein and was soluble. Attempts to increase the level of expression by altering the concentration of inducer or changing the temperature or duration of induction resulted in the production of protein in an insoluble form. An SDS–PAGE analysis demonstrated that the recombinant protein migrated with a mass of 116 kDa (Fig. 2), which was consistent with the size predicted by its amino acid sequence.
Figure 2.
SDS–PAGE analysis of expression of hHcsAA in E.coli. Induction of expression was carried out as described in the Results in the presence of 0.1 mM IPTG. Equal amounts of cells before and after induction were analyzed by 5–18% SDS–PAGE followed by Coomassie staining. Lane 1, BL21(DE3)/pET29b hHcsA clone #1 cells before induction. Lane 2, BL21(DE3)/pET29b hHcsA clone #1 cells after IPTG induction. Lane 3, BL21(DE3)/pET29b hHcsA clone #6 cells before induction. Lane 4, BL21(DE3)/pET29b hHcsA clone #6 cells after IPTG induction.
Purification of recombinant hHcsA
We have developed a purification protocol for recombinant hHcsA based on the purification procedures used for E.coli DnaB protein and yHcsA (27,31). An extract was prepared from the induced E.coli cells harboring the pET29b-hHcsA/clone #6 recombinant plasmid by sonication in buffer A. The cell lysate was centrifuged at 10 000 g for 30 min (fraction I). The hHcsA was then precipitated from fraction I using differential ammonium sulfate fractionation at 0.2 g/ml (NH4)2SO4. The precipitated protein was then dissolved in buffer B, adjusted to the conductivity of buffer B-100 and loaded onto a POROS HQ/H column (Perceptive Biosystems Inc., Boston, MA). The protein was eluted using a gradient from 100 to 500 mM NaCl in buffer B. Analysis of the fractions by SDS–PAGE and ATPase assay across the fractions indicated that the hHcsA protein was eluted at 400 mM NaCl. The peak fractions were pooled, adjusted to the conductivity of buffer C-50 and loaded onto a POROS HS/H column. Analysis of the fractions indicated that hHcsA did not bind to HS/H column and was collected in the unbound flow-through fractions. At this stage, the helicase appeared to be 80–85% pure. The final step in the purification procedure was size exclusion HPLC (SEHPLC) and is described in detail in the next section. The hHcsA protein showed many similarities to DnaB helicase with respect to its behavior during purification.
Oligomeric structure of hHcsA
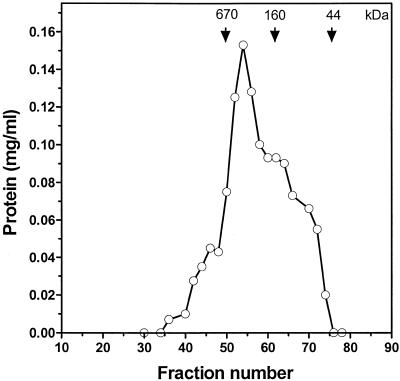
The oligomeric structure of a DNA helicase is an important parameter that may have possible implications in its mechanism of action. The native molecular weight of hHcsA was estimated from SEHPLC as described below. The hHcsA fraction IV was adjusted to 50 mM NaCl in buffer C containing 0.1 mM ATP and 5 mM MgCl2 and then fractionated on a Superdex HR-200 size exclusion chromatography column (Pharmacia Biotech, Piscataway, NJ) equilibrated with the same buffer. The column fractions were then analyzed for protein, ATPase activity and by SDS–PAGE. The results are shown in Figure 3. The ATPase activity and SDS–PAGE clearly indicated a multimeric oligomeric structure. The elution volume corresponded to a molecular weight of ∼600 kDa, as determined by a least squares analysis of molecular weight standards. This is suggestive of a possible hexameric oligomeric structure given a denatured molecular weight of 116 kDa. The oligomeric structure was not observed to be salt sensitive and similar results were obtained with fractionation carried out in buffer containing 250 mM NaCl. The peak obtained in the presence of high salt was less broad than that observed with low salt, which may suggest that the hexameric structure is stabilized by high salt.
Figure 3.
Size exclusion HPLC of hHcsA. Fractionation of recombinant helicase on Supadex HR-200 (Pharmacia) column. Gel filtration standards were from Bio-Rad Laboratories (Richmond, CA) and the proteins were as follows: thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa) and equine myoglobin (17 kDa). (A) Analysis of protein concentration of the fractions. (B) SDS–PAGE analysis of the fractions.
Characterization of ATPase activity
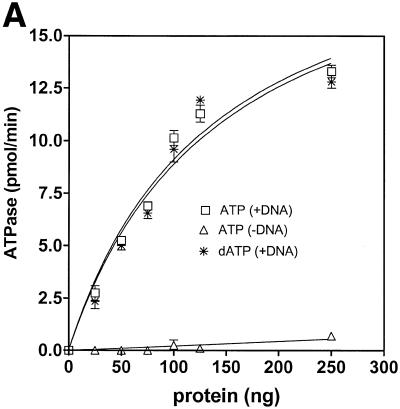
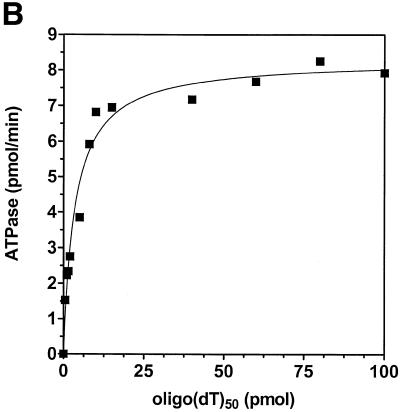
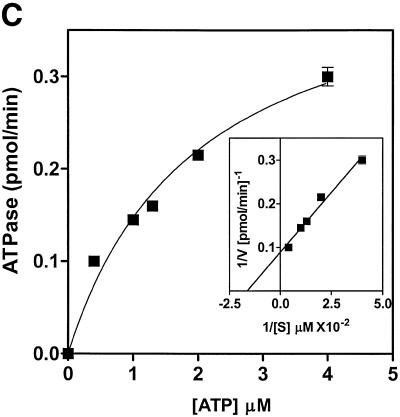
Analysis of ATPase and dATPase activities showed that human helicase A was able to hydrolyze either nucleotide with comparable efficiency (Fig. 4A). ATPase activity was DNA-dependent with ∼5-fold stimulation in ATP hydrolysis in the presence of a DNA cofactor. Half-maximal stimulation was observed at 1 nmol/ml (as nucleotide) of oligo(dT)50 (Fig. 4B and data not shown). The kinetic parameters of ATP hydrolysis for hHcsA were Km = 61 µM and Vmax = 11.3 pmol/min (Fig. 4C).
Figure 4.
Characterization of the ATPase activity of hHcsA. (A) Protein titration of hHcsA in a standard ATPase/dATPase assay in the presence and absence of 200 pmol of M13mp19 ssDNA. (B) Titration of DNA stimulation of the ATPase activity with oligo(dT)50. (C) Kinetic analysis of the ATPase. ATPase activity (V) versus ATP concentration ([S]) in the presence of 200 pmol of M13mp19 ssDNA. Inset, double reciprocal plot (1/V versus 1/[S]) of the ATPase activity in the presence of 200 pmol M13mp19 ssDNA. The data points represent the mean of three separate experiments with SD ± 4.0%.
Characterization of helicase activity
The recombinant helicase A was able to carry out DNA unwinding in a standard helicase assay utilizing a 50 bp partial duplex substrate. A dose–response curve indicated that ∼500 ng of hHcsA protein was required to unwind 30% of the input substrate at 37°C (Fig. 5A). This was significant since human proteins produced in a prokaryotic system are rarely active.
Figure 5.
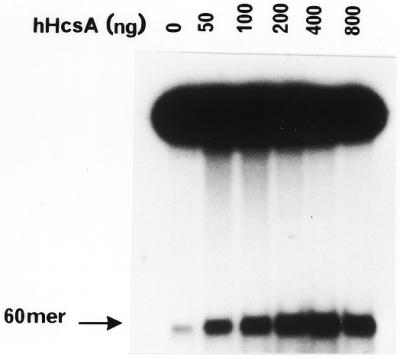
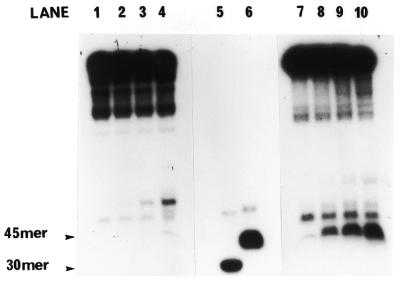
In vitro helicase activity of hHcsA expressed in E.coli. (A) A standard helicase assay was carried out utilizing a 50 bp duplex substrate and increasing amounts of purified hHcsA (fraction V). Inhibition at higher protein concentrations was presumably due to salt. (B) Analysis of the polarity of translocation. A time-course analysis of helicase activity (100 ng HcsA fraction III) using the directionality substrate, as described in Materials and Methods, was carried out in the presence of 250 ng RPA. Lanes 1–5 correspond to assay points in which the 30mer duplex is 5′-end-labeled, while lanes 6–10 correspond to assay points in which the 45mer duplex is 3′-end-labeled. Lane 1, 0 min; lane 2, 1 min; lane 3, 2 min; lane 4, 5 min; lane 5, heat denatured substrate labeled at the 30mer duplex end; lane 6, heat denatured substrate labeled at the 45mer duplex end; lane 7, 0 min; lane 8, 1 min; lane 9, 2 min; lane 10, 5 min.
The polarity of translocation was examined using a linear substrate containing 5′ and 3′ duplex ends as shown in Figure 5B. The hHcsA was found to have a 5′→3′ polarity of movement. This polarity is identical to that observed for the DnaB helicase and yHcsA. Helicase activity could be stimulated by ssDNA-binding proteins. DNA unwinding was enhanced in the presence of human and yeast RPA; however, the extent of DNA unwinding in the presence of E.coli ssDNA-binding protein was essentially that observed by hHcsA alone (data not shown).
Expression of human helicase A during various phases of the cell cycle
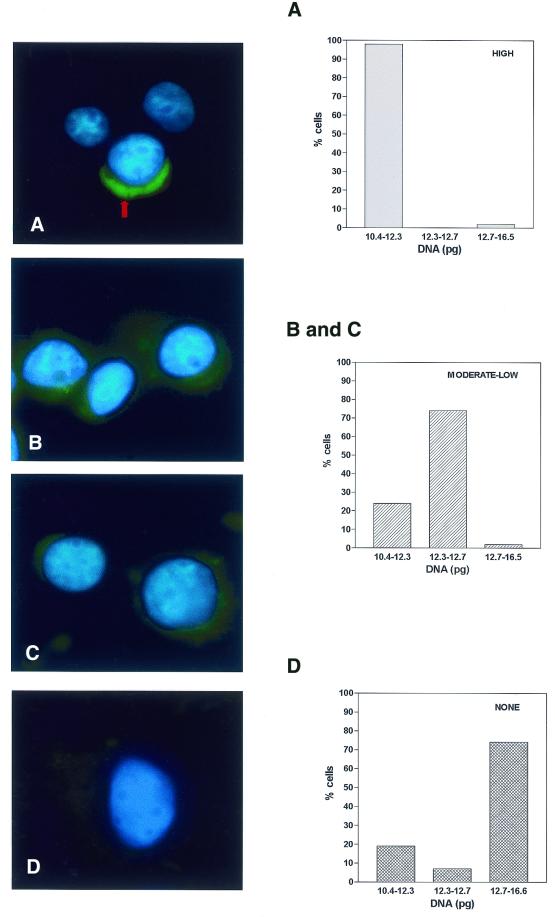
In an effort to obtain information regarding expression of the protein, we examined the levels of hHcsA mRNA throughout the cell cycle using FISH. HeLa cells were grown until they were semi-confluent, then cells were collected and fixed to coated glass microslides using a Cytospin (Shandon-Lipshaw). As described in the Materials and Methods, cells were fixed and probed with a DIG-labeled oligonucleotide complementary to a unique sequence within hHcsA. Detection of oligonucleotide hybridization was carried out using FITC-conjugated sheep anti-DIG (Boehringer Mannheim) antibody. The cells were mounted in a Vectashield mounting solution containing DAPI as DNA counterstain. Representative images obtained from these studies are shown in Figure 6. Two interesting features emerged from these fluorescent studies. The signal corresponding to hHcsA mRNA was restricted to a cap-shaped region at the periphery of the nucleus and was generally absent from other areas of the cytoplasm. In addition, the intensity of signal varied from cell to cell within the population. Correlation of signal intensity with the appearance of nucleoli (as revealed by DAPI staining) suggested that the signal intensity increased in cells that were in late G1 and S phase. Cells that were likely to be in M or G2 phase displayed little or no hHcsA-specific signal.
Figure 6.
Detection of hHcsA mRNA by FISH. HeLa cells affixed to microslides were probed with oligonucleotide homologous to hHcsA as described in Materials and Methods. The DNA content of the corresponding cell was determined using quantitative Feulgen staining. Based on this, an assignment was made as to the position of a given cell within the cell cycle. Left, images shown are representative of the three categories of signal intensities observed by immunofluorescence microscopy. (A) High level of cross-reactivity; (B and C) moderate to low level of signal intensity; (D) no signal intensities observed. Right, DNA content of the total population of cells for which images were obtained. Based on the quantitative Feulgen staining (29), cells were assigned to phases of the cell cycle as follows: G1, 10.4–12.3; S, 12.3–12.7; G2/M, 12.7–16.6.
In order to accurately correlate the expression of this gene product with the various stages of the cell cycle, the DNA content of these cells on the slides was analyzed using the CAS 200, as described previously (29). This system utilizes a stoichiometric Feulgen staining reaction coupled with microscopic densitometry to quantify the total DNA content of a single cell. Unlike fluorescence-activated cell sorting, this method can be carried out on single cells that are already affixed to slides, and a particular nucleus can be concomitantly analyzed by FISH. Analysis of randomly selected nuclei on the slide demonstrated that the cell population was completely asynchronous. Cells possessing high hHcsA-specific signal intensities were exclusively found to have nuclei with DNA content consistent with the late G1 phase of the cell cycle, while moderate levels of hHcsA-specific signal were observed in cells at early S phase. Cells in which no hHcsA-specific signal was detected had a DNA content consistent with late S and G2/M phases. Therefore, levels of hHcsA mRNA were maximal in late G1 phase and were progressively decreased during transit through S phase. The HcsA mRNA was absent during G2/M phases of the cell cycle.
DISCUSSION
Identification of a novel human hexameric DNA helicase
We have identified a human DNA helicase gene (GenBank accession no. L14754) that is homologous to the yeast HCS1 gene (yHcsA) as well as a DNA helicase from C.elegans (F3G6.1). The nematode helicase A (nHcsA) and human helicase A (hHcsA) were found to have extensive (∼40–50%) homology with yHcsA. An alignment of yHcsA, nHcsA and hHcsA sequences is shown in Figure 1. The fact that these helicases are separated by considerable evolutionary distance and retained similar structural and functional features point to a possible significance of this homology.
Cloning and expression of human helicase A
Using PCR technology, the gene for hHcsA was cloned from a HeLa cDNA library. To verify that this clone did indeed express the full-length gene product, the protein was expressed in E.coli using a T7 expression system. As shown in Figure 2, the isolated cDNA encoded the anticipated 116 kDa protein and, thus, distinguishes it from other human DNA helicases described over the past decade (17–22). The protein was expressed in a soluble form although the overall level of expression was low as compared to other lower eukaryotic constructs (31). Higher expression levels may be obtained in eukaryotes, for example baculovirus or COS systems. Human helicase A behaved chromatographically in a manner identical to that of the E.coli DnaB protein, and the protein was active in both in vitro ATPase and helicase assays (Figs 4 and 5). Using SEHPLC we were able to determine the native molecular weight of the protein, which would give important insights into the probable oligomeric structure of the protein. Many helicases characterized to date have been reported as being dimers or hexamers. As shown in Figure 3, hHcsA had an elution volume consistent with a molecular weight of ∼600 kDa. This is probably indicative of a hexameric structure, and rules out the possibility that hHcsA exists in a monomeric, dimeric or trimeric state. As such, hHcsA can be placed in the growing group of hexameric DNA helicases, which includes E.coli DnaB protein, T7 DNA helicase, yHcsA and the SV40 T-antigen RuvB protein (1).
ATP hydrolysis by hHcsA
With the availability of homogenous preparations of the helicase, it was possible to characterize the protein in terms of its enzymatic properties. With respect to its ATPase activity, the recombinant enzyme was DNA-dependent, and ∼5-fold less nucleotide hydrolysis was observed in the absence of a DNA cofactor (Fig. 4A). The rate of ATP hydrolysis (11.3 pmol/min/mg) appeared less relative to other helicases such as DnaB or yHcsA; however, the affinity of ATP binding, Km = 61 µM is comparable to other hexameric DNA helicases (Fig. 4C).
DNA unwinding activity of hHcsA
The helicase was fully active in a standard helicase assay. The protein was capable of unwinding a 50 bp duplex, and the amount of unwinding increased linearly with the amount of protein (Fig. 5A). Analysis of the polarity of translocation demonstrated that hHcsA was 5′→3′ (Fig. 5B). This is the same polarity of translocation observed with several other DNA helicases, including the DnaB protein of E.coli, yHcsA, the bacteriophage T7 gene 4 protein and human DNA helicases HDH IV and VIII (2,21,23,27,33).
FISH analysis of hHcsA mRNA levels
In an effort to gain further information regarding the cellular function of this helicase, the localization of hHcsA mRNA was examined using FISH combined with quantitative analysis of nuclear DNA content. Our analysis demonstrated that the level of mRNA, as inferred from the relative degree of fluorescence signal intensities, varied between cells. Furthermore, hHcsA mRNA was localized to a cap-shaped extranuclear region (Fig. 6). Using a stoichiometric Feulgen staining reaction and microscopic densitometry, cell cycle status of individual cells was determined. Cells displaying maximum levels of hHcsA mRNA were found only at late G1 phase, and moderate to low levels of mRNA were detected in cells at S phase of the cell cycle. We did not detect mRNA in cells at the G2/M phase.
In conclusion, our studies have identified a novel human DNA helicase and defined several of its important characteristics, including (i) it is a hexameric helicase, and its oligomerization is not salt sensitive; (ii) it is well conserved throughout evolution, from yeast to human; (iii) its ATPase activity is DNA-dependent and a 5-fold stimulation in the rate of hydrolysis is observed in the presence of DNA cofactor; (iv) it has a 5′→3′ polarity of movement along the duplex DNA; and (v) the expression pattern of its mRNA appear to be cell cycle-dependent.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank the Nucleic Acid Facility of the Kimmel Cancer Center at Thomas Jefferson University for DNA sequence determination and acknowledge the technical expertise of the Medical Media Services at Thomas Jefferson University. This work was supported by a grant from the National Institute of General Medical Sciences (R01GM36002) of the National Institutes of Health to S.B.
References
- 1.Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd Edn. W.H. Freeman and Co., San Francisco, CA.
- 2.Lebowitz J.H. and McMacken,R. (1986) The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem., 261, 4738–4748. [PubMed] [Google Scholar]
- 3.Baker T.A., Funnell,B.E. and Kornberg,A. (1987) Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J. Biol. Chem., 262, 6877–6885. [PubMed] [Google Scholar]
- 4.Bernstein J.A. and Richardson,C.C. (1989) Characterization of the helicase and primase activities of the 63-kDa component of the bacteriphage T7 gene 4 protein. J. Biol. Chem., 264, 13066–13073. [PubMed] [Google Scholar]
- 5.Dong F. and von Hippel,P.H. (1996) The ATP-activated hexameric helicase of bacteriophage T4 (gp41) forms a stable primosome with a single subunit of T4-coded primase (gp61). J. Biol. Chem., 271, 19625–19631. [DOI] [PubMed] [Google Scholar]
- 6.Venkatesan M., Silver,L.L. and Nossal,N.G. (1982) Bacteriophage T4 gene 41 protein required for the synthesis of RNA primers, is also a DNA helicase. J. Biol. Chem., 257, 12426–12434. [PubMed] [Google Scholar]
- 7.Prakash S., Sung,P. and Prakash,L. (1993) DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet., 27, 33–70. [DOI] [PubMed] [Google Scholar]
- 8.Sung P., Prakash,L., Matson,S.W. and Prakash,S. (1987) RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc. Natl Acad. Sci. USA, 84, 8951–8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung P., Guzder,S.N., Prakash,L. and Prakash,S. (1996) Negative superhelicity promotes ATP-dependent binding of yeast RAD3 protein to ultraviolet-damaged DNA. J. Biol. Chem., 271, 10821–10826. [PubMed] [Google Scholar]
- 10.Bass B.L. and Weintraub,H. (1987) A developmentally regulated activity that unwinds RNA duplexes. Cell, 48, 607–613. [DOI] [PubMed] [Google Scholar]
- 11.Rebagliati M.R. and Melton,D.A. (1987) Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell, 48, 599–605. [DOI] [PubMed] [Google Scholar]
- 12.Abramson R.D., Dever,T.E., Lawson,T.G., Ray,B.K., Thach,R.E. and Merrick,W.C. (1987) The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem., 262, 3826–3832. [PubMed] [Google Scholar]
- 13.Geider K. and Hoffmann-Berling,H. (1981) Proteins controlling the helical structure of DNA. Annu. Rev. Biochem., 50, 233–260. [DOI] [PubMed] [Google Scholar]
- 14.DePamphilis M.L. (1993) Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem., 62, 29–63. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis M.L. (1993) Origins of DNA replication that function in eukaryotic cells Curr. Opin. Cell Biol., 5, 434–441. [DOI] [PubMed] [Google Scholar]
- 16.Tuteja N. and Tuteja,R. (1996) DNA helicases: the long unwinding road. Nat. Genet., 13, 11–12. [DOI] [PubMed] [Google Scholar]
- 17.Hirota Y. and Lahti,J.M. (2000) Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res., 28, 917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karow J.K., Chakraverty,R.K. and Hickson,I.D. (1997) The Bloom’s syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- 19.Tuteja N., Rahman,K., Tuteja,R. and Falaschi,A. (1993) Human DNA helicase V, a novel DNA unwinding enzyme from HeLa cells. Nucleic Acids Res., 21, 2323–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuteja N., Rahman,K., Tuteja,R., Ochem,A., Skopac,D. and Falaschi,A. (1992) DNA helicase III from HeLa cells: an enzyme that acts preferentially on partially unwound DNA duplexes. Nucleic Acids Res., 20, 5329–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuteja N., Rahman,K., Tuteja,R. and Falaschi,A. (1991) DNA helicase IV from HeLa cells. Nucleic Acids Res., 19, 3613–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo Y.S. and Hurwitz,J. (1993) Isolation of helicase α, a DNA helicase from HeLa cells stimulated by a fork structure and single-stranded DNA-binding proteins. J. Biol. Chem., 268, 10282–10295. [PubMed] [Google Scholar]
- 23.Costa M., Ochem,A., Staub,A. and Falaschi,A. (1999) Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res., 27, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brill S.J. and Stillman,B. (1989) Yeast replication factor-A functions in the unwinding of the SV40 origin of replication. Nature, 342, 92–95. [DOI] [PubMed] [Google Scholar]
- 25.Wiemann S., Voss,H., Schwager,C., Rupp,T., Stegemann,J., Zimmermann,J., Grothues,D., Sensen,C., Erfle,H., Hewitt,N. et al. (1993) Sequencing and analysis of 51.6 kilobases on the left arm of chromosome XI from Saccharomyces cerevisiae reveals 23 open reading frames including the FAS1 gene. Yeast, 9, 1343–1348. [DOI] [PubMed] [Google Scholar]
- 26.Biswas E.E., Biswas,S.B. and Bishop,J.M. (1986) The dnaB protein of E. coli: mechanism of nucleotide binding, hydrolysis and modulation by dnaC protein. Biochemistry, 25, 7368–7374. [DOI] [PubMed] [Google Scholar]
- 27.Biswas E.E., Ewing,C.M. and Biswas,S.B. (1993) Characterization of the DNA-dependent ATPase and a DNA unwinding activity associated with the yeast DNA polymerase α complex. Biochemistry, 32, 3020–3026. [DOI] [PubMed] [Google Scholar]
- 28.Biswas S.B. and Biswas,E.E. (1987) Regulation of dnaB function in DNA replication in Escherichia coli by dnaC and lP protein. J. Biol. Chem., 262, 7831–7838. [PubMed] [Google Scholar]
- 29.Nagele R.G., Freeman,T., McMorrow,L., Thomson,Z., Kitson-Wind,K. and Hsin-yi,L. (1999) Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J. Cell Sci., 112, 525–535. [DOI] [PubMed] [Google Scholar]
- 30.Biswas E.E. and Biswas,S.B. (1999) Mechanism of DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding and oligomerization. Biochemistry, 38, 10919–10928. [DOI] [PubMed] [Google Scholar]
- 31.Biswas E.E., Chen,P.-H. and Biswas,S.B. (1993) DNA helicase associated with DNA polymerase α: isolation by a modified immunoaffinity chromatography. Biochemistry, 32, 13393–13398. [DOI] [PubMed] [Google Scholar]
- 32.Biswas E.E., Fricke,W.E., Chen,P.-H. and Biswas,S.B. (1997) Yeast DNA helicase A: cloning, expression, purification and enzymatic characterization. Biochemistry, 36, 13277–13284. [DOI] [PubMed] [Google Scholar]
- 33.Notarnicola S.M., Park,K., Griffith,J.D. and Richardson,C.C. (1995) A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J. Biol. Chem., 270, 20215–20224. [DOI] [PubMed] [Google Scholar]