Abstract
Pulsed-field gel electrophoresis (PFGE) characterization of 335 temporally and spatially matched clinical, bovine, and human Salmonella enterica subsp. enterica isolates revealed 167 XbaI PFGE patterns. These isolates were previously classified into 51 serotypes and 73 sequence types, as determined by multilocus sequence typing. Discriminatory power of PFGE (Simpson's index, D = 0.991) was considerably higher than that of multilocus sequence typing (D = 0.920) or serotyping (D = 0.913). Although 128 PFGE types each only represented a single isolate, 8 PFGE types represented >4 isolates, including (i) three serotype Enteritidis and Heidelberg patterns that were only identified among human isolates, (ii) two PFGE patterns (each representing serotypes Bardo and Newport) that were significantly more common among bovine isolates as compared with human isolates; (iii) two PFGE types that each includes two serotypes (4,5,12:i:- and Typhimurium; Thompson and 1,7:-:1,5); and (iv) one PFGE type that includes eight Typhimurium isolates from humans and cattle. Characterization of isolates collected over multiple farm visits indicated that given specific PFGE types persisted over time on 11 farms. On an additional seven farms, isolates with a given sequence type represented multiple PFGE type, which typically only differed by <3 bands, suggesting PFGE type diversification during strain persistence. Sixteen PFGE types were isolated from 2 or more farms, including two widely distributed serotype Newport-associated PFGE types each found on 10 farms. In six instances two or three human isolates collected in the same county in the same or consecutive months represented the same subtypes, suggesting small human case clusters. PFGE-based characterization and surveillance of human and animal isolates can provide improved understanding of Salmonella diversity and epidemiology, including identification of possible host-associated and common, widely distributed PFGE types.
Introduction
Salmonella is one of the most common causes of reported bacterial foodborne illnesses worldwide (Swaminathan et al., 2006a). Although the species Salmonella enterica consists of six subspecies, S. enterica subsp. enterica serotypes are responsible for the vast majority of human salmonellosis cases (Brenner et al., 2000; Uzzau et al., 2000). Subtyping methods applied to Salmonella include serotyping (Brenner et al., 2000; Wiedmann, 2002) according to the Kauffmann–White Scheme (Grimont and Weil, 2007), which differentiates Salmonella into over 2500 recognized serotypes, as well as a variety of other phenotypic and genotypic approaches, including phage typing (Liesegang et al., 2002; Lukinmaa et al., 2006), DNA sequencing-based subtyping methods (Sukhnanand et al., 2005; Alcaine et al., 2006; Seo et al., 2006; Woo and Lee, 2006), ribotyping (Ridley et al., 1998; Fontana et al., 2003), pulsed-field gel electrophoresis (PFGE) (Fakhr et al., 2005; Ribot et al., 2006; Sahlstrom et al., 2006; Seo et al., 2006; Zamperini et al., 2007), multilocus variable number of tandem repeat analysis (MLVA) (Cho et al., 2008; Malorny et al., 2008), and DNA microarray analysis (Garaizar et al., 2002; Scaria et al., 2008). PFGE is the most widely used molecular subtyping method for Salmonella (Barrett et al., 2006) and is used routinely by the Centers for Disease Control and Prevention (CDC) and state health departments in the United States, as well as in Canada, Latin America, Asia, and Europe through the U.S. and International PulseNet system (Swaminathan et al., 2001, 2006b). Although considerable data on diversity of Salmonella PFGE types and discriminatory ability of PFGE among isolates from human clinical cases are available (Agasan et al., 2002; Camps et al., 2005; CDC, 2008a, 2008b, 2008c; Grandesso et al., 2008; Lomonaco et al., 2008), only few studies (Wedel et al., 2005; Oloya et al., 2009) have evaluated and compared Salmonella PFGE type diversity among temporally and geographically matched human and animal clinical isolates. Comparative data of Salmonella subtype and PFGE diversity among humans with clinical disease and animals that represent potential sources and reservoirs for zoonotic and foodborne transmission are critical to facilitate epidemiological and source-tracking studies and to identify potentially host-restricted subtypes. We thus characterized 335 temporally and geographically matched human and bovine Salmonella isolates collected from clinical human and bovine cases in New York State and a neighboring state, Vermont, by using the standard PulseNet PFGE typing protocol and compared PFGE data with previously published multilocus sequence typing (MLST) and serotype data (Alcaine et al., 2006). A specific hypothesis to be tested in this study was that PFGE would allow for identification of host-associated PFGE types, which could be characterized in future studies aimed to further characterize host specificity determinants in Salmonella.
Materials and Methods
Salmonella isolates
A total of 335 spatially and temporally matched nontyphoidal S. enterica subsp. enterica isolates were used in this study. Human Salmonella isolates (n = 178), collected in 2004, were obtained from the Wadsworth Center, New York State Department of Health; Salmonella isolates from cattle with clinical symptoms of salmonellosis (n = 157), collected from 64 different farms located in New York State and Vermont in 2004, were obtained from the New York State Animal Health Diagnostic Laboratory (details of all isolates are provided in Supplemental Table S1, available online at www.liebertonline.com). Although the majority of Salmonella isolates used here had previously been described and characterized using MLST and serotyping, five Salmonella isolates (one bovine and four human isolates) used in this study were not included in our previous study (Alcaine et al., 2006). In addition, five human isolates included in the previous study by Alcaine et al. (2006) were not included here. Identical isolate designations (e.g., FSL S5-430) are used in the study reported here and the previous study by Alcaine et al. (2006). Serotyping of bovine and human isolates was performed using standard methods (Ewing, 1972) as previously detailed (Alcaine et al., 2006). Sequence typing using a three-gene MLST scheme (targeting manB, fimA, and mdh) was also performed as previously reported (Sukhnanand et al., 2005; Alcaine et al., 2006). Sequence-type (ST) designations were assigned to be consistent with previous articles by our group that used the same MLST scheme (Alcaine et al., 2006).
PFGE analysis
PFGE was performed according to the CDC PulseNet protocol (Ribot et al., 2006) using a CHEF-Mapper (Bio-Rad Laboratories, Hercules, CA). Electrophoresis conditions were an initial switch time of 2.16 sec, a final switch time of 63.8 sec, and a run time of 21 h. The CDC Salmonella ser. Branderup isolate H9812 was used as the reference strain (Hunter et al., 2005). Pictures of PFGE gels were taken with the Gel/ChemiDoc system (Bio-Rad Laboratories). For 10 Salmonella isolates representing all 6 serotype Kentucky isolates as well as serotypes Infantis (2 isolates), Oranienburg (1 isolate), and Havana (1 isolate), which could not be typed by the routine CDC PulseNet protocol, addition of 50 μM thiourea to the running buffer (Murase et al., 2004) was necessary to yield clear, interpretable PFGE patterns.
Analysis and comparison of PFGE types was performed using the BioNumerics Software package (Applied Maths 1998–2004, Austin, TX). Similarity analysis was performed using the Dice coefficient and clustering was performed using the unweighted pair group method by arithmetic mean. PFGE types were assigned short unique numerical identifiers (i.e., PFGE type numbers); in addition, PFGE types for all isolates were coded according to CDC PulseNet codes for naming PFGE patterns (Swaminathan et al., 2001) with initials of New York Cornell University (NYCU); for example, “NYCU.JPXX01.0001” would represent PFGE pattern 1 for Salmonella ser. Typhimurium (“JPX”) with enzyme XbaI (“X01”); these longer identifiers allow for comparison across studies and are provided in Supplemental Table S1 and PathogenTracker.
Statistical analysis
Statistical analyses on the distribution of PFGE types among human and bovine isolates were performed using all PFGE types that contained >4 isolates; PFGE types that represented ≤4 isolates were classified as rare PFGE types. The frequency distributions of PFGE types for isolates from human and bovine clinical cases were compared using the chi-square test of independence or Fisher's exact test; Fisher's exact test was performed for comparisons where one or more of the expected values was <5. p-Values lower than 0.05 were considered statistically significant. All statistical analyses were conducted with Statistical Analysis Systems (SAS) 9.1 (SAS Institute, Cary, NC).
Simpson's index of diversity (D) was calculated as described (Hunter and Gaston, 1988) to assess the differentiation of Salmonella isolates by serotyping, PFGE, MLST, or combinations of two or three subtyping methods.
Spatial analysis
A New York State and Vermont map from MapViewer software (MapViewer package version 6.0; Golden Software, Golden, CO) was used to observe counties for farms where bovine Salmonella isolates were obtained.
Access to detailed isolate information
All isolate information, including isolate source, gene sequence data, allele assignments, antibiotic resistance profiles, and PFGE images, is publicly available in the Pathogen Tracker website (www.pathogentracker.net). All supplemental materials are available online at www.liebertonline.com.
Results
PFGE characterization of bovine Salmonella isolates obtained over multiple farm visits
Among the 64 farms where Salmonella isolates were obtained for this study, isolates were obtained during multiple (at least two) visits on 20 farms (Table 1). Initial characterization of these isolates by MLST (Alcaine et al., 2006) had shown re-isolation of Salmonella with the same serotype and ST on 18 farms, including one farm (farm 261; Table 1) where two STs were isolated during multiple visits. For isolates from 11 farms (including isolates with ST17 on farm 261), PFGE provided for the same subtype grouping as MLST, confirming re-isolation of a given subtype over two or more sample collection dates (e.g., farm 329, see Table 1).
Table 1.
Salmonella Isolate Information for Clinical Bovine Isolates Collected from Farms at Multiple Visits
| Farm ID | No. of farm visits with Salmonella-positive samples | ST/serotype (no. of isolates)a | PFGE type no (no. of isolates) |
|---|---|---|---|
| Farms where PFGE supported persistence of a given Salmonella subtype | |||
| 510 | 20 | ST11/Newport (21) ST6/4,5,12:i:- (1) |
121 (20), 122 (1) 94 (1) |
| 261 | 22 | ST6/4,5,12:i:- (18) ST17/Kentucky (5) ST6/Typhimurium (1) |
89 (5), 90 (1), 91 (1), 94 (10), 95 (1) 96 (5) 94 (1) |
| 223 | 15 | ST60/Infantis (15) | 107 (13), 108 (1), 109 (1) |
| 329 | 5 | ST9/Montevideo (1) ST44/Muenster (3) ST62/Thompson (1) |
119 (1) 7 (3) 157 (1) |
| 186 | 4 | ST75/Adelaide (1) ST8/Typhimurium (3)b |
44 (1) 104 (3) |
| 524 | 5 | ST6/4,5,12:i: (1) ST11/Newport (4) |
90 (1) 126 (1), 127 (3) |
| 152 | 4 | ST11/Newport (4) | 126 (4) |
| 490 | 4 | ST11/Newport (4) | 126 (1), 127 (2), 129 (1) |
| 163 | 3 | ST60/Infantis (1) ST11/Newport (2) |
114 (1) 126 (2) |
| 259 | 3 | ST44/Muenster (3) | 2 (1), 4 (1), 6 (1) |
| 488 | 3 | ST11/Bardo (1) ST11/Newport (3) |
126 (1) 126 (3) |
| 584 | 3 | ST2/Agona (2) ST6/Typhimurium (1) |
165 (1), 166 (1) 64 (1) |
| 97 | 2 | ST8/Typhimuriumc (2) | 104 (2) |
| 125 | 2 | ST6/Typhimurium (2) | 79 (2) |
| 208 | 2 | ST6/Typhimurium (2) | 66 (2) |
| 303 | 2 | ST11/Newport (2) | 126 (2) |
| 320 | 2 | ST11/Newport (2) | 126 (2) |
| 764 | 2 | ST6/Typhimurium (2) | 60 (2) |
| Farms where isolates from different sampling dates represented distinct subtypes | |||
| 105 | 2 | ST11/Newport (1) ST8/Typhimurium (1) |
121 (1) 102 (1) |
| 415 | 2 | ST9/Montevideo (1) ST6/Typhimurium (1) |
119 (1) 70 (1) |
Sequence and serotype data were obtained from Alcaine et al. (2006).
Serotyping identified one isolate with serotype Typhimurium var. 5 (previously known as Typhimurium Copenhagen) among the three Typhimurium isolates from farm 186.
Serotyping identified one isolate with serotype Typhimurium var. 5 (previously known as Typhimurium Copenhagen) among the two serotype Typhimurium isolates from farm 97.
PFGE, pulsed-field gel electrophoresis; ST, sequence type.
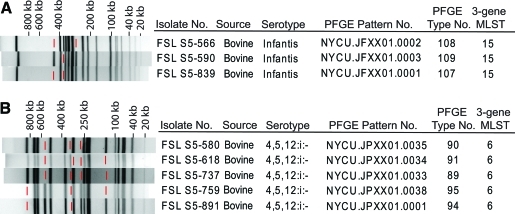
In seven instances (i.e., for isolates from farms 510, 261, 223, 524, 490, 259, and 584), PFGE differentiated Salmonella isolates with identical STs into two or more PFGE types (Table 1). In six of these instances (i.e., for all but farm 261) PFGE types differentiated among isolates with the same ST differed by <3 bands from each other (Fig. 1 and Supplemental Fig. S1, available online at www.liebertonline.com). For the three farms (farms 510, 261, and 223) where >10 isolates with the same ST were available, PFGE typically identified one predominant PFGE type and one or more less frequent PFGE types (Table 1). For example, 15 Salmonella ser. Infantis ST 60 isolates from farm 223 represented 13 isolates with PFGE pattern 107 and 1 isolate each with PFGE pattern 108 and 109. These three patterns differed by <3 bands from each other; for example, pattern 107 differed from patterns 108 and 109 by two and one bands, respectively (Fig. 1A). Among 18 Salmonella ser. 4,5,12:i:- isolates from farm 261, which all shared ST6, PFGE differentiated 5 different subtypes (Fig. 1B). These five PFGE types represented two groups: (i) PFGE types 89, 90, and 91 and (ii) PFGE types 94 and 95 (Fig. 1B). In addition to PFGE types 89 and 91, which differed by only two bands and were isolated over an initial 62-day period (6/25/2004 to 8/26/2004), PFGE type 90, which differed by one and two bands, respectively, from PFGE types 89 and 91, was also isolated only during the same time period. PFGE types 94 and 95, which differed by only a single band, were subsequently isolated for a 99-day period (8/31/2004 to 12/08/2004) on the same farm (Fig. 1B and Supplemental Fig. S1). PFGE type 90, isolated during the initial time period, and PFGE type 94, isolated during the second period, differed by only three bands, suggesting the possibility that the subtypes initially circulating on this farm represented the ancestors for the subtypes found during the second period.
FIG. 1.
Example of closely related PFGE patterns representing isolates, with identical STs, obtained from a given farm. Panels (A) and (B) show PFGE patterns for isolates from farms 261 and 223, respectively. Red lines indicate bands missing in a given PFGE patterns (as compared with other PFGE patterns found among isolates from the same farm). PFGE, pulsed-field gel electrophoresis.
Overall, for the seven farms where isolates with a given ST presented multiple closely related PFGE types, on five farms (i.e., 261, 510, 223, 524, and 490) identical PFGE types were still isolated on multiple sample collection dates. Times between first and last isolation of closely related PFGE types (i.e., PFGE types that shared the same ST and differed ≤3 bands) on these farms ranged from 1 to 243 days with a mean of 58 days (see Supplemental Table S2, available online at www.liebertonline.com). For example, on farm 510 isolates with serotype Newport, ST11, and PFGE type 121 were obtained on 20 sampling dates (between 8/4/2004 and 12/16/2004; 134 days between first and last isolation), whereas one isolate represented a PFGE type (type 122), which differed by one band from PFGE type 121, was isolated once on 10/29/2004 (Supplemental Fig. S1).
As re-isolation of Salmonella isolates with the same subtype indicates persistence of a subtype, for a given farm, only one isolate representing each unique combined subtype (based on serotype, MLST, and PFGE type) was included in the statistical analysis and Simpson's index of diversity calculations. This approach avoids over-representation of a subtype due to re-sampling in multiple visits of a given farm. For example, for farm 510, isolates with a combined subtype of serotype Newport, ST 11, and PFGE type 121 were obtained from specimens collected during 20 different visits (Table 1). However, only one of the isolates with this combined subtype from farm 510 was used for statistical analysis. Therefore, out of 157 bovine Salmonella isolates, a total of 91 cattle isolates were used in statistical analysis reported below.
PFGE type diversity and discriminatory power
The 335 Salmonella isolates characterized were differentiated into 167 XbaI PFGE types, 73 STs (as determined by a three-gene MLST scheme) (Alcaine et al., 2006), and 51 serotypes (Table 2). Although MLST and serotype data for most of these isolates have been reported previously (Alcaine et al., 2006), PFGE data for these isolates have not been previously reported. Eleven of the 14 most common serotypes represented multiple STs (Table 3); all 14 of these serotypes represented multiple PFGE types (i.e., 3 to 32 different PFGE types per serotype; see Table 3). Thus, a number of Salmonella serotypes and STs were differentiated further by PFGE. For example, 16 Salmonella serotype 4,5,12:i:- isolates with ST6 (out of a total of 17 serotype 4,5,12:i:- isolates) were differentiated into 9 different PFGE types. Overall, PFGE showed the highest discriminatory power, among the Salmonella isolates characterized, as determined by Simpson's index of discrimination (D = 0.991), followed by MLST (D = 0.920) and serotyping (D = 0.913). Subtype diversity among human Salmonella isolates was higher than among bovine isolates, regardless of subtyping methods (Table 2). Simpson's index of diversity values reported here cannot be directly compared with those reported by Alcaine et al. (2006) as the isolate sets used in these two sets were similar but not identical.
Table 2.
Serotype, Multilocus Sequence Typing, and Pulsed-Field Gel Electrophoresis Type Diversity Among Clinical Bovine and Human Salmonella Isolates
| |
No. of subtypes found among |
Simpson's index of diversity scores among |
|||||
|---|---|---|---|---|---|---|---|
| Human isolates only | Bovine isolates only | Both human and bovine isolates | Total | Human isolates | Bovine isolates | Total | |
| Serotype (SrT) | 35 | 5 | 11 | 51 | 0.933 | 0.823 | 0.913 |
| MLST | 57 | 6 | 10 | 73 | 0.941 | 0.805 | 0.920 |
| PFGE | 116 | 44 | 7 | 167 | 0.991 | 0.968 | 0.991 |
| SrT+PFGEa | 117 | 48 | 7 | 172 | 0.991 | 0.974 | 0.992 |
| MLST+PFGEa | 119 | 45 | 6 | 170 | 0.992 | 0.968 | 0.991 |
| SrT+MLST+PFGEa | 119 | 49 | 6 | 174 | 0.992 | 0.974 | 0.992 |
Isolates were also assigned unique subtypes based on combinations of (i) serotype (SrT) and PFGE type data, (ii) MLST and PFGE type data, and (iii) serotype (SrT), MLST, and PFGE type data.
MLST, multilocus sequence typing.
Table 3.
Distribution of Sequence Types and Pulsed-Field Gel Electrophoresis Types Among Common Salmonella Serotypesa
| |
No. of isolates from |
Among isolates, no. of different |
||
|---|---|---|---|---|
| Serotype | Cattle | Humans | STs found | PFGE types found |
| Typhimurium | 23 | 29 | 5 | 32 |
| Newport | 29 | 18 | 6 | 22 |
| Enteritidis | 0 | 26 | 2 | 8 |
| 4,5,12:i:- | 7 | 10 | 2 | 9 |
| Heidelberg | 0 | 10 | 3 | 4 |
| Montevideo | 3 | 6 | 5 | 6 |
| Thompson | 3 | 6 | 2 | 3 |
| Agona | 5 | 3 | 2 | 7 |
| Muenster | 7 | 1 | 1 | 6 |
| Infantis | 4 | 3 | 1 | 7 |
| Mbandaka | 1 | 4 | 3 | 5 |
| Saintpaul | 0 | 5 | 2 | 4 |
| Javiana | 0 | 4 | 2 | 4 |
| Urbana | 0 | 4 | 1 | 3 |
Salmonella serotypes that were found ≥4 times among the Salmonella isolate set used in this study were considered common Salmonella serotypes and are included in this table.
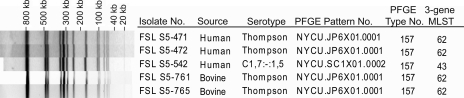
Although PFGE typing showed higher discriminatory power than serotyping and MLST, there were some instances where isolates with a given PFGE type represented multiple serotypes or STs. There were five PFGE types that each were found among isolates representing closely related serotypes (Table 4). Three PFGE types were each differentiated into two different STs: (i) two isolates with PFGE type 66 (all representing serotype Typhimurium) that were differentiated in to ST6 and ST7, (ii) four isolates with PFGE type 158 (all representing serotype Thompson) that were differentiated into ST43 and ST62, and (iii) five isolates with PFGE type 157 that were differentiated into ST43 (one isolate with serotype 1,7:-:1,5) and ST 62 (serotype Thompson) (Fig. 2 and Table 4). Combined analysis of all three subtyping methods yielded a total of 174 different combined subtypes with a Simpson's index of diversity (D = 0.992; Table 2) only marginally larger than that of PFGE alone (D = 0.991). Among these 174 combined subtypes, 119 were found among human isolates only, 49 were found among bovine isolates only, and 6 were found among both human and bovine isolates.
Table 4.
Pulsed-Field Gel Electrophoresis Patterns Shared by Salmonella Isolates with Closely Related Serotypes
| PFGE pattern | Serotype [ST]a | Antigens | Source (no. of isolates) |
|---|---|---|---|
| 89 | 4,5,12:i:- [6] | 4,5,12:i:- | Human (6), bovine (1) |
| Typhimurium [6] | 4,5,12:i:1,2 | Bovine (1) | |
| 94 | 4,5,12:i:- [6] | 4,5,12:i:- | Bovine (2) |
| Typhimurium [6] | 4,5,12:i:1,2 | Bovine (1) | |
| 121 | Newport [11] | 6,8:e,h:1,2 | Bovine (9) |
| Bardo [11] | 8:e,h:1,2 | Bovine (1) | |
| 126 | Newport [11] | 6,8:e,h:1,2 | Human (2), bovine (10) |
| Bardo [11] | 8:e,h:1,2 | Bovine (1) | |
| 157 | Thompson [62] | 6,7:k:1,5 | Human (2), bovine (2) |
| 1,7:-:1,5 [43] | 1,7:-:1,5 | Human (1) |
STs are shown in brackets.
FIG. 2.
Example of an XbaI PFGE type that was found among isolates representing two different Salmonella serotypes. PFGE type 157 was found among isolates representing two sequence types (ST43 and ST62) as well as two serotypes (1,7:-:1,5 and Thompson).
Distribution of PFGE types among human and bovine Salmonella isolates
Among the 167 PFGE types, 44 PFGE types were only found among bovine clinical isolates and 116 PFGE types were only found among human isolates. Only seven PFGE types (i.e., 60, 66, 72, 89, 126, 157, and 168) were obtained from both bovine and human clinical cases (Fig. 2). Four of these PFGE types (i.e., 60, 89, 126, and 157) represented >4 isolates each (see Table 5); for example, PFGE pattern 60 was shared by five human and three bovine Salmonella ser. Typhimurium isolates, whereas PFGE type 89 was shared by six human and one bovine Salmonella ser. 4,5,12:i:- isolates, as well as one bovine Salmonella ser. Typhimurium isolate (Table 5). Three PFGE types found among both human and bovine isolates were less common (<4 isolates per PFGE type); PFGE types 66 and 72 each represented one bovine and one human Salmonella ser. Typhimurium isolate, whereas PFGE type 168 represented two human and one bovine isolate.
Table 5.
Distribution of Common Pulsed-Field Gel Electrophoresis Patterns (i.e., Patterns Found Among >4 Isolates) Among Human and Bovine Salmonella Isolates
| |
No. of isolates from |
|
|
|---|---|---|---|
| PFGE type (serotype) | Humans | Cattle | p-Valuea |
| 27 (Enteritidis) | 9 | 0 | 0.029* |
| 32 (Enteritidis) | 6 | 0 | 0.076 |
| 57 (Heidelberg) | 7 | 0 | 0.055 |
| 60 (Typhimurium) | 5 | 3 | 0.824 |
| 89 (4,5,12:i:-, Typhimurium) | 6 | 2b | 0.287 |
| 121 (Newport, Bardo) | 0 | 10 | <0.0001** |
| 126 (Newport, Bardo) | 2 | 11 | <0.0001** |
| 157 (C 1,7:-:1,5, Thompson) | 3 | 2 | 0.768 |
| Rare PFGE typesc | 140 | 63 | 0.089 |
| Total | 178 | 91 | |
p-Values refer to comparisons of the frequency of a given PFGE type among human and bovine isolates, as determined by Fisher's exact test; p-values <0.05 indicate that a given PFGE is not independently distributed among human and bovine isolates; Symbols * and ** indicate p-values <0.05 and <0.001, respectively.
The two PFGE type 89 isolates from cattle, which represented different serotypes (4,5,12:i:- and Typhimurium), were obtained from the same farm.
The category “Rare PFGE types” includes the isolate numbers for all PFGE types that were found among ≤4 isolates.
Categorical analysis of the distribution of the eight PFGE types that represented >4 isolates (as well as one category that includes all isolates for PFGE types classified as rare [≤4 isolates]) showed that PFGE types were not independently distributed among human and bovine isolates (p-value <0.0001; Monte Carlo estimation of exact test). Subsequent categorical analyses of the distribution of the eight individual PFGE type that each represented >4 isolates (using Fisher's exact tests and individual 2 × 2 tables) showed that PFGE types 121 and 126 (both associated with serotypes Newport and Bardo; see Table 5) were significantly over-represented among bovine isolates (p < 0.001 for each). PFGE type 126 represented 2 human and 11 bovine isolates, which had been obtained from 10 different farms in 7 counties in New York State and Vermont, whereas PFGE type 121 represented 10 bovine isolates, which had been obtained from 10 different farms in 6 counties in New York State, but was not found among the human isolates. Only one PFGE type (type 27) was significantly (p < 0.05) overrepresented among human clinical isolates as compared with bovine isolates (Table 5).
Evidence of temporal and spatial clusters of Salmonella subtypes
Among the 178 human isolates, we identified six instances where two or three isolates with the same subtype (i.e., same serotype, MLST, and PFGE type) had been obtained from human patients in the same county in the same or consecutive months (see Table 6), possibly indicating small temporal and geographical case clusters. PFGE types linked to four of these clusters (i.e., PFGE types 14, 47, 86, and 151) were not isolated outside these clusters (Table 6 and Supplemental Table S1). On the other hand, PFGE type 27 (which represented a cluster with two cases) was found among another seven human isolates (thus representing 5.1% of all human isolates), whereas PFGE type 157 (which represented another cluster with two cases) was also found among two bovine isolates (see Supplemental Table S1).
Table 6.
Possible Spatial and Temporal Clusters of Human Salmonella Cases as Determined by Pulsed-Field Gel Electrophoresisa
| Isolate no. | Date of isolation | County | Serotype | MLST | PFGE pattern (pattern frequency among 178 human isolates) |
|---|---|---|---|---|---|
| FSL S5-529 | 9/15/2004 | Erie | Anatum | 25 | 151 (1.7%)b |
| FSL S5-530 | 9/14/2004 | Erie | Anatum | 25 | 151 (1.7%)b |
| FSL S5-540 | 9/22/2004 | Erie | Anatum | 25 | 151 (1.7%)b |
| FSL S5-369 | 12/22/2003 | Monroe | Saintpaul | 38 | 86 (1.1%)b |
| FSL S5-405 | 12/23/2003 | Monroe | Saintpaul | 38 | 86 (1.1%)b |
| FSL S5-376 | 12/31/2003 | Nassau | Enteritidis | 14 | 27 (5.1%) |
| FSL S5-377 | 12/31/2003 | Nassau | Enteritidis | 14 | 27 (5.1%) |
| FSL S5-471 | 5/4/2004 | Nassau | Thompson | 62 | 157 (1.7%) |
| FSL S5-472 | 5/3/2004 | Nassau | Thompson | 62 | 157 (1.7%) |
| FSL S5-456 | 4/22/2004 | Orleans | Schwarzengrund | 4 | 14 (1.1%)b |
| FSL S5-458 | 4/30/2004 | Orleans | Schwarzengrund | 4 | 14 (1.1%)b |
| FSL S5-388 | 1/11/2004 | Schenectady | Urbana | 52 | 47 (1.1%)b |
| FSL S5-410 | 2/28/2004 | Schenectady | Urbana | 52 | 47 (1.1%)b |
Two or more human cases were considered a possible cluster if isolates from cases in a given county shared the same PFGE type and were obtained in the same or consecutive months.
A PFGE pattern was only found among the isolates representing a possible spatial and temporal cluster listed in this table.
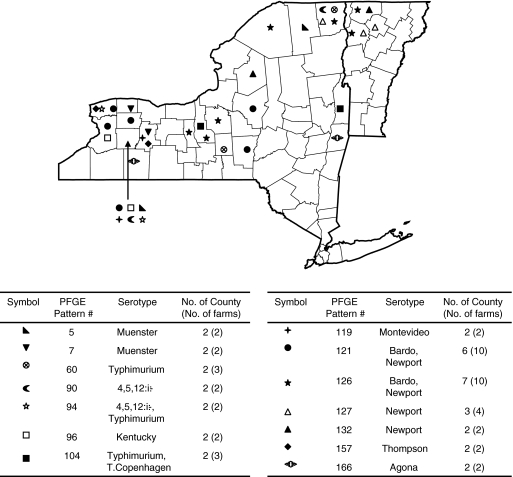
Analysis of the spatial distribution of bovine isolates that shared the same PFGE pattern showed that (i) nine PFGE types (PFGE types 5, 7, 60, 90, 94, 104, 132, 157, and 166) represented isolates from at least two farms in two different nonadjacent counties and that (ii) two PFGE types (PFGE types 121 and 126) represented isolates from 10 farms in 6 and 7 counties, suggesting that these are commonly found and widely distributed subtypes (Fig. 3). In addition, three PFGE types (PFGE types 96, 119, and 127) were collected from multiple farms in two or three adjacent counties, possibly representing spatial clusters. Preliminary analyses of the geographical location of isolates with PFGE types that are shared between humans and cattle found no instance where human and bovine isolates with matching PFGE types were obtained from the same county (see Supplemental Fig. S2, available online at www.liebertonline.com); our data set was too small for more formal spatial cluster analyses.
FIG. 3.
Geographical location of farms where bovine Salmonella isolates with PFGE types that were found among isolates from two or more farms were obtained. The map shows both New York State and Vermont and county borders within each state; location of farms is indicated at the county level; placement of the symbol does not represent actual locations of a farm within a county.
Discussion
A total of 335 clinical human and bovine Salmonella isolates, collected from New York and a neighboring state (Vermont), were characterized by PFGE to provide a better understanding of the genetic relationship, diversity, and epidemiology of human- and cattle-associated Salmonella. Combined with previously reported MLST and serotype data for virtually the same isolates set (Alcaine et al., 2006), our data indicate that (i) PFGE provides for highly discriminatory subtyping of human and bovine Salmonella isolates, particularly as compared with serotyping and MLST; (ii) although the majority of Salmonella PFGE types are uncommon, some PFGE types are widely distributed and some seem associated with specific hosts; (iii) PFGE typing can identify temporal and spatial clusters of human and bovine Salmonella cases that may represent small outbreaks; and (iv) specific Salmonella PFGE types can persist on dairy farms, but may diversify over time.
PFGE provides higher discrimination of human and bovine Salmonella isolates than serotyping and MLST
Our finding that PFGE provides for more discriminatory subtyping than serotyping and MLST is consistent with previous studies (Fakhr et al., 2005; Harbottle et al., 2006; Cooke et al., 2008), even though a number of these previous studies have focused on comparing discriminatory power of different subtyping methods among one or a few specific Salmonella serotypes. For example, Harbottle et al. (2006) reported that 81 Salmonella ser. Newport isolates from humans, feed, and foods were differentiated into 12 STs (using a seven-gene MLST scheme) and 43 XbaI PFGE types. In another study, 85 bovine clinical Salmonella ser. Typhimurium isolates all represented the same ST based on a four-gene sequence typing scheme, but were differentiated into 50 XbaI PFGE types (Fakhr et al., 2005). Similarly, other researchers reported that PFGE provided more discriminatory power for differentiation of Salmonella isolates as compared with ribotyping (Ridley et al., 1998; Fontana et al., 2003) and randomly amplified polymorphic DNA (RAPD) (Hudson et al., 2000; Lim et al., 2005). Although an increasing number of data (Lindstedt et al., 2004; Cho et al., 2008; Davis et al., 2009) indicate that MLVA can provide improved subtype discrimination particularly for highly clonal Salmonella serotypes (e.g., Enteritidis, Newport, and Typhimurium), our data indicate that due to its high discriminatory power among diverse serotypes, PFGE still remains the best choice for initial population-based subtype surveillance of Salmonella, particularly since all MLVA schemes described so far are serotype specific (Lindstedt et al., 2004; Davis et al., 2009) and cannot be applied to isolates representing diverse serotypes.
Although the majority of Salmonella PFGE types are uncommon, some PFGE types are widely distributed and some seem associated with specific hosts
Although many PFGE types found here only represented a single isolate, eight PFGE types represented >4 isolates (not counting re-isolation of a specific PFGE type on a given farm). It was not surprising that, among these common PFGE types, two PFGE types associated with serotype Enteritidis and one associated with serotype Heidelberg were only found among human but not among bovine isolates, as it is well known that these serotypes are predominantly associated with poultry and poultry products (DuPont, 2007; Gantois et al., 2009). Interestingly, two PFGE types (both representing serotypes Newport and Bardo, which are closely related serotypes) were not only commonly found, but also significantly overrepresented among bovine isolates, including one PFGE type (121) that represented 10 bovine and no human isolates. Although one may hypothesize that these two PFGE represent host-adapted strains (possible adapted to bovine hosts), further phenotypic and epidemiological studies will be needed to test this hypothesis. This hypothesis is, though, consistent with previous MLST studies, which have shown that the ST presented by these two PFGE types (i.e., ST 11) is associated with bovine cases (Sukhnanand et al., 2005; Alcaine et al., 2006). Other previous studies have also shown that some Salmonella serotypes include host-adapted subtypes; for example, Salmonella ser. Typhimurium phage type DT40 was shown to represent an avian-adapted Salmonella ser. Typhimurium phage type (Rabsch et al., 2002). We also identified a small number (i.e., seven) PFGE types that represented both human and bovine isolates, consistent with a recent PFGE study (Oloya et al., 2009) that identified eight PFGE patterns found among both human and animal isolates. Comprehensive subtype characterization of human and animal Salmonella isolates thus may help identify host specific or host-adapted Salmonella subtypes. Although host-specific subtypes are defined as those that only infect very specific host species (e.g., Salmonella ser. Typhi), host adapted subtypes preferentially infect one host species but can also cause disease in other host species (e.g., such as Salmonella ser. Typhimurium). Identification of host-specific or host-adapted Salmonella subtypes will improve not only our understanding of Salmonella biology, but also the ability to track outbreak sources and to perform subtype-based source attribution (Hald et al., 2004).
PFGE typing can identify temporal and spatial clusters of Salmonella cases that may represent small outbreaks
PFGE data can be critical for detection of human salmonellosis outbreaks (CDC, 2008a, 2008b, 2008c, 2009; Lomonaco et al., 2008; Smith et al., 2008) and clusters, as also supported by detection of six single-county clusters of human salmonellosis cases in this study. Although epidemiological data would be needed to determine whether these clusters represent true single-source outbreaks, the observed patterns and the fact that four clusters represented PFGE types that were not identified outside a given cluster suggest that at least some of these clusters represent small outbreaks, possibly linked to localized sources, for example, restaurants (CDC, 2008b), group events where food is served (Camps et al., 2005), or even nonfood sources (CDC, 2008a). PFGE-based identification of single-source disease clusters and source tracking by PFGE is complicated by the fact that some PFGE types may be common and widely distributed (Woo, 2005; Lindqvist and Pelkonen, 2007). Identification of such a common PFGE type in multiple human or animal cases or among clinical cases and a possible source is more likely to be by chance and may sometimes not represent a causal relationship. Establishment of causal relationships in these cases will require strong epidemiological linkages and/or the use of additional, more sensitive subtyping methods such as MLVA (Hyytia-Trees et al., 2006; Cho et al., 2008; Malorny et al., 2008), microarray (Garaizar et al., 2002; Reen et al., 2005; Scaria et al., 2008), or whole-genome sequencing (Cooke et al., 2008). Although PFGE thus represents a sensitive subtyping approach that is suitable for surveillance, data on PFGE type frequency among different sources (e.g., human clinical cases, animals, and foods) are needed to (i) evaluate the relative significance of a subtype match and (ii) decide when additional subtyping methods may be needed to establish causal relationships with a high confidence (recognizing that, in all cases, epidemiological data supporting linkages are also needed) (Gerner-Smidt et al., 2006).
Specific Salmonella PFGE types can persist on dairy farms, but may diversify over time
PFGE data further supported prior MLST-based findings (Alcaine et al., 2006), on the same isolate sets, which suggested persistence of specific Salmonella subtypes on different farms. One cannot, though, completely exclude re-introduction of a PFGE type on a given farm, for example, from wildlife or other farms, as the cause of re-isolation of a specific subtype on a given farm. Overall, our data are consistent with a number of studies that also have provided evidence for Salmonella persistence on farms and in flocks (Vanselow et al., 2007; Pedersen et al., 2008). For example, Ogilvie (1986) reported that, on one dairy farm, an asymptomatic cow shed Salmonella ser. Typhimurium in her milk over a 36-day period (Ogilvie, 1986). In another study, Vanselow et al. (2007) reported that Salmonella ser. Typhimurium, which caused severe salmonellosis among dairy cows and calves, persisted in a dairy herd over approximately 2 years (Vanselow et al., 2007). Another study reported long-term persistence of specific Salmonella ser. Newport PFGE types in two dairy herds (Cobbold et al., 2006). Interestingly, Van Kessel et al. (2007) reported that Salmonella ser. Cerro isolates persisted in a dairy farm for almost two years without causing clinical consequences among the herd (Van Kessel et al., 2007). Besides Salmonella ser. Newport and ser. Typhimurium, we also found evidence that Salmonella ser. 4,5,12:i:-, ser. Infantis, and ser. Kentucky persisted in dairy herds over time.
Interestingly, in a number of farms Salmonella subtypes identified by MLST as persisting (Alcaine et al., 2006) represented multiple XbaI PFGE types that typically differed by ≤3 bands. Although these findings suggest that Salmonella isolates diversify during persistence, one cannot entirely exclude that, at least in some farms, a Salmonella subtype closely related to a PFGE type already present was introduced independently. Overall, our findings are consistent with reports that human salmonellosis outbreaks (Laconcha et al., 1998; CDC, 2002) as well as outbreaks associated with other bacterial pathogens (Graves et al., 2005) can be caused by multiple closely related PFGE types, which presumably diversified over the duration of an outbreak (or during persistence in the outbreak source). Diversification over short time periods most likely occurs due to plasmid losses or gains or changes in prophages or other mobile genetic elements (Baggesen et al., 1997; Barrett et al., 2006). Combined, these studies indicate that the so-called three-band rule, which states that isolates that differ by ≤3 bands may be so closely related that they can share a recent common ancestor and that they can be considered related (given appropriate epidemiological evidence) (Tenover et al., 1995), may sometimes need to be applied when investigating salmonellosis outbreaks.
Conclusions
Our data reported here clearly indicate the value of comprehensive PFGE characterization of human- and animal-associated Salmonella isolates, not only for surveillance and outbreak detection, but also for gaining an improved understanding of Salmonella biology, ecology, and transmission. Future molecular subtype studies on coclustering of human- and bovine-associated Salmonella cases with identical subtypes will be critical to further improve our understanding of the transmission of this pathogen, including possible direct zoonotic transmission. Utilizing the full potential of PFGE will require continued development of large subtype databases (such as PulseNet) (Swaminathan et al., 2001) that encompass isolates from different source populations and are publicly accessible. Our data also indicate that XbaI PFGE sometimes may be, what could be considered, too discriminatory for subtype differentiation of Salmonella by yielding different PFGE patterns for isolates that share a very recent common ancestor. In this case, additional use of a less discriminatory subtyping method, for example, MLST, may sometimes facilitate appropriate interpretation of PFGE data. On the other hand, XbaI PFGE also may sometimes be not discriminatory enough by classifying isolates from different farms and sources with no epidemiological linkages into identical PFGE types. In these cases, PFGE typing with additional enzymes (Zamperini et al., 2007; CDC, 2009) and/or use of more discriminatory subtyping methods, for example, MLVA (Cho et al., 2008; Malorny et al., 2008; Davis et al., 2009), may be required.
Supplementary Material
Acknowledgments
This work was supported by Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. NO1-A1-30054. We thank Eric B. Fugett for help with laboratory techniques used in this study.
Disclosure Statement
No competing financial interests exist.
References
- Agasan A. Kornblum J. Williams G, et al. Profile of Salmonella enterica subsp. enterica (subspecies I) serotype 4,5,12:i:- strains causing food-borne infections in New York City. J Clin Microbiol. 2002;40:1924–1929. doi: 10.1128/JCM.40.6.1924-1929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaine SD. Soyer Y. Warnick LD, et al. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl Environ Microbiol. 2006;72:7575–7585. doi: 10.1128/AEM.01174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggesen DL. Wegener HC. Madsen M. Correlation of conversion of Salmonella enterica serovar enteritidis phage type 1, 4, or 6 to phage type 7 with loss of lipopolysaccharide. J Clin Microbiol. 1997;35:330–333. doi: 10.1128/jcm.35.1.330-333.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TJ. Gerner-Smidt P. Swaminathan B. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog Dis. 2006;3:20–31. doi: 10.1089/fpd.2006.3.20. [DOI] [PubMed] [Google Scholar]
- Brenner FW. Villar RG. Angulo FJ, et al. Salmonella nomenclature—guest commentary. J Clin Microbiol. 2000;38:2465–2467. doi: 10.1128/jcm.38.7.2465-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps N. Dominguez A. Company M, et al. A foodborne outbreak of Salmonella infection due to overproduction of egg-containing foods for a festival. Epidemiol Infect. 2005;133:817–822. doi: 10.1017/S0950268805004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Outbreak of multidrug-resistant Salmonella Newport-United States, January–April 2002. Morb Mortal Wkly Rep. 2002;51:545–548. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Multistate outbreak of human Salmonella infections associated with exposure to turtles—United States, 2007–2008. Morb Mortal Wkly Rep. 2008a;57:69–72. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Multistate outbreak of Salmonella infections associated with frozen pot pies—United States, 2007. Morb Mortal Wkly Rep. 2008b;57:1277–1280. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Salmonella Litchfield outbreak associated with a hotel restaurant—Atlantic City, New Jersey, 2007. Morb Mortal Wkly Rep. 2008c;57:775–779. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Multistate outbreak of Salmonella infections associated with peanut butter and peanut butter containing products—United States, 2008–2009. Morb Mortal Wkly Rep. 2009;58:85–90. [PubMed] [Google Scholar]
- Cho S. Whittam TS. Boxrud DJ, et al. Allele distribution and genetic diversity of VNTR loci in Salmonella enterica serotype Enteritidis isolates from different sources. BMC Microbiol. 2008;8:146–157. doi: 10.1186/1471-2180-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold RN. Rice DH. Davis MA, et al. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J Am Vet Med Assoc. 2006;228:585–591. doi: 10.2460/javma.228.4.585. [DOI] [PubMed] [Google Scholar]
- Cooke FJ. Brown DJ. Fookes M, et al. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J Bacteriol. 2008;190:8155–8162. doi: 10.1128/JB.00636-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA. Baker KN. Call DR, et al. Multilocus variable-number tandem-repeat method for typing Salmonella enterica serovar Newport. J Clin Microbiol. 2009;47:1934–1938. doi: 10.1128/JCM.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPont HL. The growing threat of foodborne bacterial enteropathogens of animal origin. Clin Infect Dis. 2007;45:1353–1361. doi: 10.1086/522662. [DOI] [PubMed] [Google Scholar]
- Ewing WH. The genus Salmonella. In: Edwards RE, editor; Ewing WH, editor. Identification of Enterobacteriaceae. 3rd. Minneapolis: Burgess Publishing Co.; 1972. pp. 146–256. [Google Scholar]
- Fakhr MK. Nolan LK. Logue CM. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J Clin Microbiol. 2005;43:2215–2219. doi: 10.1128/JCM.43.5.2215-2219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J. Stout A. Bolstorff B, et al. Automated ribotyping and pulsed-field gel electrophoresis for rapid identification of multidrug-resistant Salmonella serotype Newport. Emerg Infect Dis. 2003;9:496–499. doi: 10.3201/eid0904.020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantois I. Ducatelle R. Pasmans F, et al. Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol Rev. 2009;33:718–738. doi: 10.1111/j.1574-6976.2008.00161.x. [DOI] [PubMed] [Google Scholar]
- Garaizar J. Porwollik S. Echeita A, et al. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J Clin Microbiol. 2002;40:2074–2078. doi: 10.1128/JCM.40.6.2074-2078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner-Smidt P. Hise K. Kincaid J, et al. PulseNet USA: a five-year update. Foodborne Pathog Dis. 2006;3:9–19. doi: 10.1089/fpd.2006.3.9. [DOI] [PubMed] [Google Scholar]
- Grandesso F. Jourdan-da Silva N. Le Hello S, et al. Excess of infections due to a multi-drug sensitive Salmonella enterica serotype Typhimurium in France in June 2008. Euro Surveill. 2008;13(pii):19022. [PubMed] [Google Scholar]
- Graves LM. Hunter SB. Ong AR, et al. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J Clin Microbiol. 2005;43:2350–2355. doi: 10.1128/JCM.43.5.2350-2355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont P. Weil F. Antigenic Formulae of the Salmonella Serovars. Paris: World Health Organization Centre for Reference and Research on Salmonella, Pasteur Institute; 2007. [Google Scholar]
- Hald T. Vose D. Wegener HC, et al. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24:255–269. doi: 10.1111/j.0272-4332.2004.00427.x. [DOI] [PubMed] [Google Scholar]
- Harbottle H. White DG. McDermott PF, et al. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J Clin Microbiol. 2006;44:2449–2457. doi: 10.1128/JCM.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CR. Quist C. Lee MD, et al. Genetic relatedness of Salmonella isolates from nondomestic birds in Southeastern United States. J Clin Microbiol. 2000;38:1860–1865. doi: 10.1128/jcm.38.5.1860-1865.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR. Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SB. Vauterin P. Lambert-Fair MA, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytia-Trees E. Smole SC. Fields PA, et al. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157) Foodborne Pathog Dis. 2006;3:118–131. doi: 10.1089/fpd.2006.3.118. [DOI] [PubMed] [Google Scholar]
- Laconcha I. Lopez-Molina N. Rementeria A, et al. Phage typing combined with pulsed-field gel electrophoresis and random amplified polymorphic DNA increases discrimination in the epidemiological analysis of Salmonella enteritidis strains. Int J Food Microbiol. 1998;40:27–34. doi: 10.1016/s0168-1605(98)00007-5. [DOI] [PubMed] [Google Scholar]
- Liesegang A. Davos D. Balzer JC, et al. Phage typing and PFGE pattern analysis as tools for epidemiological surveillance of Salmonella enterica serovar Bovismorbificans infections. Epidemiol Infect. 2002;128:119–130. doi: 10.1017/s0950268801006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. Lee KH. Hong CH, et al. Comparison of four molecular typing methods for the differentiation of Salmonella spp. Int J Food Microbiol. 2005;105:411–418. doi: 10.1016/j.ijfoodmicro.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Lindqvist N. Pelkonen S. Genetic surveillance of endemic bovine Salmonella Infantis infection. Acta Vet Scand. 2007;49:15–23. doi: 10.1186/1751-0147-49-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt BA. Vardund T. Aas L, et al. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J Microbiol Methods. 2004;59:163–172. doi: 10.1016/j.mimet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Lomonaco S. Nucera D. Griglio B, et al. Real-time subtyping via PFGE reveals potential epidemiological relatedness among human salmonellosis cases in Northern Italy. Lett Appl Microbiol. 2008;47:227–234. doi: 10.1111/j.1472-765x.2008.02393.x. [DOI] [PubMed] [Google Scholar]
- Lukinmaa S. Nakari UM. Liimatainen A, et al. Genomic diversity within phage types of Salmonella enterica ssp. enterica serotypes Enteritidis and Typhimurium. Foodborne Pathog Dis. 2006;3:97–105. doi: 10.1089/fpd.2006.3.97. [DOI] [PubMed] [Google Scholar]
- Malorny B. Junker E. Helmuth R. Multi-locus variable-number tandem repeat analysis for outbreak studies of Salmonella enterica serotype Enteritidis. BMC Microbiol. 2008;8:84–91. doi: 10.1186/1471-2180-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase T. Nagato M. Shirota K, et al. Pulsed-field gel electrophoresis-based subtyping of DNA degradation-sensitive Salmonella enterica subsp enterica serovar Livingstone and serovar Cerro isolates obtained from a chicken layer farm. Vet Microbiol. 2004;99:139–143. doi: 10.1016/j.vetmic.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Ogilvie TH. The persistent isolation of Salmonella typhimurium from the mammary gland of a dairy cow. Can Vet J. 1986;27:329–331. [PMC free article] [PubMed] [Google Scholar]
- Oloya J. Doetkott D. Khaitsa ML. Antimicrobial drug resistance and molecular characterization of Salmonella isolated from domestic animals, humans, and meat products. Foodborne Pathog Dis. 2009;6:273–284. doi: 10.1089/fpd.2008.0134. [DOI] [PubMed] [Google Scholar]
- Pedersen TB. Olsen JE. Bisgaard M. Persistence of Salmonella Senftenberg in poultry production environments and investigation of its resistance to desiccation. Avian Pathol. 2008;37:421–427. doi: 10.1080/03079450802216561. [DOI] [PubMed] [Google Scholar]
- Rabsch W. Andrews HL. Kingsley RA, et al. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun. 2002;70:2249–2255. doi: 10.1128/IAI.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen FJ. Boyd EF. Porwollik S, et al. Genomic comparisons of Salmonella enterica serovar Dublin, Agona, and Typhimurium strains recently isolated from milk filters and bovine samples from Ireland, using a Salmonella microarray. Appl Environ Microbiol. 2005;71:1616–1625. doi: 10.1128/AEM.71.3.1616-1625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot EM. Fair MA. Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Ridley AM. Threlfall EJ. Rowe B. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1998;36:2314–2321. doi: 10.1128/jcm.36.8.2314-2321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlström L. de Jong B. Aspan A. Salmonella isolated in sewage sludge traced back to human cases of salmonellosis. Lett Appl Microbiol. 2006;43:46–52. doi: 10.1111/j.1472-765X.2006.01911.x. [DOI] [PubMed] [Google Scholar]
- Scaria J. Palaniappan RU. Chiu D, et al. Microarray for molecular typing of Salmonella enterica serovars. Mol Cell Probes. 2008;22:238–243. doi: 10.1016/j.mcp.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YS. Lee SH. Shin EK, et al. Pulsed-field gel electrophoresis genotyping of Salmonella gallinarum and comparison with random amplified polymorphic DNA. Vet Microbiol. 2006;115:349–357. doi: 10.1016/j.vetmic.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Smith KE. Medus C. Meyer SD, et al. Outbreaks of salmonellosis in Minnesota (1998 through 2006) associated with frozen, microwaveable, breaded, stuffed chicken products. J Food Prot. 2008;71:2153–2160. doi: 10.4315/0362-028x-71.10.2153. [DOI] [PubMed] [Google Scholar]
- Sukhnanand S. Alcaine S. Warnick LD, et al. DNA sequence-based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. J Clin Microbiol. 2005;43:3688–3698. doi: 10.1128/JCM.43.8.3688-3698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B. Barrett TJ. Hunter SB, et al. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7:382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B. Gerner-Smidt P. Barrett T. Focus on Salmonella. Foodborne Pathog Dis. 2006a;3:154–156. doi: 10.1089/fpd.2006.3.154. [DOI] [PubMed] [Google Scholar]
- Swaminathan B. Gerner-Smidt P. Ng LK, et al. Building PulseNet International: an interconnected system of laboratory networks to facilitate timely public health recognition and response to foodborne disease outbreaks and emerging foodborne diseases. Foodborne Pathog Dis. 2006b;3:36–50. doi: 10.1089/fpd.2006.3.36. [DOI] [PubMed] [Google Scholar]
- Tenover FC. Arbeit RD. Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel-electrophoresis—criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S. Brown DJ. Wallis T, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125:229–255. doi: 10.1017/s0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kessel JS. Karns JS. Wolfgang DR, et al. Longitudinal study of a clonal, subclinical outbreak of Salmonella enterica subsp. enterica serovar Cerro in a U.S. dairy herd. Foodborne Pathog Dis. 2007;4:449–461. doi: 10.1089/fpd.2007.0033. [DOI] [PubMed] [Google Scholar]
- Vanselow BA. Hum S. Hornitzky MA, et al. Salmonella Typhimurium persistence in a Hunter Valley dairy herd. Aust Vet J. 2007;85:446–450. doi: 10.1111/j.1751-0813.2007.00224.x. [DOI] [PubMed] [Google Scholar]
- Wedel SD. Bender JB. Leano FT, et al. Antimicrobial-drug susceptibility of human and animal Salmonella typhimurium, Minnesota, 1997–2003. Emerg Infect Dis. 2005;11:1899–1906. doi: 10.3201/eid1112.050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M. Molecular subtyping methods for Listeria monocytogenes. J AOAC Int. 2002;85:524–531. [PubMed] [Google Scholar]
- Woo YK. Finding the sources of Korean Salmonella enterica serovar Enteritidis PT 4 isolates by pulsed-field gel electrophoresis. J Microbiol. 2005;43:424–429. [PubMed] [Google Scholar]
- Woo YK. Lee SH. Genetic diversity of multi-resistant Salmonella enterica serotype Typhimurium isolates from animals and humans. J Microbiol. 2006;44:106–112. [PubMed] [Google Scholar]
- Zamperini K. Soni V. Waltman D, et al. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:- from poultry is a variant Typhimurium serovar. Avian Dis. 2007;51:958–964. doi: 10.1637/7944-021507-REGR.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.