Abstract
The focus of this study was Salmonella enterica serotype Cerro, a potentially emerging pathogen of cattle. Our objectives were to document the within-herd prevalence of Salmonella Cerro among a sample of New York dairy herds, to describe the antimicrobial resistance patterns and pulsed-field gel electrophoresis types of the isolates, and to elucidate the status of this serotype as a bovine pathogen. Data were collected prospectively from dairy herds throughout New York that had at least 150 lactating cows and that received clinical service from participating veterinarians. Following enrollment, Salmonella surveillance consisted of both environmental screening and disease monitoring within the herd. Herds positive by either environmental or fecal culture were sampled during three visits to estimate the within-herd prevalence of Salmonella. Among 57 enrolled herds, 44 (77%) yielded Salmonella-positive samples during the study period. Of these, 20 herds (46%) were positive for Salmonella Cerro. Upon follow-up sampling for estimation of prevalence, Cerro was identified in 10 of the 20 herds; the median within-herd Cerro prevalence was 17%, with a maximum of 53%. Antimicrobial resistance ranged from zero to nine drugs, and eight (40%) of the Cerro-positive farms generated drug-resistant isolates. Eight XbaI pulsed-field gel electrophoresis types were represented among 116 isolates tested, although 89% of these isolates shared the predominant type. Among herds with clinical cases, cattle that had signs consistent with salmonellosis were more likely to test positive for Cerro than apparently healthy cattle, as estimated by a logistic regression model that controlled for herd as a random effect (odds ratio: 3.9). There is little in the literature concerning Salmonella Cerro, and published reports suggest an absence of disease association in cattle. However, in our region there has been an apparent increase in the prevalence of this serotype among cattle with salmonellosis. Other Salmonella serotypes important to bovine health have emerged to become leading causes of human foodborne disease, and close monitoring of Cerro is warranted.
Introduction
Salmonella enterica is a zoonotic pathogen that poses a considerable threat to public health, resulting in approximately 1.4 million illnesses, 16,000 hospitalizations, and between 400 and 600 deaths annually in the United States alone (Mead et al., 1999; Voetsch et al., 2004). People generally become infected with Salmonella through foodborne exposure or direct contact with infected animals (Mead et al., 1999; L Plym and Wierup, 2006). Human infections may be asymptomatic or result in varying degrees of clinical disease (Jones et al., 2008), ranging from self-limiting acute enteritis to sepsis and death. The prevalence of multidrug resistance among Salmonella strains has increased over the past two decades (Glynn et al., 1998; Dunne et al., 2000; Gupta et al., 2003; Davis et al., 2007), causing an increase in treatment failures and hospitalization rates (Helms et al., 2002, 2004; Varma et al., 2005a, 2005b). Over 2500 Salmonella serotypes have been identified to date, but relatively few are responsible for a large proportion of clinical infections (Jones et al., 2008). Preliminary Centers for Disease Control and Prevention (CDC) FoodNet data from 2008 show that 10 Salmonella serotypes comprised 73% of the laboratory-confirmed cases of disease, based on surveillance in 10 states (CDC, 2009b).
Salmonella is also an important cause of clinical illness in both calves and adult dairy cattle. Salmonellosis can be a costly disease for producers on account of treatment expenses, mortality, reduced milk yield, and weight loss within the herd (Peters, 1985; Huston et al., 2002b). Clinical signs of salmonellosis in cattle may include diarrhea, fever, anorexia, dehydration, decreased milk production, abortion, and evidence of endotoxemia, although many infections remain subclinical (Divers and Peek, 2008). As in people, however, relatively few serotypes are recognized as causing the majority of clinical disease. For example, a recent field study on the incidence of salmonellosis among 831 dairy herds in the northeastern United States found that just seven Salmonella serotypes accounted for 87% of the cases, namely Newport, Typhimurium (including the Copenhagen variant), Infantis, 4,5,12:i:-, Agona, Muenster, and Kentucky (Cummings et al., 2009). In contrast, the serotypes most commonly isolated from clinically healthy cattle differ from those that most frequently cause disease. According to the three most recent National Animal Health Monitoring System (NAHMS) Dairy reports (1996, 2002, and 2007), Salmonella serotypes Meleagridis, Montevideo, and Mbandaka were consistently among the most prevalent serotypes isolated from healthy lactating cows (CEAH, 2009). Clearly there is a great diversity in Salmonella serotypes shed by dairy cattle, both in terms of serotype numbers and their pathogenicity.
The focus of this study was S. enterica serotype Cerro, an infrequent human pathogen (CDC, 2008) whose role in causing clinical disease in cattle is unclear (Huston et al., 2002a; Peek et al., 2004; Van Kessel et al., 2007). Our objectives were to document the within-herd prevalence of Salmonella Cerro among a sample of dairy herds throughout New York, to describe the antimicrobial resistance patterns and pulsed-field gel electrophoresis (PFGE) types of the Cerro isolates, and to further clarify the status of this serotype as a bovine pathogen.
Materials and Methods
Study design
As part of a larger project to evaluate the prevalence of fecal Salmonella shedding among dairy herds with clinical versus subclinical infections (Cummings et al., 2010), data were collected prospectively from a convenience sample of herds throughout New York that had at least 150 lactating cows and that received clinical service from participating veterinarians. Following enrollment, Salmonella surveillance consisted of both environmental screening and disease monitoring within the herd, for a period of at least 12 months. Environmental surveillance involved the repeated collection of samples from four locations per herd for Salmonella culture (cow housing, calf housing, manure storage area, and sick pen); the targeted interval between sample collections was monthly. In addition, veterinarians submitted fecal samples from suspected clinical cases for Salmonella culture. The diagnostic criteria provided to the veterinarians included diarrhea with blood, mucus, or foul odor, fever of at least 103°F, depression, and decreased appetite, as well as sudden death in the absence of specific clinical signs or death following a course of diarrhea. To encourage the submission of samples from every clinical suspect animal, all shipping and laboratory costs were covered by the study. A positive culture result arising by either surveillance method prompted a series of three herd visits (at 4–8-week intervals) for cattle sampling by project personnel, with the goal of estimating the within-herd prevalence of Salmonella. The number of animals sampled at each visit ranged from 50 to 70, depending on herd size (<500 lactating cows: 50 animals sampled, 500–1000 lactating cows: 60, and >1000 lactating cows: 70). A subset of each sample was comprised of preweaned calves, that is, a total of 10, 15, and 20 calves made up the samples of 50, 60, and 70 animals, respectively. A conscious effort was made to sample cattle from each pen on the farm, and animals within a given pen were sampled systematically (every nth animal) to the extent possible. No attempt was made to collect samples from the same cattle during the subsequent herd visits, though some animals may have been sampled again by chance.
Sample collection and processing
Environmental samples were collected using sterile 4 × 4 inch gauze pads saturated in double-strength skim milk, which had been placed beforehand into a sterile flip-top container. For each of the four sampling locations per farm, four different gauze pads were used to collect samples and were subsequently pooled into a single flip-top container. Locations sampled in the cow housing area included four sites on the floor within high-traffic sections of the barn. Calf housing samples consisted of either four swabs of the floor in group housing areas or four swabs of the bedding in individual hutches or pens. Manure storage areas were sampled by sticking an instrument deep into the lagoon or slurry pit and then swabbing it. Sick pen samples consisted of either four swabs of the floor in group pens or four swabs of the bedding in individual sick pens. All environmental samples were maintained at 4°C until processing; samples were shipped to the research laboratory for bacteriologic culture.
Fecal samples from suspected clinical cases were collected by veterinarians via rectal retrieval, with a new glove being used to collect each sample. Approximately 10 g of fecal matter was placed into a Para-Pak bottle (Meridian Bioscience, Cincinnati, OH) and sealed. These samples were shipped to the Animal Health Diagnostic Center (College of Veterinary Medicine, Cornell University, Ithaca, NY) for bacteriologic culture.
Fecal samples obtained by project personnel during the three monthly visits were collected via rectal retrieval, again with a new glove being used to collect each sample. Approximately 10 g of fecal matter was placed into a Para-Pak bottle and sealed. All of these samples were transported to the research laboratory for bacteriologic culture.
Standard culture methods were used to isolate Salmonella from fecal samples. Fecal swab specimens from each sample container were enriched in 10 mL tetrathionate broth (Difco, Detroit, MI) containing 0.2 mL of iodine solution; the mixture was incubated at 42°C for 18–24 hours. After incubation, the sample–broth mixture was streaked onto brilliant green agar with novobiocin (Becton Dickinson and Company, Franklin Lakes, NJ) and xylose lysine tergitol 4 (XLT-4) selective media, and both plates were incubated at 37°C for 18–24 hours. Red colonies (lactose nonfermenting bacteria) on brilliant green agar with novobiocin and black colonies (H2S-producing bacteria) on XLT-4 were inoculated into Kligler iron agar slants and then incubated at 37°C for 18–24 hours. XLT-4 plates without suspected colonies were reincubated at 37°C for an additional 18–24 hours before checking again for characteristic black colonies. Colonies on Kligler iron agar slants that exhibited the biochemical properties of Salmonella were then serogrouped by slide agglutination using standard protocols. Those colonies that were positive by slide agglutination were then identified as Salmonella using the Sensititre Automated Microbiology System's A80 panel (TREK Diagnostic Systems, Cleveland, OH). Confirmed Salmonella isolates were sent to the United States Department of Agriculture, Animal and Plant Health Inspection Service (APHIS) National Veterinary Services Laboratories in Ames, Iowa, for serotyping using standard protocols.
Antimicrobial susceptibility testing
Antimicrobial susceptibility of Salmonella isolates was determined by use of the broth dilution method. Minimum inhibitory concentrations (MICs) were established for each isolate against a panel of up to 15 antimicrobial agents (Sensititre; TREK Diagnostic Systems). The panel used for Salmonella organisms isolated via environmental and follow-up sampling included 15 drugs (amikacin, amoxicillin/clavulanic acid, ampicillin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, tetracycline, and trimethoprim/sulfamethoxazole); the panel used for clinical isolates included 11 drugs (ampicillin, ceftiofur, chlortetracycline, enrofloxacin, florfenicol, gentamicin, neomycin, oxytetracycline, spectinomycin, sulfadimethoxine, and trimethoprim/sulfamethoxazole). Clinical and Laboratory Standards Institute (CLSI) guidelines were used to interpret MIC values when available (CLSI, 2008). Otherwise, MIC values were interpreted using National Antimicrobial Resistance Monitoring System breakpoints (CDC, 2009a). Isolates were classified as being resistant or susceptible to each agent; those isolates with intermediate susceptibility were categorized as being susceptible. Quality control was performed weekly using four strains of bacteria: Escherichia coli ATCC 25922, Staphylococcus aureus 29213, Enterococcus faecalis 29212, and Pseudomonas aeruginosa 27853. The MIC ranges for quality control recommended by the CLSI were used, and results were accepted if the MIC values were within expected ranges for these bacterial strains.
PFGE
PFGE was performed on a representative sample of study isolates, using the standard CDC PulseNet protocol for Salmonella subtyping (Ribot et al., 2006). Our goal was to select one isolate per farm per sample date per source (clinical suspect animal, asymptomatic animal tested via follow-up sampling, or environment) per antimicrobial resistance pattern; when there were a number of isolates with the same resistance profile from the same farm/date/source, a random number generator was utilized to select one. XbaI was used as the restriction enzyme. XbaI-digested S. enterica serotype Braenderup (CDCH9812) DNA was used as a reference size standard (Hunter et al., 2005). Electrophoresis was performed for 21 hours using the CHEF Mapper apparatus (Bio-Rad Laboratories, Hercules, CA). Pattern images were captured with a Bio-Rad Gel Doc and Quantity One 1-D Analysis software (Bio-Rad Laboratories). PFGE patterns were then analyzed and compared using the BioNumerics version 4.5 software (Applied Maths, Saint-Matins-Latem, Belgium). Similarity clustering analyses were performed in BioNumerics based on the Dice correlation coefficient with a tolerance of 1.5%, using the unweighted pair group method with arithmetic mean algorithm. PFGE patterns differing by one or more bands were considered different.
Data analysis
Study herds were considered positive for Salmonella Cerro either if this serotype was isolated from one or more environmental samples or if there was at least one laboratory-confirmed clinical case. The distribution of Cerro isolates was characterized by herd and sample type. Descriptive analysis of antimicrobial resistance and PFGE data was performed. Finally, statistical methods including chi-squared testing and logistic regression analysis were used to assess the importance of Salmonella Cerro as a pathogen in cattle, both at the herd and animal level. All data analyses were performed in SAS (version 9.1; SAS Institute, Cary, NC), and p-values of <0.05 were considered significant.
Results
Thirty-four veterinarians representing 11 veterinary practices participated in this study. A total of 62 dairy farms were enrolled, although five farms withdrew their involvement. Among the remaining 57 study herds, the median herd size was 875 female dairy cattle (range: 245–7412). Forty-four herds (77.2%) yielded Salmonella-positive samples over the course of the study period. Of these, 20 herds (45.5%) in 13 counties across New York were positive for Salmonella Cerro (Table 1). All 20 of these herds yielded Cerro isolates from the environment, and 6 also had one or more clinical cases of salmonellosis (as identified by the herd veterinarian) that were positive for this serotype. Upon follow-up sampling for estimation of within-herd prevalence, Cerro was identified in 10 of the 20 herds; 5 had clinical disease attributed to salmonellosis, and 5 had positive environmental samples but no clinical cases. The within-herd Cerro prevalence ranged from 3.3% to 53.1%, with a median of 17.4%. This median within-herd prevalence among Cerro-positive herds was significantly greater (Wilcoxon rank sum p-value = 0.007) than the median within-herd prevalence of fecal Salmonella shedding among herds that were positive for other serotypes (4.1%). The estimated prevalence in three of the Cerro-positive herds was approximately 50% (45%, 51%, and 53%).
Table 1.
Summary of Results from 20 New York Dairy Herds That Were Positive for Salmonella enterica Serotype Cerro
| Herd | Cerro-positive environmental samples | Cerro-positive clinical cases | Within-herd Cerro prevalence (%) |
|---|---|---|---|
| 1 | + | + | 53.1 |
| 2 | + | + | 30.6 |
| 3 | + | + | 20.0 |
| 4 | + | + | 14.7 |
| 5 | + | + | 7.8 |
| 6 | + | + | — |
| 7 | + | − | 51.3 |
| 8 | + | − | 45.3 |
| 9 | + | − | 9.2 |
| 10 | + | − | 4.0 |
| 11 | + | − | 3.3 |
| 12 | + | − | — |
| 13 | + | − | — |
| 14 | + | − | — |
| 15 | + | − | — |
| 16 | + | − | — |
| 17 | + | − | — |
| 18 | + | − | — |
| 19 | + | − | — |
| 20 | + | − | — |
A total of 31 Salmonella serotypes were isolated during the study period. Cerro was the predominant serotype among herds with confirmed clinical cases as well as herds with positive environmental samples only, accounting for 56.3% (655/1163) of all isolates. Cerro was also the most commonly isolated serotype in the three individual sampling categories (environment: 44.1%, 171/388; clinical cases: 59.2%, 71/120; follow-up sampling: 63.1%, 413/655). Of the six herds with Cerro-positive clinical cases, three yielded Cerro as the only serotype from environmental, clinical suspect, and follow-up sampling. Two herds also generated Salmonella Kentucky from these sample types. In the final herd, both Salmonella Cerro and Salmonella Montevideo were isolated from the environment, and follow-up samples yielded primarily Salmonella Cerro (33 out of 38 positive cattle) but also three Salmonella Newport isolates and a single isolate each of Salmonella Typhimurium (Copenhagen) and Salmonella Thompson; however, Cerro was the only serotype isolated from clinical cases.
Antimicrobial resistance among Salmonella Cerro isolates ranged from zero (pan-susceptible) to nine drugs. Resistance to at least one antimicrobial agent was found in 5.5% (36/651) of the isolates on which susceptibility testing was performed, whereas 94.5% were pan-susceptible. Eight (40.0%) of the Cerro-positive herds in our study generated drug-resistant isolates from cattle, the environment, or both. The antimicrobial drugs to which isolates were most commonly resistant included sulfadimethoxine (15 isolates), sulfisoxazole (12), tetracycline (11), ampicillin (9), ceftiofur (8), and amoxicillin/clavulanic acid (8). Chi-squared testing revealed that drug resistance was significantly more common among isolates from clinically ill cattle (21.2%, 14/66) than among either environmental isolates (4.1%, 7/171; Fisher's exact p-value < 0.0001) or isolates from healthy cattle (3.6%, 15/414; Fisher's exact p-value < 0.0001).
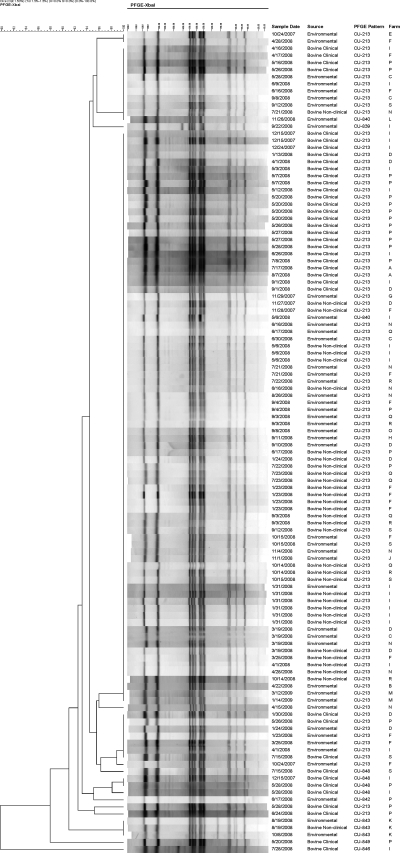
Among 116 isolates selected for PFGE typing, eight PFGE types were differentiated (Fig. 1). A total of 103 isolates shared the predominant PFGE type, whereas the number of isolates corresponding to each of the other seven types ranged from one to four. The main PFGE type was identified on 17 farms. This pattern was shared by 84.2% (32/38) of the clinical case isolates, 97.0% (32/33) of the asymptomatic cattle isolates, and 86.7% (39/45) of the environmental isolates. The other seven PFGE types differed by just one or two bands from the predominant type. Twelve of the 13 isolates with these sporadic PFGE patterns were pan-susceptible; one was resistant only to sulfadimethoxine.
FIG. 1.
XbaI pulsed-field gel electrophoresis (PFGE) patterns for 116 Salmonella enterica serotype Cerro isolates selected for typing.
The median within-herd Cerro prevalence among herds that had clinical cases (20.0%) was greater than that among herds that only had positive environmental samples (9.2%), but this difference was not statistically significant. Among herds with clinical cases, chi-squared testing showed that the prevalence of fecal Cerro shedding among clinical suspect cattle (56.6%, 69/122) was significantly higher (p-value < 0.0001) than that among apparently healthy cattle tested via follow-up sampling (25.6%, 249/974). Clinical suspect cattle were also more likely to test positive for Cerro than apparently healthy cattle as estimated by a logistic regression model that controlled for herd as a random effect (odds ratio: 3.9; p-value = 0.05).
Discussion
There is very little in the literature concerning Salmonella Cerro in cattle or other species. This study had the advantage of utilizing both environmental surveillance and disease monitoring to assess the occurrence of Salmonella Cerro within each enrolled dairy herd. These herds were located throughout New York and were characterized by a wide range of sizes and management types representative of the dairy industry in this area of the country. Another strength of this study was the longitudinal sampling approach for estimating the within-herd prevalence of Salmonella Cerro when herds were identified as being positive. To our knowledge, no field studies have identified this serotype on so many intensively sampled dairy farms, thus permitting a thorough description of its within-herd prevalence, clinical implications, and isolate characteristics.
Culture of environmental samples has been shown to be a relatively effective means of monitoring for the presence of Salmonella on dairy farms (Warnick et al., 2003). However, identifying clinical cases based on detection of suspect animals by herd owners or veterinarians may have underestimated the number of cases in this study. Further, fecal culture does not have perfect sensitivity for detecting the presence of Salmonella, and we recognize that some positive cattle were presumably missed by culturing. On the other hand, it is also plausible that some animals with a positive Salmonella culture result and compatible clinical signs were actually symptomatic because of another primary disease process.
Salmonella was identified in 77% of the study herds, and nearly half of the positive herds (20/44) yielded Salmonella Cerro. In a study of a single herd in Pennsylvania experiencing an outbreak of Salmonella Cerro, investigators found that this serotype was also isolated from other dairy herds in the same geographic area before and during their study period (Van Kessel et al., 2007). Introduction of Salmonella onto a dairy farm can occur through a variety of routes, including purchased cattle, contaminated feed or water, wild animals such as rodents and birds, human traffic, and insects (Bender, 1994; Evans and Davies, 1996; Anderson et al., 2001; Sanchez et al., 2002; Murinda et al., 2004; Nielsen et al., 2007). More information on the biosecurity procedures, feed sources, and cattle importation practices of the Cerro-positive herds would be required to ascertain the likely means of introduction and potentially establish an epidemiologic linkage.
The median within-herd Cerro prevalence was 17%, and in some herds the estimated prevalence approximated 50%. Of the 10 herds from which Salmonella Cerro was identified on follow-up sampling, 5 had experienced clinical disease compatible with a diagnosis of salmonellosis. The role of Salmonella Cerro in causing clinical illness among dairy cattle is unclear, although published reports suggest an absence of disease association in cattle. This serotype persisted in the Pennsylvania dairy herd for nearly 2 years at a high within-herd prevalence (reaching a peak of 88%) in the absence of clinical signs of salmonellosis (Van Kessel et al., 2007). Salmonella Cerro was also isolated from four dairy herds in Ohio for up to 18 months in the absence of clinical signs; however, three of these herds had experienced clinical outbreaks (diarrhea, abortion, and evidence of endotoxemia) prior to the study period, and Salmonella Cerro was isolated from some of the affected cattle in each instance (Huston et al., 2002a). Salmonella Cerro was isolated from the environment of one Wisconsin dairy farm without a history of salmonellosis, but no cattle were sampled in that study (Peek et al., 2004). The NAHMS Dairy 2007 study, based on a single sampling visit to 121 farms in 17 states, found Salmonella Cerro to be the most common serotype isolated from healthy lactating cows (CEAH, 2009).
Although many asymptomatic cattle shed Salmonella Cerro in their feces, there has been an apparent increase in the prevalence of this serotype among cattle with clinical signs of salmonellosis. Cerro was the main serotype isolated from clinical cases in this study (59%), and the prevalence of fecal Cerro shedding among clinical suspect cattle was significantly higher than that among apparently healthy cattle tested via follow-up sampling. In herds with Cerro-positive clinical cases, Cerro was the predominant or only Salmonella serotype isolated from the farm. In contrast, a recent comprehensive study on the incidence of salmonellosis among dairy herds in New York and other northeastern states, based on data collected between 2004 and 2005, found Salmonella Cerro to be a very rare isolate (Cummings et al., 2009). In that study, Cerro accounted for 0.2% of the cases, whereas Newport and Typhimurium accounted for 41% and 19%, respectively. Salmonella Cerro has also been isolated from clinically affected chickens, turkeys, pigs, and horses in recent years (CDC, 2007, 2008).
Salmonella Cerro has been a rare isolate among people in the United States with laboratory-confirmed Salmonella infections, accounting for 0.1% of the cases in both 2005 (CDC, 2007) and 2006 (CDC, 2008). Reports of human disease due to this serotype are sparse. An outbreak of Salmonella Cerro involving 29 known patients occurred in New Mexico in 1985, traced to contaminated beef jerky (CDC, 1985). This serotype has also been incriminated as a sporadic cause of infant diarrhea (Fule and Kaundinya, 1986), pyemia (Bhore et al., 1980), and osteomyelitis (Le, 1982). Finally, a rise in Salmonella Cerro isolation was documented in Italy between 1997 and 1999, although these were primarily from healthy food handlers and samples of urban sewage plant effluent (Mammina et al., 2000). Future surveillance for confirmed cases of salmonellosis will determine if Salmonella Cerro is seen increasingly among people. Other serotypes important to bovine health (Newport and Typhimurium) have emerged across the United States and become leading causes of human foodborne disease (Glynn et al., 1998; Gupta et al., 2003; CDC, 2009b); therefore, close monitoring of Cerro is warranted.
There was a low frequency of antimicrobial resistance among Cerro isolates in this study, with 95% being pan-susceptible. However, drug-resistant isolates were widespread among farms. We found drug resistance to be significantly more common among isolates from clinically ill cattle than among isolates from other sources. This is in agreement with the apparent correlation between antimicrobial resistance and the presence of clinical disease in dairy cattle. Studies of fecal Salmonella shedding among healthy cattle across the United States have shown antimicrobial resistance to be uncommon (Wells et al., 2001; Blau et al., 2005; Ray et al., 2007), but multidrug resistance was found to be highly prevalent among isolates from cattle with clinical signs of salmonellosis in the northeastern United States (Cummings et al., 2009). A similar phenomenon has also been observed in people, as infections with resistant Salmonella strains tend to be more severe and lead to higher rates of hospitalization than those caused by susceptible strains (Helms et al., 2002, 2004; Varma et al., 2005a, 2005b).
Regardless of their source, the isolates in this study displayed tremendous homogeneity with respect to PFGE types. Of the 116 isolates that were typed, 89% had an identical PFGE pattern. This particular pattern was found to be stable and conservative across herds. It remains possible that there was a disparity in virulence between isolates obtained from clinical cases as opposed to those obtained from asymptomatic cattle; however, a more discriminatory typing method or combination of methods may be needed to differentiate these isolates. Alternatively, it is conceivable that host factors impacting immune status were responsible for the discrepancy in clinical outcomes observed in this study. Such factors could include stage of lactation, concurrent illness, and plane of nutrition. The minor differences in banding patterns exhibited by the 13 isolates with sporadic PFGE types apparently did not reflect an acquisition of antimicrobial resistance genes, as 12 of these isolates were pan-susceptible.
The isolation of a predominant PFGE type from cattle throughout New York could indicate the rapid spread of a single clone on account of being a successful phenotype. PFGE homogeneity was also noted among bovine Newport–MDRAmpC isolates from Massachusetts when this serotype was discovered to be an emergent threat in the United States (Gupta et al., 2003). Evidence for widespread dissemination of a single Cerro strain over a short duration of time should further prompt concern over the potential danger to animal and human health. It is noteworthy that the New York State Department of Health has isolated Salmonella Cerro from two human patients since 2003 (in August, 2007 and August, 2008), and both isolates had a PFGE banding pattern that was identical to that of the predominant PFGE type seen among cattle in this study (unpublished data).
Conclusions
Salmonella Cerro appears to be an emerging pathogen of dairy cattle. In contrast to previously published reports, our results suggest that this serotype may be associated with extensive clinical outbreaks in dairy herds. The apparent increase in the prevalence of Salmonella Cerro among cattle with salmonellosis, coupled with evidence for rapid clonal spread, may have important implications for public health.
Acknowledgments
The authors thank the veterinarians and dairy herd owners who participated in this study. This project was supported in part by the Cornell University Zoonosis Research Unit of the Food and Waterborne Diseases Integrated Research Network, funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract number N01-AI-30054.
Disclosure Statement
No competing financial interests exist.
References
- Anderson RJ. House JK. Smith BP, et al. Epidemiologic and biological characteristics of salmonellosis in three dairy herds. J Am Vet Med Assoc. 2001;219:310–322. doi: 10.2460/javma.2001.219.310. [DOI] [PubMed] [Google Scholar]
- Bender JB. Reducing the risk of Salmonella spread and practical control measures in dairy herds. Bovine Pract. 1994;28:62–65. [Google Scholar]
- Bhore AV. Phadke SA. Joshi BN. Salmonella Cerro causing pyaemia in man—report of a case. Indian J Pathol Microbiol. 1980;23:309–311. [PubMed] [Google Scholar]
- Blau DM. McCluskey BJ. Ladely SR, et al. Salmonella in dairy operations in the United States: prevalence and antimicrobial drug susceptibility. J Food Prot. 2005;68:696–702. doi: 10.4315/0362-028x-68.4.696. [DOI] [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Salmonellosis associated with carne seca—New Mexico. MMWR Morb Mortal Wkly Rep. 1985;34:645–646. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control Prevention. Salmonella Surveillance: Annual Summary, 2005. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2007. [Google Scholar]
- [CDC] Centers for Disease Control Prevention. Salmonella Surveillance: Annual Summary, 2006. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2008. [Google Scholar]
- [CDC] Centers for Disease Control Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2006. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2009a. [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Preliminary FoodNet Data on the incidence of infection with pathogens transmitted commonly through food—10 States, 2008. MMWR Morb Mortal Wkly Rep. 2009b;58:333–337. [PubMed] [Google Scholar]
- [CEAH] Centers for Epidemiology Animal Health. Fort Collins, CO: United States Department of Agriculture; 2009. National Animal Health Monitoring System (NAHMS) Dairy 2007: Salmonella and Campylobacter on U.S. Dairy Operations, 1996–2007. [Google Scholar]
- [CLSI] Clinical and Laboratory Standards Institute. 3rd. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard. CLSI document M31-A3. [Google Scholar]
- Cummings KJ. Warnick LD. Alexander KA, et al. The incidence of salmonellosis among dairy herds in the northeastern United States. J Dairy Sci. 2009;92:3766–3774. doi: 10.3168/jds.2009-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings KJ. Warnick LD. Elton M, et al. The effect of clinical outbreaks of salmonellosis on the prevalence of fecal Salmonella shedding among dairy cattle in New York. Foodborne Pathog Dis. 2010 doi: 10.1089/fpd.2009.0481. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA. Besser TE. Eckmann K, et al. Multidrug-resistant Salmonella Typhimurium, Pacific Northwest, United States. Emerg Infect Dis. 2007;13:1583–1586. doi: 10.3201/eid1310.070536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divers TJ. Peek SF. Rebhun's Diseases of Dairy Cattle. St. Louis: Saunders Elsevier; 2008. [Google Scholar]
- Dunne EF. Fey PD. Kludt P, et al. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA. 2000;284:3151–3156. doi: 10.1001/jama.284.24.3151. [DOI] [PubMed] [Google Scholar]
- Evans S. Davies R. Case control study of multiple-resistant Salmonella Typhimurium DT104 infection of cattle in Great Britain. Vet Rec. 1996;139:557–558. [PubMed] [Google Scholar]
- Fule RP. Kaundinya DV. S. Cerro (18:z4,z23-), a rare serotype isolation from infants with diarrhoea in Ambajogai. Indian J Pathol Microbiol. 1986;29:205–207. [PubMed] [Google Scholar]
- Glynn MK. Bopp C. Dewitt W, et al. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- Gupta A. Fontana J. Crowe C, et al. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J Infect Dis. 2003;188:1707–1716. doi: 10.1086/379668. [DOI] [PubMed] [Google Scholar]
- Helms M. Simonsen J. Molbak K. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J Infect Dis. 2004;190:1652–1654. doi: 10.1086/424570. [DOI] [PubMed] [Google Scholar]
- Helms M. Vastrup P. Gerner-Smidt P, et al. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg Infect Dis. 2002;8:490–495. doi: 10.3201/eid0805.010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SB. Vauterin P. Lambert-Fair MA, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston CL. Wittum TE. Love BC. Persistent fecal Salmonella shedding in five dairy herds. J Am Vet Med Assoc. 2002a;220:650–655. doi: 10.2460/javma.2002.220.650. [DOI] [PubMed] [Google Scholar]
- Huston CL. Wittum TE. Love BC, et al. Prevalence of fecal shedding of Salmonella spp in dairy herds. J Am Vet Med Assoc. 2002b;220:645–649. doi: 10.2460/javma.2002.220.645. [DOI] [PubMed] [Google Scholar]
- Jones TF. Ingram LA. Cieslak PR, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008;198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- L Plym F. Wierup M. Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech. 2006;25:541–554. [PubMed] [Google Scholar]
- Le CT. Salmonella vertebral osteomyelitis: a case report with literature review. Am J Dis Child. 1982;136:722–724. [PubMed] [Google Scholar]
- Mammina C. Cannova L. CarfiPavia S, et al. Endemic presence of Salmonella enterica serotype Cerro in southern Italy. Eur Surveill. 2000;5:84–86. doi: 10.2807/esm.05.07.00028-en. [DOI] [PubMed] [Google Scholar]
- Mead PS. Slutsker L. Dietz V, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murinda SE. Nguyen LT. Nam HM, et al. Detection of sorbitol-negative and sorbitol-positive Shiga toxin-producing Escherichia coli, Listeria monocytogenes, Campylobacter jejuni, and Salmonella spp. in dairy farm environmental samples. Foodborne Pathog Dis. 2004;1:97–104. doi: 10.1089/153531404323143611. [DOI] [PubMed] [Google Scholar]
- Nielsen LR. Warnick LD. Greiner M. Risk factors for changing test classification in the Danish surveillance program for Salmonella in dairy herds. J Dairy Sci. 2007;90:2815–2825. doi: 10.3168/jds.2006-314. [DOI] [PubMed] [Google Scholar]
- Peek SE. Hartmann FA. Thomas CB, et al. Isolation of Salmonella spp from the environment of dairies without any history of clinical salmonellosis. J Am Vet Med Assoc. 2004;225:574–577. doi: 10.2460/javma.2004.225.574. [DOI] [PubMed] [Google Scholar]
- Peters AR. An estimation of the economic impact of an outbreak of Salmonella Dublin in a calf rearing unit. Vet Rec. 1985;117:667–668. doi: 10.1136/vr.117.25-26.667. [DOI] [PubMed] [Google Scholar]
- Ray KA. Warnick LD. Mitchell RM, et al. Prevalence of antimicrobial resistance among Salmonella on midwest and northeast USA dairy farms. Prev Vet Med. 2007;79:204–223. doi: 10.1016/j.prevetmed.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Ribot EM. Fair MA. Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Sanchez S. Hofacre CL. Lee MD, et al. Animal sources of salmonellosis in humans. J Am Vet Med Assoc. 2002;221:492–497. doi: 10.2460/javma.2002.221.492. [DOI] [PubMed] [Google Scholar]
- Van Kessel JS. Karns JS. Wolfgang DR, et al. Longitudinal study of a clonal, subclinical outbreak of Salmonella enterica subsp. enterica serovar Cerro in a U.S. dairy herd. Foodborne Pathog Dis. 2007;4:449–461. doi: 10.1089/fpd.2007.0033. [DOI] [PubMed] [Google Scholar]
- Varma JK. Greene KD. Ovitt J, et al. Hospitalization, and antimicrobial resistance in Salmonella outbreaks 1984–2002. Emerg Infect Dis. 2005a;11:943–946. doi: 10.3201/eid1106.041231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma JK. Molbak K. Barrett TJ, et al. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis. 2005b;191:554–561. doi: 10.1086/427263. [DOI] [PubMed] [Google Scholar]
- Voetsch AC. Van Gilder TJ. Angulo FJ, et al. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38(Suppl 3):S127–S134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- Warnick LD. Kaneene JB. Ruegg PL, et al. Evaluation of herd sampling for Salmonella isolation on midwest and northeast US dairy farms. Prev Vet Med. 2003;60:195–206. doi: 10.1016/s0167-5877(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Wells SJ. Fedorka-Cray PJ. Dargatz DA, et al. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J Food Prot. 2001;64:3–11. doi: 10.4315/0362-028x-64.1.3. [DOI] [PubMed] [Google Scholar]