Abstract
Taxa in the early stages of speciation may bear intraspecific allelic variation at loci conferring barrier traits in hybrids such as hybrid sterility. Additionally, hybridization may spread alleles that confer barrier traits to other taxa. Historically, few studies examine within- and between-species variation at loci conferring reproductive isolation. Here, we test for allelic variation within Drosophila persimilis and within the Bogota subspecies of D. pseudoobscura at regions previously shown to contribute to hybrid male sterility. We also test whether D. persimilis and the USA subspecies of D. pseudoobscura share an allele conferring hybrid sterility in a D. pseudoobscura bogotana genetic background. All loci conferred similar hybrid sterility effects across all strains studied, though we detected some statistically significant quantitative effect variation among D. persimilis alleles of some hybrid incompatibility QTLs. We also detected allelism between D. persimilis and D. pseudoobscura USA at a 2nd chromosome hybrid sterility QTL. We hypothesize that either the QTL is ancestral in D. persimilis and D. pseudoobscura USA and lost in D. pseudoobscura bogotana, or gene flow transferred the QTL from D. persimilis to D. pseudoobscura USA. We discuss our findings in the context of population features that may contribute to variation in hybrid incompatibilities.
Keywords: Drosophila, Speciation, Reproductive Isolation, Gene Flow, Hybrid Sterility
INTRODUCTION
The process of speciation results when gene flow is prevented between populations, creating reproductive isolation. A wide variety of traits can inhibit gene flow, and understanding the genetic differences that lead to these traits informs us about the genetic changes that create new species. More progress has been made in mapping, characterizing, and cloning genes that cause incompatibilities, such as fitness reduction, in species hybrids (reviewed in Presgraves 2010) than in traits that prevent formation of hybrids (e.g., mate preference, pollinator differences), although both have been studied extensively (Coyne and Orr 2004, Chapters 5 and 6).
However, the genetic mapping studies that lead to cloning incompatibility genes are often unreplicated. A typical speciation genetics study will identify quantitative trait loci (QTLs) differentiating one strain of one species from one strain of another species but does not test between other pairs of strains. Yet, the localized QTL may be specific to the strains studied and not actually characteristic of the entire species, and many recent studies have demonstrated intraspecies variation in QTLs (Demuth and Wade 2007; Sweigart et al. 2007; Case and Willis 2008; Good et al. 2008; Nolte et al. 2008; Reed 2008; Teeter et al. 2010). For example, Barnwell and Noor (2008) examined variation within and between populations of Drosophila pseudoobscura in species mate discrimination. While the behavioral difference between these populations was fixed, the underlying QTLs differentiating one pair of strains failed to contribute to the behavioral difference in any other pair of strains surveyed (see also Ortiz-Barrientos et al. 2004). Thus, two questions must be asked of speciation genetics QTL mapping studies: 1) can the same QTLs be detected in different pairs of strains, and if so, 2) are the effects similar in magnitude?
The Drosophila pseudoobscura/D. persimilis system has been used extensively as a model for hybrid male sterility, pioneered by Dobzhansky (1933, 1936) and continued by others (Wu and Beckenbach 1983; Orr 1987; Orr and Irving 2001; Phadnis and Orr 2009). Noor et al. (2001b) found that much of the genetic basis for hybrid sterility in this system localizes to three regions bearing fixed inversion differences between the two species. Brown et al. (2004b) and Chang and Noor (2007, 2010) found that a geographically isolated subspecies of D. pseudoobscura (D. pseudoobscura bogotana) had additional factors outside the inverted regions that conferred sterility in hybrid males between the subspecies and D. persimilis. These findings supported a model where sterility factors in hybridizing species (D. pseudoobscura USA and D. persimilis) persist in fixed inverted regions that differ between the two species. Unlike the inverted regions, most collinear (uninverted) regions of the genome are prevented from diverging if the nascent species were sympatric or are homogenized by interspecies gene flow after secondary contact if the nascent species emerged in allopatry (Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009). In contrast, non-hybridizing species (D. pseudoobscura bogotana and D. persimilis) can maintain additional sterility factors across the genome, both in inverted and collinear regions.
Here, we examine the system in greater detail, specifically the D. pseudoobscura bogotana-D. persimilis hybridization, to determine the extent of genetic variation contributing to hybrid sterility within and among these taxa. We test whether and how variation within species affects hybrid sterility by re-examining the data of Brown et al. (2004b) and collecting new data to see if the inverted regions contributed similar effects in D. persimilis/D. pseudoobscura bogotana hybrids when using different strains of the two taxa. We also replicate the experimental design of Chang and Noor (2007) in multiple strains of D. persimilis to detect variation among collinear factors contributing to hybrid sterility between D. persimilis and D. pseudoobscura bogotana. Finally, to understand the evolutionary history of hybrid sterility in this system and study between-species variation, we test for shared alleles in a collinear region of D. pseudoobscura USA and D. persimilis that confers sterility in D. persimilis/D. pseudoobscura bogotana hybrids. Demonstration of allelism (that this region has the same effect when derived from either D. persimilis or D. pseudoobscura USA) could suggest that this hybrid sterility-conferring allele introgressed from D. persimilis into D. pseudoobscura USA in collinear regions from the homogenizing gene flow described above.
MATERIALS AND METHODS
Fly stocks
The D. persimilis MSH1993 line was derived from females collected at Mount Saint Helena, California, in 1993 (Noor 1995). The D. persimilis MSH3 and MSH1 lines were derived from females collected at Mount Saint Helena, California, in 1997 (stock number 14011-0111.49). The D. persimilis Santa Cruz Island (SCI) was obtained from the stock center (number: 14011-0111.50, year: 2004). The D. pseudoobscura USA Flagstaff1993 line was derived from four different lines, each collected in Flagstaff, Arizona, in 1993 (Noor 1995). Since the collections vary both spatially and temporally, we assume that our sampling does not affect our tests of variation. The D. pseudoobscura bogotana line is a subculture of the D. pseudoobscura bogotana El Recreo line collected in 1978 (provided by H. A. Orr). Lines of D. pseudoobscura bogotana and crosses described in the re-examination of the Brown et al. (2004b) data are described therein.
Fly crosses
D. pseudoobscura bogotana females carrying a white eye mutation (hereafter bogw) were collected as virgins and maintained for at least six days. After six days, bogw females were crossed to D. persimilis MSH3, MSH1993, MSH1, or SCI males (Figure 1). F1 females were backcrossed to bogw males to generate backcross males (hereafter “BCbogw males”) for fertility assays. Only those male progeny bearing the white mutation were scored because the mutation is linked to an inversion on the left arm of the X chromosome (XL), which is known to confer sterility between the two species (Dobzhansky 1936; Orr 1987; Noor et al. 2001a; Brown et al. 2004b). Because a D. persimilis XL inversion would cause sterility in a D. pseudoobscura bogotana background, we selected white-eyed flies, which maintain a D. pseudoobscura bogotana XL inversion, allowing sterility conferred from autosomal loci to be detected.
Figure 1. Backcross Design.
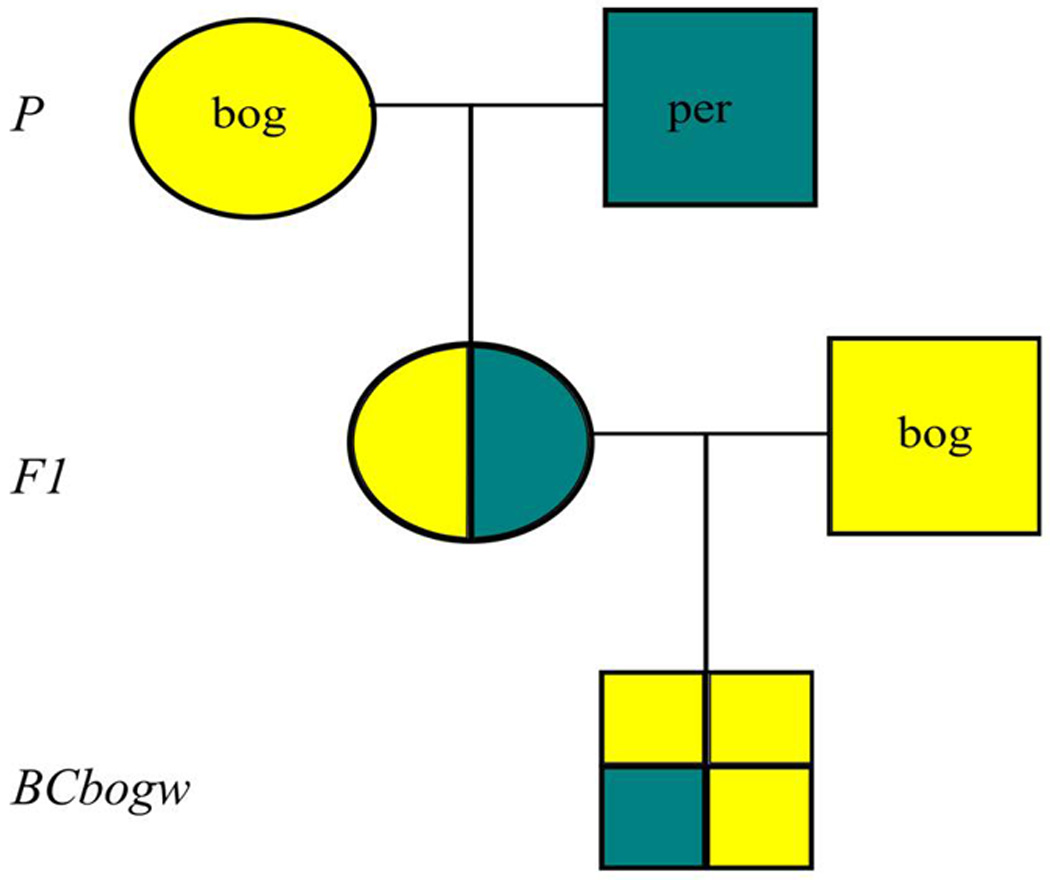
Males from each line of D. persimilis were crossed to white-eyed D. pseudoobscura bogotana (bog) females. The fertile F1 hybrid females were backcrossed to white-eyed D. pseudoobscura bogotana males. The white-eyed backcross male progeny (BCbogw) were then dissected and genotyped.
For the test for allelism of a hybrid sterility locus, regions of the D. pseudoobscura USA Flagstaff1993 2nd and 4th chromosomes which correspond to D. persimilis MSH1993 QTLs associated with hybrid male sterility when in a D. pseudoobscura bogotana background (Chang and Noor 2007), were introgressed separately into a D. persimilis MSH1993 background. These introgressed males were crossed to bogw females, and F1 females were backcrossed to bogw males to generate backcross males (BCbogw males) used for fertility assays. All crosses were performed on standard sugar/yeast/agar medium at 20° ± 1° and 85% relative humidity.
Fertility assays
BCbogw males were collected as virgins and maintained for 7 days in food-containing vials holding 1–20 males. On day 7, the fertility of each backcross male was assessed by dissection of the testes in Ringer’s solution following the method of Coyne (1984). A male was scored as “fertile” if at least one motile sperm was observed and “sterile” if no motile sperm were observed. Treating fertility as a binary trait has been shown to be conservative (Campbell and Noor 2001), although other methods of scoring fertility exist (e.g., White-Cooper 2004). All dissected BCbogw males were labeled and stored at −20°.
Microsatellite genotyping
DNA was extracted from all dissected BCbogw males following the protocol of Gloor and Engels (1992). Microsatellite genotyping was performed in two steps. First, all BCbogw males were genotyped for markers associated with each inversion that distinguishes D. pseudoobscura from D. persimilis. The markers used for this initial screen were DPSX002 or DPSX046 (left arm of X chromosome- XL), DPSX030 (XR), and DPS2022 or DPS2026 (2). These markers denoted the species identity of the inversion arrangement on these chromosome arms. Second, only those BCbogw males that were hemizygous or homozygous for the D. pseudoobscura bogotana allele at the three inversion markers (hereafter “BCbogwLim males”) were further genotyped at the three collinear QTLs identified previously (Chang and Noor 2007). Again, these males were selected because the D. pseudoobscura bogotana inversion alleles permit detection of sterility caused by autosomal loci. The markers used for genotyping at these QTLs were DPS2_27.05 and DPS2_2395c for chromosome 2, DPS3001 for chromosome 3 and two of the following for chromosome 4: DPS4G1a, DPS4G1h, DPS4033h, and DPS4611966. Primer sequences for all markers used in this study are available in supplemental table 1. PCR amplification followed a touchdown protocol: 95° for 1 min; 3 cycles of 94° for 30 sec, 56° for 30 sec, 72° for 30 sec; 3 cycles of 94° for 30 sec, 53° for 30 sec, 72° for 30 sec; and 30 cycles of 94° for 30 sec, 50° for 30 sec, 72° for 30 sec. PCR products were visualized on acrylamide gels on LiCor 4200 and 4300 DNA sequencer/analyzers.
Data Analysis
Following the protocols of Brown et al. (2004b) and Chang and Noor (2007), each individual from each cross was parsed into categories based on genotype using Microsoft Excel. Counts of the number of individuals in each category in each cross were summed. The number of fertile progeny for each genotype category was divided by the total number of progeny with that genotype to obtain the percent fertility of individuals with that genotype. Crosses of a similar type were also analyzed together (i.e. backcrosses of different lines of D. persimilis to the same line of D. pseudoobscura bogotana). The fertile individuals in each of those crosses of that type were summed and divided by the total number of progeny to calculate percent fertility of a certain genotype for all crosses together. Statistical differences in fertility between each of the crosses of a similar type were calculated using a G-test or Chi-Square (if the G-test failed). Analyses were performed using StatView 5 (SAS; Cary, NC), though chi-Square probabilities were estimated using the online calculator at: http://faculty.vassar.edu/lowry/fisher2x4.html.
RESULTS
Variation among strains in effect of inverted regions on hybrid sterility
Brown et al. (2004b) examined the fertility of backcross hybrids between one strain of D. persimilis and multiple strains of D. pseudoobscura bogotana, specifically limiting analysis to those hybrids that were homo- or hemizygous for all inversions from D. pseudoobscura bogotana. We reanalyzed the raw data they obtained (Brown et al. 2004a) before they imposed this limitation. We tested if the effects of the inverted regions on hybrid sterility are similar between crosses using different strains of D. persimilis. We split the data into two classes (discussed below) for analysis because in some of the previous research (Brown et al. 2004b; Chang and Noor 2007) and in all of our new research, selection of progeny for phenotyping and genotyping was limited to males bearing the XL chromosome arm from D. pseudoobscura bogotana. This limitation is based on the "white" visible mutation located on the XL; flies bearing an XL from D. persimilis were almost invariably sterile (see below). We focus on the fertility of backcross progeny from the cross shown in Figure 1.
Two classes of backcross progeny were analyzed: strains of D. pseudoobscura bogotana with an XL from either species (Table 1, corresponding p-valuesa), and strains with only a D. pseudoobscura bogotana-derived XL (Table 1, corresponding p-valuesb) while using a single strain of D. persimilis. We found that all backcrosses bearing a D. persimilis-derived XL had greatly reduced fertility (less than 15%), regardless of the genotypes at the other markers. Among all backcross progeny, fertility remains comparatively high in individuals with D. pseudoobscura bogotana genotypes at the XL (Table 1, lines 1–4). However, we observe a strong reduction in fertility when the XR chromosome arm is derived from D. pseudoobscura bogotana, in combination with a heterozygous second chromosome (Table 1, line 2).
Table 1. Variation in percent fertility between lines of D. pseudoobscura bogotana at each inversion genotype.
For each cross, D. persimilis MSH1993 was backcrossed to each of three D. pseudoobscura bogotana strains (described in Brown et al (2004b) and the percent fertility at each genotype is listed.
| BCbogSusa6 | BCbogSutatausa5 | BCbogERw | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Line | Genotype | % Fertility | Sample Size | % Fertility | Sample Size | % Fertility | Sample Size | p-valuea | p-valueb |
| 1 | XL=bog, XR=bog, 2=bog | 74% | 46 | 64% | 64 | 67% | 1184 | 0.271 | 0.514 |
| 2 | XL=bog, XR=bog, 2=het | 25% | 89 | 8% | 77 | 18% | 1042 | 0.003 | 0.011 |
| 3 | XL=bog, XR=per, 2=bog | 74% | 53 | 77% | 61 | 79% | 938 | 0.668 | 0.686 |
| 4 | XL=bog, XR=per, 2=het | 76% | 63 | 73% | 55 | 82% | 1036 | 0.667 | 0.165 |
The p-value listed in this column represents statistical significance of the difference between the percent fertilities of only crosses with D. pseudoobscura bogotana Susa6 and Sutatausa5.
The p-value listed in this column represents statistical significance of the difference between the percent fertilities of all three crosses (D. pseudoobscura bogotana Susa6, Sutatausa5 and ERw). Both types of analyses were performed using a G-test (see Methods). "bog": D. pseudoobscura bogotana allele, "per": D. persimilis allele, and "het": heterozygous for both D. pseudoobscura bogotana and D. persimilis alleles.
Generally, we detect few statistically significant differences in fertility among the backcross progeny classes of the same genotype derived from different strains of D. pseudoobscura bogotana. Among the different genotypes, we observe very similar reductions in fertility when particular D. persimilis inverted regions are introduced. Although a few genotypes exhibited statistically significant variation in hybrid sterility effects among strains (Table 1, line 2), the difference is in the magnitude of associated hybrid sterility rather than the absence or presence of an effect on hybrid sterility.
While the crosses from Brown et al (2004b) used the same strain of D. persimilis, but varied the strains of D. pseudoobscura bogotana, we performed an analogous cross using the D. pseudoobscura bogotana ER white strain for background but varied the D. persimilis strain from which the inverted regions were derived (strains MSH1993, MSH1, MSH3 and SCI). Only white males were selected, so we surveyed backcross hybrid males with an XL inversion derived from D. pseudoobscura bogotana. These results are presented in Table 2. Again, we observed a noticeable reduction in fertility when the XR chromosome arm is derived from D. pseudoobscura bogotana but the second chromosome is heterozygous for D. persimilis (Table 2, line 2). We detected statistically significant differences in fertility among the backcross hybrids from different strains when the XR chromosome arm was derived from D. persimilis (Table 2, lines 3–4), but again, the differences are in the magnitude of associated hybrid sterility rather than the absence or presence of an effect on hybrid sterility.
Table 2. Variation in percent fertility between lines of D. persimilis at the inversions.
For each cross, a different strain of D. persimilis was backcrossed to D. pseudoobscura bogotana ER white and the percent fertility at each genotype is listed. The p-value listed represents statistical significance of the difference between the percent fertilities of each cross, and was calculated using a G-test (see Methods). "bog": D. pseudoobscura bogotana allele, "per": D. persimilis allele, and "het": heterozygous for both D. pseudoobscura bogotana and D. persimilis alleles.
| BCbog (MSH1993) | BCbog (MSH1) | BCbog (MSH3) | BCbog (SCI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Line | Genotype | % Fertility | Sample Size | % Fertility | Sample Size | % Fertility | Sample Size | % Fertility | Sample Size | p-value |
| 1 | XL=bog, XR=bog, 2=bog | 67% | 1184 | 61% | 455 | 67% | 246 | 71% | 192 | 0.054 |
| 2 | XL=bog, XR=bog, 2=het | 18% | 1042 | 21% | 381 | 18% | 214 | 22% | 218 | 0.292 |
| 3 | XL=bog, XR=per, 2=bog | 79% | 938 | 85% | 311 | 86% | 160 | 94% | 156 | <0.001 |
| 4 | XL=bog, XR=per, 2=het | 82% | 1036 | 94% | 293 | 86% | 166 | 93% | 181 | <0.001 |
In summary, we detect generally small, though statistically significant, variance among strains of D. pseudoobscura bogotana and D. persimilis in hybrid sterility effects associated with inverted regions.
Variation among strains in hybrid sterility effect of collinear regions
Chang and Noor (2007) identified three D. persimilis autosomal QTLs outside the fixed inversions contributing to hybrid sterility when in a D. pseudoobscura bogotana background. Using the MSH1993 line of D. persimilis, we tested the effects of one of the three collinear QTLs using backcrosses involving three other lines of D. persimilis: MSH1, MSH3 and SCI, into the D. pseudoobscura bogotana background. The collinear QTL tested is located on chromosome 2 (Q2), where Q2 is 1.42 Mb long. The QTL on chromosome 3 (Q3) was located within an inversion polymorphic within the species, rendering fine-mapping this QTL impossible, and it is not examined here. The third QTL is located on the fourth chromosome (Q4) and is not tested because of its much weaker effect on hybrid sterility compared to Q2 (Chang and Noor 2007). Following Chang and Noor (2007), we focused on backcross hybrid individuals bearing the left and right arm inversions of the X chromosome (XL and XR) and the 2nd chromosome inversion derived from D. pseudoobscura bogotana.
The effects of each collinear QTL in each of the three additional lines tested, including the original, D. persimilis MSH1993, is presented in Table 3. Each QTL is represented by two flanking markers (listed in supplemental table 1). As expected, having both QTLs with D. persimilis genotypes in a D. pseudoobscura bogotana background strongly reduces fertility in all lines (Table 3, line 4; Supplemental Table 2, line 8). The very slight reduction in fertility in the presence of a D. persimilis Q4 by itself (Table 3, line 2) confirms the observation that this foreign allele can be made homozygous and maintained in a D. pseudoobscura bogotana background Chang and Noor (2010). The slight effect of a D. persimilis Q4 is amplified when in combination with a D. persimilis Q2, and is consistent with the epistatic effects discussed in Chang and Noor (2010). As with our other analysis of the inverted regions, the fertility of backcross hybrid males bearing particular genotypes are not significantly different across the strains surveyed.
Table 3. Variation in percent fertility between lines of D. persimilis at the collinear QTLs.
Each line of D. persimilis was backcrossed to D. pseudoobscura bogotana ER white, and the percent fertility at each genotype is listed for each cross. The p-value listed represents statistical significance of the difference between the percent fertilities of each cross, and was calculated using a G-test (see Methods). "bog": D. pseudoobscura bogotana allele, "per": D. persimilis allele, and "het": heterozygous for both D. pseudoobscura bogotana and D. persimilis alleles.
| BCbog (MSH1993) | BCbog (MSH1) | BCbog (MSH3) | BCbog (SCI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Line | Genotype | % Fertility | Sample Size | % Fertility | Sample Size | % Fertility | Sample Size | % Fertility | Sample Size | p-value |
| 1 | Q2=bog, Q4=bog | 91% | 191 | 83% | 84 | 79% | 39 | 87% | 45 | 0.174 |
| 2 | Q2=bog, Q4=het | 70% | 190 | 70% | 83 | 77% | 39 | 75% | 52 | 0.748 |
| 3 | Q2=het, Q4=bog | 65% | 147 | 58% | 77 | 73% | 49 | 59% | 29 | 0.335 |
| 4 | Q2=het, Q4=het | 35% | 139 | 34% | 76 | 39% | 49 | 47% | 38 | 0.531 |
Allelism in collinear regions of D. pseudoobscura USA and D. persimilis
The results from the experiments described above demonstrate only some quantitative within-species variation in the hybrid sterility effects of particular QTLs. Both D. pseudoobscura USA and D. persimilis form sterile hybrid males when crossed with D. pseudoobscura bogotana, and work reported here and elsewhere (e.g., Orr and Irving 2001) has shown that dominant, collinear, autosomal QTLs contribute to this sterility. Many studies have shown evidence for gene exchange between D. pseudoobscura USA and D. persimilis in such collinear regions (Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009), suggesting the hybrid-sterility-conferring allele may have even spread from between the two species.
We tested for allelism between D. pseudoobscura USA and D. persimilis at one collinear QTL (Q2) that confers sterility in a D. pseudoobscura bogotana genetic background. Q3 was not introgressed because of its linkage to an inversion, and Q4 demonstrated a much weaker effect on hybrid sterility between D. persimilis and D. pseudoobscura bogotana (Chang and Noor 2007). First, we introgressed approximately 1.42 Mb of a 2 Mb hybrid sterility QTL on chromosome 2 (Q2) identified in Chang and Noor (2007) from D. pseudoobscura into D. persimilis. Once made homozygous in D. persimilis, those flies were backcrossed to D. pseudoobscura bogotana. If hybrid sterility is observed to be associated with this QTL, D. pseudoobscura USA may contain the same hybrid-sterility-conferring allele as D. persimilis when in a D. pseudoobscura bogotana genetic background.
The presence of the D. pseudoobscura USA copy of the D. persimilis hybrid sterility QTL on chromosome 2 was significantly associated with hybrid sterility (Figure 2). We also examined whether the D. pseudoobscura USA alleles have the same effect on hybrid sterility at the Q2 regions as the D. persimilis alleles (comparing Figure 2 to Table 3/Supplemental Table 2). Both the D. persimilis Q2 and D. pseudoobscura USA Q2 show a greater fertility reduction when in combination with either a D. persimilis Q3 or Q4, and an even stronger reduction when all three QTLs are heterozygous. The effects of the D. pseudoobscura USA Q2 are indeed similar to the effects of the D. persimilis Q2, especially when in combination with other QTLs. BCbogw males with a D. pseudoobscura USA Q2 and two other D. persimilis MSH1993 alleles at Q3 and Q4 exhibit 53.5% fertility (Figure 2). In Table 3, BCbogw males with D. persimilis MSH1993 alleles at Q2 and Q4 exhibit 35% fertility, and in Supplemental Table 2, BCbogw males with D. persimilis MSH1993 alleles at Q2, Q3 and Q4 exhibit 30% fertility. While the D. pseudoobscura USA allele at Q2 may not reduce fertility as much as D. persimilis, the effect of the Q2 marker is still highly significant (p<0.0001) and the epistatic effects between a D. pseudoobscura USA Q2 and D. persimilis Q3/Q4 are still maintained. Therefore, the D. pseudoobscura USA allele at Q2 is associated with hybrid sterility in a D. pseudoobscura bogotana background.
Figure 2. Percent fertility of individuals with a D. pseudoobscura USA copy of QTL 2.
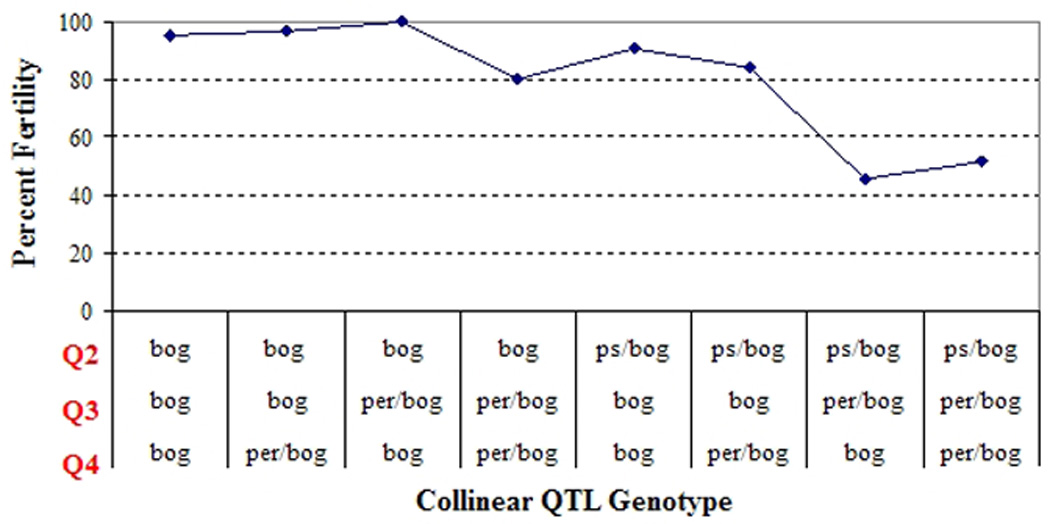
Genotypes for each QTL (Q2, Q3 and Q4) are listed vertically. Sample sizes for each genotype combination are listed below the genotypes. "bog": D. pseudoobscura bogotana allele, "per/bog": heterozygous for D. persimilis and D. pseudoobscura bogotana alleles, "ps/bog": heterozygous for D. pseudoobscura USA and D. pseudoobscura bogotana alleles.
DISCUSSION
Historically, most genetic studies mapping reproductive isolation have been unreplicated and make the unstated assumption that the mapped loci are representative across the population or species. This study tests that assumption in the D. pseudoobscura- D. persimilis system. Previous studies in this system demonstrate an association of inverted regions of the D. persimilis genome with hybrid male sterility when crossed to D. pseudoobscura USA (Noor et al. 2001a; Noor et al. 2001b) or to D. pseudoobscura bogotana (Brown et al. 2004b). In addition, Chang and Noor (2007) demonstrate an association of collinear regions of D. persimilis with hybrid male sterility when crossed to D. pseudoobscura bogotana. We test for variation in those regions responsible for hybrid male sterility, both between lines of the same species as well as between species. We find that, in this system, effects of both inverted and collinear regions on hybrid male sterility vary minimally when derived from different strains within species, suggesting little intraspecific variation in their effect on reproductive isolation. We also found a hybrid sterility QTL shared between D. pseudoobscura USA and D. persimilis when introgressed into D. pseudoobscura bogotana. These findings are discussed in turn.
Variation in hybrid incompatibilities within species
We found that the same QTLs across the different lines occasionally exhibited statistically significant variation in effect on hybrid male sterility. However, all regions exhibited statistically significant effects in the same direction; any variation detected was in magnitude of effect rather than presence or absence, unlike the differences observed in Case and Willis (2008), Nolte et al. (2008), Barnwell and Noor (2008), Good et al. (2008), and Sweigart et al. (2007). Further, the effects of these QTLs from various lines were assayed over long, non-overlapping periods of time, which may contribute environmental variance. Therefore, a lack of temporal control may have overestimated differences in effect among lines. However, only three strains of D. pseudoobscura bogotana and four strains of D. persimilis were used – inclusion of additional strains might demonstrate greater variation in hybrid sterility effects. Additionally, the effects of the inverted regions were likely averaged across many loci because of the multi-megabase length of each inversion, which may have underestimated differences in effect by averaging across many loci. This study further tested the effects of the collinear regions on hybrid male sterility, where we still detected minimal variation in sterility effects among strains within species.
Testing the presence or absence of effects of localized QTLs across lines within species is extremely important, yet historically is rarely performed. Variation in reproductive isolation factors has been demonstrated in Crepis (Hollingshead 1930), Mimulus (Sweigart et al. 2007; Case and Willis 2008), Tribolium (Demuth and Wade 2007), Drosophila (Patterson 1952, Chapter 10; Reed 2008), and mouse (Good et al. 2008).. Despite these recent studies showing variation in hybrid incompatibilities within species, many studies assume fixation of the same hybrid incompatibility alleles within species. Part of this assumption may be tied with the observation that directional selection and/or meiotic drive seem to be associated with the fixation of multiple cloned hybrid incompatibility genes (Ting et al. 1998; Presgraves et al. 2003; Barbash et al. 2004; Phadnis and Orr 2009; but see Masly et al. 2006).
Several factors may contribute to the amount of segregating variation in factors contributing to hybrid incompatibilities. These may include divergence time, amount of hybridization, amount of standing molecular genetic variation, effective population size, or disadvantage as a homozygote. In the case of D. pseudoobscura bogotana and D. persimilis, the divergence time is roughly 0.5–1.0 million years (Aquadro et al. 1991; Wang et al. 1997; Hey and Nielsen 2004; Leman et al. 2005), which is long enough to observe limited variation in hybrid incompatibility alleles within-species. Also, D. pseudoobscura bogotana and D. persimilis do not hybridize since they are on different continents, and D. pseudoobscura bogotana has a very small effective population size and little standing genetic variation (Machado et al. 2002). Given the long divergence time and low effective population size, this system may be less likely than others to bear extensive standing genetic variation for hybrid incompatibilities.
Allelism in hybrid incompatibilities between species
A previous study demonstrated the significant effect of D. persimilis Q2 on hybrid male sterility when in a D. pseudoobscura bogotana background (Chang and Noor 2007), and here we demonstrate a significant effect of a D. pseudoobscura USA Q2 allele on hybrid male sterility in a D. pseudoobscura bogotana background. The D. pseudoobscura USA region may contain distinct hybrid-sterility-conferring loci that are not identical to those in D. persimilis. However, such loci are not ubiquitous: Chang and Noor (2010) introgressed a large collinear region of chromosome 2 between D. p. bogotana and D. persimilis and found no effect on hybrid sterility. Further, our introgression of Q2 interacts precisely the same way as the D. persimilis region with a D. p. bogotana genetic background (though there may be additional interactions between the D. pseudoobscura USA Q2 and D. pseudoobscura USA Q4 that we were unable to tease out). That said, the effects of a D. pseudoobscura USA allele at Q2 is perhaps slightly reduced compared to the effects of a D. persimilis allele at Q2 (see results in Chang and Noor 2010). Therefore, our results are suggestive of allelism between D. pseudoobscura USA and D. persimilis at Q2, and the effect of the D. pseudoobscura allele appears to be weaker than the D. persimilis allele, but multiple linked factors cannot be completely ruled out.
Several scenarios could explain the possible allelism between D. pseudoobscura USA and D. persimilis at the hybrid-sterility-conferring Q2 locus. The simplest explanation is that this allele may represent the ancestral form, and a subsequent change within the lineage leading to D. pseudoobscura bogotana eliminated this effect, hence allowing for the other changes in D. pseudoobscura bogotana that led to the incompatibility. Serial changes within one lineage resulting in hybrid lethality have been suggested to have occurred in D. mauritiana (Cattani and Presgraves 2009). If these alleles are ancestral, perhaps the observed variation in strength of Q2 was caused by functional divergence of the incompatibility alleles in D. persimilis and D. pseudoobscura USA.
Another intriguing possibility, for which there is some limited support, is that the sterility-conferring allele arose in D. persimilis, and gene flow between D. persimilis and D. pseudoobscura USA in the collinear regions led to allelism at Q2 after the split of D. pseudoobscura USA and D. pseudoobscura bogotana. These species are known to hybridize in the wild (Dobzhansky 1973), molecular data suggest interspecies introgression has occurred in or near the Q2 region (Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009), and this introgression appears to have affected the genetic architecture of hybrid sterility in this system (Brown et al. 2004b; Chang and Noor 2007). Machado et al. (2007) also demonstrated extensive gene flow and higher variation in collinear regions on the 2nd chromosome (not only near Q2), as well as in both arms of the X chromosome, which collectively make up approximately 60% of the genome.
The D. persimilis Q2 allele could have introgressed into D. pseudoobscura USA following hybridization. This explanation is supported by evidence from Kulathinal et al. (2009), who demonstrated that at least some gene flow occurred in collinear regions between D. pseudoobscura USA and D. persimilis after the divergence of the two D. pseudoobscura subspecies.
We know of no demonstrations in which introgression between two species led to hybrid sterility with respect to a third, but a vaguely similar situation where subdivisions within one lineage (e.g. D. pseudoobscura USA and bogotana) have experienced divergence from each other via reproductive isolation with a different lineage (e.g. D. persimilis) was observed in green-eyed tree frogs (Litoria genimaculata). Premating isolation between the northern and southern lineages of Litoria genimaculata led to divergence not only from each other but also between an isolate of the southern lineage found in the northern region (Hoskin et al. 2005). To distinguish between a loss of ancestral Q2 in D. pseudoobscura bogotana and introgression of Q2 from D. persimilis into D. pseudoobscura USA, the causative loci would need to be cloned and sequenced.
This study has demonstrated that some minor within-species variation in strength of hybrid male sterility exists within D. persimilis and D. pseudoobscura bogotana; however, even the minor variation we observe is likely overestimated due to environmental variation in experimental setup. However, despite the negative finding here, QTL mapping studies should continue to test relevant QTLs in multiple lines. We have also determined that a collinear region on chromosome 2 in D. persimilis is allelic to the same region in D. pseudoobscura USA. This study raises the question of whether this region is involved not only in speciation between D. persimilis and D. pseudoobscura bogotana, but also between the two D. pseudoobscura subspecies. If so, perhaps other regions are more closely related between D. pseudoobscura USA and D. persimilis than either are to D. pseudoobscura bogotana.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank C. Barnwell, S. Bennett, B. Manzano-Winkler, and A. Graham for technical assistance, and N. Barton, I. Liu, K. Pelak, members of the Noor laboratory, and two anonymous reviewers for critical comments on the manuscript. This work was funded by National Science Foundation grant 0715484 as well as National Institutes of Health grants GM076051 and GM086445.
LITERATURE CITED
- Aquadro CF, Weaver AL, Schaeffer SW, Anderson WW. Molecular evolution of inversions in Drosophila pseudoobscura - the amylase gene region. Proc. Natl. Acad. Sci U.S.A. 1991;88:305–309. doi: 10.1073/pnas.88.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Awadalla P, Tarone AM. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2004;2:839–848. doi: 10.1371/journal.pbio.0020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell CV, Noor MAF. Failure to replicate two mate preference QTLs across multiple strains of Drosophila pseudoobscura. J. Hered. 2008;99:653–656. doi: 10.1093/jhered/esn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Burk LM, Henagan LM, Noor MAF. Data from: A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution. 2004a doi: 10.1111/j.0014-3820.2004.tb00469.x. [DOI] [PubMed] [Google Scholar]
- Brown KM, Burk LM, Henagan LM, Noor MAF. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution. 2004b;58:1856–1860. doi: 10.1111/j.0014-3820.2004.tb00469.x. [DOI] [PubMed] [Google Scholar]
- Campbell R, Noor MAF. Assessing hybrid male fertility in Drosophila species: Correlation between sperm motility and production of offspring. Dros. Inf. Serv. 2001;84:6–9. [Google Scholar]
- Case AL, Willis JH. Hybrid male sterility in Mimulus (phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution. 2008;62:1026–1039. doi: 10.1111/j.1558-5646.2008.00360.x. [DOI] [PubMed] [Google Scholar]
- Cattani MV, Presgraves DC. Genetics and lineage-specific evolution of a lethal hybrid incompatibility between Drosophila mauritiana and its sibling species. Genetics. 2009;181:1545–1555. doi: 10.1534/genetics.108.098392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AS, Noor MAF. The genetics of hybrid male sterility between the allopatric species pair Drosophila persimilis and D. pseudoobscura bogotana: dominant sterility alleles in collinear autosomal regions. Genetics. 2007;176:343–349. doi: 10.1534/genetics.106.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AS, Noor MAF. Epistasis modifies the dominance of loci causing hybrid male sterility in the Drosophila pseudoobscura species group. Evolution. 2010;64:253–260. doi: 10.1111/j.1558-5646.2009.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA. Genetic basis of male sterility in hybrids between 2 closely related species of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 1984;81:4444–4447. doi: 10.1073/pnas.81.14.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, Mass.: Sinauer Associates; 2004. [Google Scholar]
- Demuth JP, Wade MJ. Population differentiation in the beetle Tribolium castaneum. II. Haldane's rule and incipient speciation. Evolution. 2007;61:694–699. doi: 10.1111/j.1558-5646.2007.00049.x. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. On the sterility of the interracial hybrids in Drosophila pseudoobscura. Proc. Natl. Acad. Sci. U.S.A. 1933;19:397–403. doi: 10.1073/pnas.19.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Is there gene exchange between Drosophila pseudoobscura and Drosophila persimilis in their natural habitats. Am. Nat. 1973;107:312–314. [Google Scholar]
- Gloor GB, Engels WR. Single-fly DNA preps for PCR. Dros. Inf. Serv. 1992;71:148–149. [Google Scholar]
- Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead L. A lethal factor in Crepis effective only in an interspecific hybrid. Genetics. 1930;15:114–140. doi: 10.1093/genetics/15.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskin CJ, Higgie M, McDonald KR, Moritz C. Reinforcement drives rapid allopatric speciation. Nature. 2005;437:1353–1356. doi: 10.1038/nature04004. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman SC, Chen YG, Stajich JE, Noor MAF, Uyenoyama MK. Likelihoods from summary statistics: Recent divergence between species. Genetics. 2005;171:1419–1436. doi: 10.1534/genetics.104.040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MAF. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: The case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Masly JP, Jones CD, Noor MAF, Locke J, Orr HA. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- Nolte VD, Weigel D, Schlotterer C. The impact of shared ancestral variation on hybrid male lethality - a 16 codon indel in the Drosophila simulans Lhr gene. J. Evol. Biol. 2008;21:551–555. doi: 10.1111/j.1420-9101.2007.01486.x. [DOI] [PubMed] [Google Scholar]
- Noor MA. Speciation driven by natural selection in Drosophila. Nature. 1995;375:674–675. doi: 10.1038/375674a0. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Almendarez Y, Reiland J, Smith KR. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution. 2001a;55:512–521. doi: 10.1554/0014-3820(2001)055[0512:tgoria]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. U.S.A. 2001b;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Genetics of Male and Female Sterility in Hybrids of Drosophila pseudoobscura and Drosophila persimilis. Genetics. 1987;116:555–563. doi: 10.1093/genetics/116.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Irving S. Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics. 2001;158:1089–1100. doi: 10.1093/genetics/158.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Counterman BA, Noor MAF. The genetics of speciation by reinforcement. PLoS Biol. 2004;2:2256–2263. doi: 10.1371/journal.pbio.0020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JT. Evolution in the genus Drosophila. New York, NY: Macmillan; 1952. [Google Scholar]
- Phadnis N, Orr HA. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 2009;323:376–379. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- Reed LK, LaFlamme BA, Marko TA. Genetic architecture of hybrid male sterility in Drosophila: analysis of intraspecies variation for interspecies isolation. PLoS One. 2008;3:e3076. doi: 10.1371/journal.pone.0003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart AL, Mason AR, Willis JH. Natural variation for a hybrid incompatibility between two species of Mimulus. Evolution. 2007;61:141–151. doi: 10.1111/j.1558-5646.2007.00011.x. [DOI] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, Buerkle CA, Nachman MW, Tucker PK. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution. 2010;64:472–485. doi: 10.1111/j.1558-5646.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- Wang RL, Wakeley J, Hey J. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics. 1997;147:1091–1106. doi: 10.1093/genetics/147.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H. Spermatogenesis: analysis of meiosis and morphogenesis. In: Henderson DS, editor. Drosophila Cytogenetics Protocols. Totowa, NJ: Humana Press; 2004. pp. 45–75. [DOI] [PubMed] [Google Scholar]
- Wu CI, Beckenbach AT. Evidence for extensive genetic differentiation between the sex-ratio and the standard arrangement of Drosophila pseudoobscura and Drosophila persimilis and identification of hybrid sterility factors. Genetics. 1983;105:71–86. doi: 10.1093/genetics/105.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


