Abstract
Background. Mathematical models of hepatitis C virus (HCV) during therapy may elucidate mechanisms of action for antiviral therapy. In genome-wide association studies, IL28B gene polymorphisms are highly predictive of therapeutic clearance of HCV.
Methods. We collected sera from 20 chronically infected HCV participants at 13 points during the first 28 days of therapy. We assessed the presence of the C allele at single-nucleotide polymorphism rs12979860 using the ABI TaqMan allelic discrimination kit. We estimated dynamic parameters from the entire population using the Neumann model for HCV infection. Statistical methods for repeated nonlinear measures compared model parameters by established predictors of response.
Results. The frequencies of IL28B genotypes were 6 (C/C), 11 (C/T), and 3 (T/T). The mean log decline in HCV RNA from 0 to 48 hours was more rapid among C/C genotype participants compared with C/T or T/T genotype participants (1.4 vs 0.7; P = .07), and from 2 days to 14 days (1.6 vs 0.7; P = .04). In the multivariate model, the C/C genotype predicted a steeper second-phase decline when adjusted for race (P = .01).
Conclusions. The presence of the C/C genotype at IL28B rs12979860 exerts its antiviral effect by increasing the infected hepatocyte death rate. This suggests that an immune-mediated mechanism is responsible.
BACKGROUND
Chronic hepatitis C virus (HCV) infection affects an estimated 3 million Americans and 170 million people worldwide [1]. Although therapy with peginterferon-alfa and ribavirin is curative in approximately 50% of all patients, its use is dampened by the high cost and toxic side effect profile. Virologic relapse, in which a patient achieves undetectable levels of HCV RNA during therapy but then has a recrudescence after stopping therapy, occurs in 25%–33% of the treated cases [2–4].
Mathematical models have been used to infer the mechanisms of action of both interferon and ribavirin [5, 6], as well as to guide the optimal dosing [7] and duration of therapy [8]. They provide insight into the in vivo biology by giving estimates of viral production and clearance. Importantly, a good in vitro model of antiviral efficacy was long lacking, so the estimates of antiviral efficacy from models allowed for comparison of different drug regimens. The mathematical model of Neumann describes a biphasic decline in HCV RNA after treatment with an interferon-based regimen [6]. Following the initial dose of interferon, there is a “first phase” of rapid viral decline, between 0.5 to 2.0 log, during the next 24 to 48 hours. The model explains this initial decline primarily as a result of free virion clearance (c) and the degree of antiviral effectiveness in blocking viral production (ε). After this initial rapid decay, the viral level declines more slowly, entering a “second phase” over the next 14 days. In this second phase, viral decline is primarily dependent on the blocking of new virions (ε) and the death of infected hepatocytes (ε).
Recently, it has been discovered that a genetic polymorphism at rs12979860, near the IL28B gene on chromosome 19, is the strongest predictor of response to peginterferon/ribavirin therapy [9]. Moreover, it is predictive of spontaneous virologic clearance in acute infections [10]. Preliminary data suggest that patients with the C/C genotype are more likely to achieve a rapid virologic response; however, it is not known how this genetic polymorphism enables or prevents a virologic cure [11].
The purpose of the current study was twofold: to study the interplay between the host genetic polymorphisms at SNP rs12979860 and the early response to peginterferon and ribavirin therapy among those chronically infected with hepatitis C; and, secondly, to determine how dynamic parameters in the Neumann model were influenced by traditional predictors of response and the rs12979860 polymorphism.
PATIENTS AND METHODS
Patient Population
Patients aged ≥18 years with difficult-to-treat, chronic hepatitis C infection were recruited from the Hepatitis and Liver Clinic at the Harborview Medical Center, a large public hospital run by the University of Washington (Seattle), from 1 January 2007 to 1 September 2009. For the purposes of this study, chronic infection was defined as a detectable HCV RNA level without a prior percutaneous exposure in the preceding 6 months. Participants were required to have genotype 1 infection or HIV coinfection with any HCV genotype, to be abstinent from drugs and alcohol for the preceding 12 months, to have no suicide attempt or psychiatric hospitalization in the preceding 12 months, and to be able to take full dose peginterferon and ribavirin initially. Treatment-naive and experienced patients were included. In addition, patients were enrolled if they met standard criteria for starting peginterferon and ribavirin therapy. Participants were excluded if they had decompensated cirrhosis. The protocol was approved by the University of Washington Institutional Review Board and conforms to the ethical guidelines of the 1975 Declaration of Helskinki; all patients participated with informed consent.
Blood Sampling and Clinical Evaluations
Each participant had a baseline liver biopsy and was offered treatment according to the National Institutes of Health (NIH) consensus statement and the American Association for the Study of Liver Disease (AASLD) guidelines [12, 13]. Each participant had a baseline HCV RNA level 7 days prior to treatment initiation and then was admitted overnight to the University of Washington General Clinical Research Center for treatment initiation and blood sampling. Blood samples were obtained immediately prior to the first dose of peginterferon-alfa 2a 180 μg subcutaneously and ribavirin 1000–1200 mg/d orally and then at 6, 8, 12, and 24 hours. Participants were discharged and returned for subsequent serum samples at 48, 72, and 96 hours; 7, 9, and 11 days; and then every week up to the 12-week mark. If there was not at least a 2 log decline in HCV RNA by week 12, the participant was discontinued on therapy. Otherwise, participants returned for blood sampling monthly until the completion of 48 weeks of therapy. We evaluated adherence according to the reported number of missed doses in the last month, checked by pharmacy refill information. All patients received their medications through the hospital pharmacy. An independent data and safety committee monitored the development of adverse events, particularly the development of anemia while on therapy, given the frequent phlebotomy. We ceased blood draws if the hematocrit level fell below 30% or if it was below 32% and the patient was symptomatic. We permitted the use of erythropoietin, which occurred at the discretion of the treating physician.
HCV RNA Quantification
We processed participants’ sera within 20 minutes of collection using standard collection and processing techniques. The samples were then immediately placed in a −20°C or −80°C freezer. We determined serum HCV RNA levels using the real-time PCR technique and analyte-specific reagents (LightCycler, Roche). The level of detection was 60 IU/mL. Patient samples were run in 2 batches (the first 5 weeks and weeks 6–48) to prevent viral degradation during prolonged freezing.
Determination of rs12979860 Polymorphism
Genotyping of baseline peripheral blood mononuclear cells was performed at Duke University using the ABI TaqMan allelic discrimination kit and the ABI7900HT Sequence Detection System (Applied Biosystems), as described previously [10].
Participants were reported to have the C/C, C/T, or T/T genotype.
Mathematical Model and Statistical Analysis for the Estimation of HCV Kinetics
The model for HCV virions and infected cell dynamics presented by Neumann [6] was used to describe viral decay after treatment. The model is composed of a system of 3 differential equations that describe viral kinetics during therapy. The model describes states for uninfected cells, T, productively infected cells, I, and free virus, V:
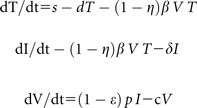 |
Two parameters of interest are δ and c, which allow estimation of the decay rates of infected cells (dI/dt) and free virus (dV/dt), respectively. In addition, the parameters ϵ and η describe the efficacy of the treatment regimen in suppressing viral production by infected cells, and efficacy of the treatment regimen in preventing infection of uninfected target cells, respectively.
It is generally assumed that prior to the start of treatment the entire system is in steady state; ie, prior to treatment, all 3 states T, I, and V are in steady state. It is also generally assumed that the uninfected hepatocyte population, T, remains in steady state over the time interval of interest, ie, posttreatment. Finally, the parameter η has very little effect on the model solution for free virus, V(t) (personal communication, Chia Wang); therefore, this parameter was set to .5 for all analyses. Sensitivity analyses were conducted for a range of values of η from 0 to 1, and, as expected, different values of η did not substantially change any of the parameter estimates of interest or study conclusions.
The parameters c, δ, and ϵ were first estimated for each individual using nonlinear least squares and Splus version 8.0. Data from the first 80 days of treatment when viral load was detectable were used to estimate model parameters. Because of the heterogeneous nature of the study population, comparison of parameters by covariates of interest, such as ancestry and IL28B genotype, was conducted using aggregate data from the entire patient population and generalized nonlinear regression, which allows for adjustment for potential confounding factors as well as correlation due to repeated measures from the same patient [14]. This statistical approach, in contrast to the more commonly used 2-stage approach, improves power to detect differences in kinetic parameters by utilizing all observed data in making statistical inferences In this type of analysis, estimates of average values of the 3 parameters of interest for each level of the covariate(s) are obtained, as well as a P value indicating whether there is a significant difference in each of the parameters by covariate level. In this population level analysis, η was set constant at .5. In the univariate analysis, each parameter (c,δ,ϵ) was estimated for each level of a single covariate, such as IL28B genotype. In multivariate analysis, differences in all 3 parameters were evaluated by >1 covariate, such as IL28B genotype and ancestry, simultaneously, to adjust for the contribution of both factors. The change in viral load from baseline to 48 hours (approximate first-phase decay) and from 48 hours to 2 weeks (approximate second-phase decay) after initiation of therapy was compared by IL28B genotype using linear regression and generalized estimating equations (GEE).
RESULTS
Baseline Characteristics
The cohort was predominantly male, middle-aged, and from diverse ethnic and racial backgrounds (Table 1). The C/T genotype was most common (n = 11, 55%), followed by the C/C (n = 6, 30%) and the T/T genotypes (n = 3, 15%). All 20 patients completed at least 4 weeks of therapy; however, 2 patients stopped for psychosocial reasons after 6 weeks (both patients were Hispanic). Also, 2 participants failed to have an early virologic response, and therefore their treatment was terminated at 12 weeks. The remaining patients completed the recommended 48 weeks of therapy. All participants received >80% of the peginterferon and ribavirin.
Table 1.
Baseline Characteristics of Study Participants (N = 20)*
| Mean (SD) | |
| Age, y | 49 (8) |
| Body mass index | 29 (5) |
| HCV RNA, IU/mL | 6.49 (0.6) |
| ALT, U/mL | 81 (45) |
| Frequency (%) | |
| Male | 17 (85) |
| Race | |
| White | 13 (65) |
| Black | 6 (30) |
| Native American | 1 (5) |
| Ethnicity | |
| Hispanic | 5 (25) |
| Not Hispanic | 15 (75) |
| HCV genotype | |
| 1a | 13 (65) |
| 1b | 6 (30) |
| 3 | 1 (5) |
| Fibrosis level | |
| 1 | 2 (10) |
| 2 | 15 (75) |
| 3 | 3 (15) |
| IL28-B genotype | |
| C/C | 6 (30) |
| C/T | 11 (55) |
| T/T | 3 (15) |
| HIV-positive | 3 (15) |
| Steatosis | |
| >33% | 0 (0) |
NOTE. SD, standard deviation; HCV, Hepatitis C virus; ALT; alanine transaminase; HIV, Human immunodeficiency virus.
*3 patients had been previously treated, 2 with standard interferon and ribavirin, the other with 2 weeks of an investigational polymerase inhibitor.
HCV Response to Treatment, Based on IL28B Polymorphism
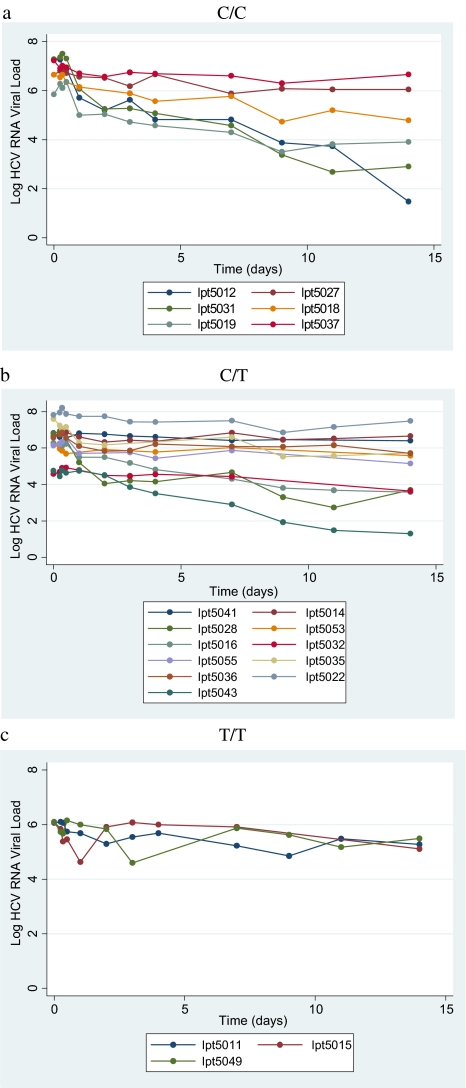
HCV RNA decline based on rs12979860 genotype status is shown in Figures 1 and 2. The steepest first-phase decline occurred in those participants with the C/C genotype (mean HCV RNA decline = 1.4 log after 48 hours of therapy). This value was greater than the pooled group of C/T and T/T genotype patients (mean, 0.7 log; P = .07). The T/T genotype group had the flattest decline (mean, .3 log; P = .01). The differences became more pronounced in the second phase, with a statistically significant greater decline for the C/C genotype (mean HCV RNA decline from 48 hours to 14 days, 1.7 log), compared with participants with either the C/T or T/T genotypes (mean, .7; P = .04). The second-phase decay is generally thought to represent the decay of infection cells; our results suggest that infected cell death between 2 and 14 days posttreatment is significantly increased in patients with C/C genotype compared with those with the C/T or T/T genotype.
Figure 1.
Fourteen-day Hepatitis C virus RNA decline according to genotype.
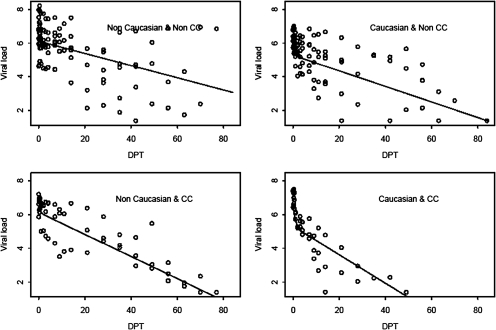
Figure 2.
Hepatitis C virus RNA decline on peginterferon and ribavirin therapy according to race and IL28B genotype. DPT, Days Posttreatment Start.
HCV RNA Dynamics During Antiviral Therapy and Mathematical Modelling
We used the mathematical model developed by Neumann to fit the decrease in HCV RNA level during therapy. The parameters of δ, c, and ϵ were estimated and compared by categories of the covariate listed in Table 2. The HCV RNA decline over time in each patient, incorporating the model fit, is available in Supplement 1. The influence of established covariates, such as gender, fibrosis level, HIV status, body mass index, and race, on these parameters was determined. There were too few participants in the prior treatment group or nongenotype 1 HCV for meaningful comparisons. In the univariate model, race significantly affected the free virion decay rate (c), but was not significant after adjusting for the rs12978960 genotype. In the bivariate model, patients with European ancestry had greater efficacy (ϵ) and a faster hepatocyte death rate (δ), although this did not meet statistical significance. Interestingly, the univariate model did not find that the rs12978960 genotype affected any of the model parameters. However, on the bivariate analysis, which adjusted for ancestry, the rs12978960 genotype significantly affected the infected cell death rate (δ). In that analysis, participants with the C/C genotype had a much faster infected cell death rate, compared with those with either the C/T or T/T genotype.
Table 2.
Influence of Covariates on Parameter Estimates
| Covariate or effect | Effect levels | Parameter |
||||||
| c (free virion decay/day) |
Delta (δ, infected hepatocyte decay/day) |
Epsilon (ε) |
||||||
| Estimates | P value | Estimates | P value | Estimates | P value | |||
| Race* | Race alone | Non-European ancestry | 1.16 | .02 | .13 | .10 | .75 | .07 |
| European ancestry | 2.87 | .07 | .98 | |||||
| Race adjusted for genotype | Non-European ancestry | 1.1 | .33 | .11 | .08 | .78 | .07 | |
| European ancestry | 1.88 | .05 | .97 | |||||
| IL28B Genotype* | Genotype alone | T/C or T/T | 1.51 | .52 | .10 | .21 | .84 | .35 |
| C/C | 1.95 | .14 | .94 | |||||
| Genotype adjusted for race | T/C or T/T | 1.1 | .52 | .11 | <.01 | .78 | .28 | |
| C/C | 1.67 | .3 | .16 | |||||
| Fibrosis stage | ≤2 | 2.07 | .32 | .09 | .35 | .95 | .11 | |
| >2 | .89 | .15 | .71 | |||||
| Body mass index | >30 | 1.58 | .43 | .09 | .67 | .81 | .15 | |
| ≤30 | .63 | .10 | .95 | |||||
| Sustained virologic response | Nonresponder | 1.16 | .07 | .08 | .20 | .73 | .06 | |
| Responder | 3.07 | .13 | .99 | |||||
| Gender | Male | 2.08 | .21 | .09 | .58 | .93 | .49 | |
| Female | 1.04 | .12 | .85 | |||||
| HIV status | Not infected | 1.91 | .83 | .10 | .24 | .90 | .60 | |
| Infected | 2.09 | .06 | .96 | |||||
NOTE. ϵ = efficacy of suppressing production of new virions, expressed as % inhibition of viral production; c = free virion clearance rate per day; δ = infected cell clearance rate per day; HIV, human immunodeficiency virus.
*These effects were tested both alone (in univariate models) and with each other (bivariate model). All other effects were tested in univariate models only.
Ancestry and the rs12979860 genotype exerted a synergistic effect on early viral kinetics (Figure 2). The steepest HCV RNA declines occurred in patients who were of European ancestry and the C/C genotype (Figure 2), followed by non-European ancestry, C/C genotype patients.
DISCUSSION
In this ethnically and racially diverse cohort of difficult-to-treat HCV-infected patients, we found that participants with the C/C genotype had a significantly steeper second-phase decline corresponding to infected cell death rate. Participants with C/C genotype also had a more rapid first-phase decline compared with those with T/T genotype. Although not statistically significant, there was a trend toward European ancestry and an association with the efficacy of suppressing the production of new virions, even after accounting for the IL28 genotype.
Infected hepatocytes can decay from a variety of causes, including natural cell death, immune-mediated decay, and noncytolytic cure. Without histological data during therapy, it is difficult to ascertain the etiology of the infected hepatocyte decay. However, we believe that an immune-mediate mechanism is responsible, based on several studies. Preliminary evidence suggests that the rs12979860 polymorphism is very close to the IL28B gene [9], perhaps in the promoter or enhancer region of the gene. The polymorphism is also associated with natural clearance, suggesting that innate immunity is engaged [10]. The IL28B gene produces interferon lambda, a recently discovered type III interferon, which is expressed after viral infection of the liver and signals through a similar janus kinase–signal transducer and activator of transcription (JAK-STAT) pathway as interferon-α [15–17]. The JAK-STAT pathway activates interferon-sensitizing genes (ISG) such as 2’,5’-oligoadenylate synthetase (OAS) and mitogen-activated protein (MAP) kinases, which are known to cause apoptosis, growth inhibition, and viral replication [18].
Interestingly, Marcello et al found that HCV subgenomic RNA replication in vitro treated with IFN-λ showed a steady increase in ISGs, whereas those HCV-infected cell cultures treated with IFN-α showed a rapid peak and then decline in ISGs [19]. Furthermore, IFN-λ can act as an “antiviral boost” to IFN-α, by enhancing the antiviral effect [20]. In light of the current findings and these in vitro results, we hypothesize that patients of European ancestry are inherently more sensitive to IFN-α, as reflected in the trend toward improved efficacy in suppressing virus production (ϵ), but that participants of non-European ancestry are able to still achieve viral clearance through activation of the backup IFN-λ pathway.
The results are similar to previous studies that have found that δ (second phase) was the best predictor of antiviral response and that δ is lower in African-Americans [21–23]. Layden-Almer compared the viral kinetics in African-Americans and found that they had a lower ϵ and δ, compared with those of white participants [21]. Our data suggest that the rs12979860 polymorphism explains this difference in δ. However, there may be other racial factors that are associated with a difference in ϵ. Of note, the T allele occurs more frequently in African-Americans, but the frequency explains only 50% of the variability in response, when compared with those participants with European ancestry [9]. Thompson et al similarly found that patients with European ancestry were more likely than African-Americans to have more rapid kinetics, even in the absence of the C/C genotype [11]. A recent study identified 4 other gene regions associated with natural clearance (TNFSF18, TANK, HAVCR1, and IL18BP); these regions may show differential expression in various races undergoing therapy [24]. In a study of Europeans taking peginterferon or standard interferon, the second-phase decay rate was most predictive of SVR [23]. Finally, in a multinational study of high dose induction with interferon, the infected cell decay rate was again the strongest predictor of viral cure [22].
There are several unique strengths to this study. First, our cohort was heterogenous racially and ethnically, and included 3 HIV-positive patients, which makes the results more generalizable, especially to the American population. Second, we did not exclude any data in our modeling. Finally, the statistical approach to parameter estimation was rigorous, and it allows for evaluation of a diverse population of study participants. Many researchers evaluating viral dynamics in a population of patients fit the data to patients one at a time and conduct statistical analysis on the estimated dynamics parameters (this method is often referred to as the 2-stage approach). In this work, data from the aggregate population was used with nonlinear regression analysis to simultaneously estimate and compare parameters by subgroups of interest (eg, IL28 genotype). The use of regression methods with the aggregate data increases power to detect differences in model parameters and allows for adjusted analysis in our diverse population. Thus, we were able to estimate the effect of the IL28 genotype adjusted for patient ancestry. Our analysis demonstrates that genotype and ancestry must be considered simultaneously, something that is possible only when aggregate data and regression methods are used. A weakness of the study was the assumption that there is constant effectiveness of antiviral therapy. Some authors have questioned the validity of this assumption [25].
In summary, the presence of the C/C genotype at IL28B rs12979860 exerts its antiviral effect by increasing the infected hepatocyte death rate. This suggests that an immune-mediated mechanism is responsible. Functional studies of the polymorphism will more clearly elucidate the mechanism of antiviral response.
Funding
Funding for this study was provided by the National Institutes of Health National Center for Research Resources (grants UL1 RR025014; K23 RR02206 to J. S.; R01 AI055343 to S. H.).
Acknowledgments
The patient care was conducted at the University of Washington Clinical Research Center, a core resource of the Institute of Translational Health Sciences.
The authors wish to thank Wan Chong Qiu and Erica Seddig for their assistance in collecting and processing participant specimens.
References
- 1.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet. 2001;358:958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, et al. APRICOT Study Group. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. New Engl J Med. 2004;351:438–50. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 5.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–4. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 6.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-α. therapy. Science. 1998;282:103–7. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 7.Lam NP, Neumann AU, Gretch DR, Wiley TE, Perelson AS, Layden TJ. Dose-dependent acute clearance of hepatitis C genotype 1 virus with interferon α. Hepatology. 1997;26:226–31. doi: 10.1002/hep.510260130. [DOI] [PubMed] [Google Scholar]
- 8.Neumann AU, Lam NP, Dahari H, et al. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J Infect Dis. 2000;182:28–35. doi: 10.1086/315661. [DOI] [PubMed] [Google Scholar]
- 9.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 10.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson AJ, Muir AJ, Sulkowski MS, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–9. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 12.National Institutes of Health. National Institutes of Health Consensus Development Conference statement: Management of hepatitis C: 2002—June 10–12 2002. Hepatology. 2002;36(5 Suppl 1):S3–20. doi: 10.1053/jhep.2002.37117. [DOI] [PubMed] [Google Scholar]
- 13.Strader DB, Wright TL, Thomas DL, Seeff LB. American Association for the Study of Liver Disease. Diagnosis, management, and treatment of hepatitis C. Hepatology. 2004;39:1147–71. doi: 10.1002/hep.20119. [DOI] [PubMed] [Google Scholar]
- 14.Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. 1st ed. New York: Springer Verlag; 2000. [Google Scholar]
- 15.Kotenko SV, Gallagher G, Baurin VV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 16.Sheppard P, Kinsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine rececptor IL-28R. Nat Immunol. 2003;4:63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 17.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-λ is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:1–23. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maher SG, Sheikh F, Scarzello AJ, et al. IFN alpha and IFN lambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther. 2008;7:1109–15. doi: 10.4161/cbt.7.7.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–98. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, Butera M, Nelson DR, Liu C. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J. 2005;2:80. doi: 10.1186/1743-422X-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Layden-Almer JE, Ribiero RM, Wiley T, Perelson AS, Layden TJ. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003;37:1343–50. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 22.Rosen HR, Ribeiro RM, Weinberger L, et al. Early hepatitis C viral kinetics correlate with long-term outcome in patients receiving high dose induction followed by combination interferon and ribavirin therapy. J Hepatol. 2002;37:124–30. doi: 10.1016/s0168-8278(02)00114-9. [DOI] [PubMed] [Google Scholar]
- 23.Zeuzeum S, Herrmann E, Lee JH, et al. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon alpha2a. Gastroenterology. 2001;120:1438–47. doi: 10.1053/gast.2001.24006. [DOI] [PubMed] [Google Scholar]
- 24.Mosbruger TL, Duggal P, Goedert JJ, et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis. 2010;201:1371–80. doi: 10.1086/651606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shudo E, Ribeiro RM, Perelson AS. Modelling hepatitis C virus kinetics during treatment with pegylated interferon alpha-2b: Errors in the estimation of viral kinetic parameters. J Viral Hepat. 2008;15:357–62. doi: 10.1111/j.1365-2893.2007.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]