Introduction
There is a tremendous history of capitalizing on the biodiversity of our natural environment for molecules to treat diseases and regulate host physiology (e.g. penicillin, acetylsalicylic acid). However, until recently we have failed to probe the therapeutic potential of our microbial symbionts: a biodiverse population of bacteria, archaea, viruses, protists, and fungi that reside on and within us. Recently there has been an increased realization that the microbiota significantly impact many of our physiological and immunological processes (Fig. 1), suggesting that there is great opportunity for therapeutic discoveries.
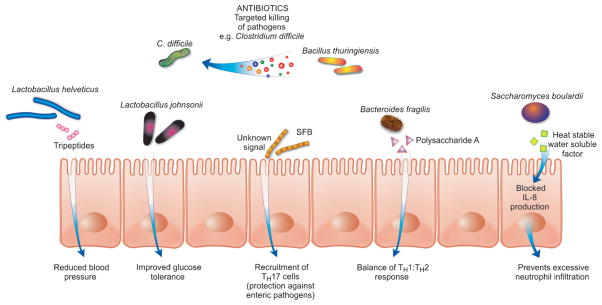
Figure 1.
Some of the many mechanisms through which gut microbes and their signals can affect host health. SFB, segmented filamentous bacteria.
The potential of the microbiota to affect the host is vast, considering its impressive genetic composition. The collective genomes of our resident microbes, referred to as the metagenome, represent a gene set which is estimated to be 150-fold greater than the human genetic complement [1]. Many of these genes are the product of co-evolution and have been selected on the basis of improving host fitness rather than the fitness of the microbe itself. As we begin to understand previously unidentified properties and molecules of the microbiota, we will be able to “mine” these for potential therapeutic benefit. Here, we discuss some of the avenues of discovery that present opportunities for further exploitation. Possibilities discussed include strategies for excluding pathogens, manipulation of the immune system and regulation of non-intestinal sites.
Modulating nutritional homeostasis
The resident microbiota of the human gastrointestinal (GI) tract encode many biochemical pathways that humans have not evolved, facilitating the break-down of proteins and indigestible polysaccharides into essential amino acids, vitamins, and short chain fatty acids [2]. The generation of some of these metabolites is a collaborative effort between distant microbial community members. However, alliance is not always beneficial, as specific mechanisms to exploit nutrients enable individual community members to thrive in this competitive environment [1, 3, 4]. A recently described “minimal gut bacterial genome” represents some of the genes that are key to survival in the gut [1]. Further characterization of these genes will facilitate the development of strategies to modify the microbiota for therapeutic purposes.
Just as the resident members of the microbiota must compete, enteric pathogens must vie with the microbiota for nutrients to join and dominate this highly dense yet dynamic community. Enteric pathogens such as enterohemorrhagic Escherichia coli (EHEC), Salmonella enterica serovar Typhimurium, Campylobacter jejuni, and Vibrio cholerae are just some pathogens that have developed strategies to compete, utilize, and/or induce virulence gene expression upon encountering unique metabolites found within the GI tract [5–8]. Some pathogens directly interact with the host to generate beneficial molecules for their survival. For example, acute gut inflammation caused by S. Typhimurium infection generates a respiratory electron acceptor (tetrathionate) that provides a competitive growth advantage to the pathogen [9]. While the nutrient environment, which is modulated by the resident microbiota, influences enteric pathogenicity, changes in the composition of the microbiota could also lead to downstream changes in the nutrient environment and the enteric colonization potential of the gut [10]. Therefore, strategies to manipulate the composition of the intestinal microbiota to limit some of these pathogen-enhancing nutrients within the GI tract could restrict growth and virulence of bacterial disease. Additionally, more direct measures to modulate levels of specific nutrients could be employed, which could involve chemical chelation or supplementation.
Just as some nutrients found within the GI tract promote the growth and virulence of some pathogens, other microbiota-generated metabolites are known to be protective against enteric pathogen colonization. One example includes acetate produced by Bifidobacterium, which inhibits translocation of EHEC toxins from the gut to the blood [11]. Future therapies to restrict virulence by specific enteric pathogens could employ the use of targeted vitamin pills designed to release their pathogen inhibiting metabolites in specific regions of the GI tract. It is important to note that the consequences of perturbing the nutrient and/or micro-biota composition are unknown. Thorough studies will be required to ensure that manipulating the intestinal nutrient environment is fully beneficial. As the nutrient environment in the GI tract is modulated by the host micro-biota, understanding how the micro-biota composition controls metabolite sources that pathogens encounter will certainly lead to unique strategies to control colonization and virulence.
Antibiotic discovery
Besides nutrients, microbes in the GI tract encounter thousands of small molecules that are not readily used as carbon and energy sources. This includes molecules produced by the host as well as the micro-biota and, in many cases, these molecules are important players in the maintenance of homeostasis in the intestinal environment (e.g. hormones). They can act as signals that carry messages from microbial or host cells to specific targets and act in competition and cooperation. Microbial signals are structurally diverse, and many classes of important chemical mediators have been described in both prokaryotes and eukaryotes. Some signals come from unexpected sources: antibiotics are known to act as microbial signals to regulate gene expression at subinhibitory concentrations [12]. Small signaling molecules are active at very low concentrations (picomolar to nanomolar ranges). This raises the possibility that many signaling molecules discovered to date have distinct activities at higher concentrations. Indeed, some signaling molecules have been shown to possess antibiotic activity when used at higher concentrations [13]. In the complex environment of the GI tract, it is likely that many of the small molecules used for signaling will show antibiotic activity. In fact, a Bacillus thuringiensis strain isolated from a human fecal sample has recently been shown to produce a potent antimicrobial peptide (thuricin) with a narrow spectrum of activity against Clostridium difficile [14]. In light of the current scarcity of new antibiotics and the development of microbial drug resistance, the gut may represent an underappreciated source of new molecules that could be used to fight infectious agents.
Harvesting immune regulators
Microbial signals are also required for proper immune development and throughout life to maintain immunity, as microbial depletion quickly results in immune deficits [15]. Bacteria have been shown to regulate immune function via bacterial components, metabolic products, and secreted proteins [11, 16, 17]. The effects of microbiota on the immune system can be both inductive and inhibitory; therefore, they could provide a means to exogenously control the immune system in multiple directions.
Many diseases, such as allergy and inflammatory bowel diseases (IBDs), are associated with an imbalanced or uncontrolled immune response. Can we define the microbial signals that direct the immune system so as to treat disease? Therapies that block the over expression of inflammatory cytokines that are seen in IBD are effective. An unidentified secreted signal, produced by a member of the microbiota that is depleted in IBD patients, has shown promise in maintaining this balance in animal models of the disease [17]. Additionally, several other microbial peptides and proteins have been shown to have anti-inflammatory properties. Therapeutic potential for treating neutrophil-associated inflammatory diseases may already exist with Saccharomyces boulardii, as it produces and secretes a heat-stable, water-soluble factor that blocks the neutrophil recruiting signal, IL-8 [18].
Microbes can also provide immuno-stimulatory signals to help prevent pathogen colonization. For example, segmented filamentous bacteria (SFB) induce a TH17 response, which protects the host from pathogen infection [19]. The signal or mechanism involved has yet to be defined, but could certainly be a useful preventative therapy. Another signal that induces protection is a quorum-sensing molecule produced by Bacillus subtilis that activates cytoprotective heat shock proteins, preventing oxidant-induced epithelial injury and loss of barrier function [20]. It has also been shown that a bacterial cell surface polysaccharide (Polysaccharide A from Bacteroides fragilis) is sufficient to correct the T-helper cell imbalance in germ-free mice [16].
Treating and preventing diseases outside of the GI tract
Alterations in the intestinal microbiota have been shown to have an impact on organs outside of the GI tract. Complex diseases such as obesity, autism, diabetes, and allergies have been linked to imbalances in the intestinal microbiota composition [21]. The increasing knowledge in this field has unveiled a new resource that could be mined for compounds to help counter these diseases.
The microbiome plays an essential role in energy balance. The Firmicutes-Bacteroidetes ratio in the gut has been shown to correlate with obesity in mice [22]. The obesity-associated microbiome harbors a substantially greater number of genes encoding for polysaccharide-degrading enzymes [22]. In this example, identifying specific members of the Bacteroidetes phylum that are related to a leaner phenotype would be of great potential to control obesity, perhaps through the administration of probiotics. Alternatively, polysaccharide-degrading enzymes linked to a better absorption of nutrients in obese mice could be helpful for treatment of mal-nourishment in developing countries or metabolic diseases. In the quest for treatment of hyperglycemia, oral administration of a specific probiotics strain (Lactobacillus johnsonii La1) was shown to improve glucose tolerance affecting blood glucose regulation via alteration of the autonomic nervous system [23].
There is also a correlation between the microorganisms living in the gut and the development of atopic diseases in children. Differences in gut colonization with Lactobacillus spp., Bifidobacterium spp., and C. difficile were shown to correlate with predisposition to the development of allergies [21]. Probiotic strains (L. reuteri and L. fermentum) were shown to have a positive impact on the control of atopic diseases, such as eczema and atopic dermatitis in children [24, 25]. Treatments against these diseases could be considered by defining the bacterial structures necessary for protection.
Alterations in the gut microbiota have also been linked to late-onset autism, particularly related to the presence of Clostridium spp. [26]. Several hypotheses for how these bacteria might cause such a complex disease have been made, including the production of neurotoxins and self-recognizing antibodies that attack neuron-associated proteins [26]. Unraveling the mechanisms that cause neurological diseases will be fundamental in moving toward a treatment.
A probiotic bacterium (L. helveticus) has been shown to produce specific tri-peptides (isoleucyl-prolyl-proline and valyl-prolyl-proline) with hypotensive effects through proteolytic activity on milk-based proteins [27]. These bacteria possess strong proteinase activity that produce peptides that inhibit angiotensin-converting enzyme, thus reducing blood pressure [27]. An example of how this information might be translated to treatment is the administration of these peptides to lower blood pressure in hypertensive patients.
Above are some examples of how the microbiota and its products might affect diseases of organs distant from the gut and how we might capitalize on this knowledge to identify new strategies for treatment of these multifactorial disorders. Understanding the mechanisms behind these interactions is an essential step in this search. Although significant challenges arise from studying such a complex community, identification of bacterial species, and their genes and products would open new avenues of treatment and prevention for many diseases.
Exploratory studies to unveil new therapeutic avenues
As mentioned above, there are many examples of host-microbe interactions that display potential for manipulation for human benefit. In most cases, crossing the barrier between basic research and drug discovery will rely on identification of microbial species involved in health or disease states or microbial molecules that mediate these interactions. Identification of components of the mammalian intestinal micro-biome has come a long way with the development of culture-independent techniques for community analysis. Using untargeted sequencing of rRNA genes isolated from whole communities, one can compare significant changes in microbial composition between healthy and disturbed communities. Individual microbial species associated with health can then be used as probiotics (assuming they can be cultured), whereas species associated with disease can be targeted for elimination.
Besides the administration of whole organisms that function as probiotics, isolated microbial components may also be used and, as mentioned above, promising results have been obtained to treat IBD. Identifying molecules in the human intestine that could be used as therapeutics will also require high-throughput studies of the intestinal ecosystem. However, in this case the focus will be on the molecules themselves and not the microbes. Although this type of analysis can still be technically challenging and cumbersome, new techniques of small molecule mass spectrometry have been developed to allow the detection and identification of thousands of compounds in complex biological samples in a short period of time [28]. For example, Jansson et al. [29] used mass-spectrometry-based metabolomics to identify chemicals in the GI tract of healthy subjects and IBD patients and showed that many metabolites strongly correlated with health status (e.g. fatty acid, bile acid, and amino acid metabolites). Also, our group has recently used high-throughput metabolomics to identify changes in the intestinal metabolome caused by microbiota disturbance due to antibiotics and enteric infection [30, 31]. Such studies can be used to identify small molecules associated with health and disease. This can be done with a view of developing therapeutics that target not only intestinal ailments but also diseases of remote organs. Wikoff et al. [32] have recently shown that the intestinal microbiota has a significant impact on the metabolic composition of blood. For instance, plasma levels of sulfate and glycine-conjugated metabolites, tryptophan and other indole-containing molecules as well as the hormone serotonin were significantly different between germ-free and conventional mice. This suggests that some molecules whose synthesis can be affected by the intestinal microbiota may be promising targets for the development of therapeutics that could act in other organs or even at a systemic level. Although the studies mentioned above show great promise in this field, more concrete associations still need to be made to demonstrate the potential of microbial molecules as new therapeutics. This will require a concerted effort for the definitive identification of the microbial species and the small molecules involved.
Acknowledgments
L. C. M. A and R. B. R. F. are funded by the Canadian Institute of Health Research (CIHR). K. M. K. is supported by the National Institute of Health under Ruth L. Kirschstein National Research Service Award FAIO85761A. B. B. F. is an HHMI International Research Scholar and The University of British Columbia Peter Wall Distinguished Professor. Work in his laboratory is funded by operating grants from the CIHR and the Canadian Crohn’s and Colitis Foundation.
Abbreviations
- EHEC
enterohemorrhagic Escherichia coli
- GI
gastrointestinal
- IBD
inflammatory bowel disease
- SFB
segmented filamentous bacteria
Footnotes
There are no conflicts of interest.
References
- 1.Qin JJ, Li RQ, Raes J, Arumugam M, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Possemiers S, Grootaert C, Vermeiren J, Gross G, et al. The intestinal environment in health and disease – recent insights on the potential of intestinal bacteria to influence human health. Curr Pharm Des. 2009;15:2051–65. doi: 10.2174/138161209788489159. [DOI] [PubMed] [Google Scholar]
- 3.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acid Sci USA. 2009;106:5859–64. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell A, Romano GH, Groisman B, Yona A, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–4. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 5.Velayudhan J, Jones MA, Barrow PA, Kelly DJ. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun. 2004;72:260–8. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schild S, Tamayo R, Nelson EJ, Qadri F, et al. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–77. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Suyemoto M, Garner CD, Cicconi KM, et al. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol. 2008;190:4233–41. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, et al. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–30. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 9.Winter SE, Thiennimitr P, Winter MG, Butler BP, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–9. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda S, Toh H, Hase K, Oshima K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 12.Goh EB, Yim G, Tsui W, McClure J, et al. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–30. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosoda K, Shimomura H, Hayashi S, Yokota K, et al. Steroid hormones as bactericidal agents to Helicobacter pylori. FEMS Microbiol Lett. 2011;318:68–75. doi: 10.1111/j.1574-6968.2011.02239.x. [DOI] [PubMed] [Google Scholar]
- 14.Rea MC, Sit CS, Clayton E, O’Connor PM, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107:9352–7. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandl K, Plitas G, Mihu CN, Ubeda C, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–7. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Sokol H, Pigneur B, Watterlot L, Lakhdari O, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sougioultzis S, Simeonidis S, Bhaskar KR, Chen X, et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-kappaB-mediated IL-8 gene expression. Biochem Biophys Res Commun. 2006;343:69–76. doi: 10.1016/j.bbrc.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov II, Atarashi K, Manel N, Brodie EL, et al. Induction of intestinal Th17cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiya M, Musch MW, Nakagawa Y, Hu S, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 23.Yamano T, Tanida M, Niijima A, Maeda K, et al. Effects of the probiotic strain Lactobacillus johnsonii strain La1 on autonomic nerves and blood glucose in rats. Life Sci. 2006;79:1963–7. doi: 10.1016/j.lfs.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 24.Abrahamsson TR, Jakobsson T, Bottcher MF, Fredrikson M, et al. Probiotics in prevention of IgE-associated eczema: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:1174–80. doi: 10.1016/j.jaci.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Weston S, Halbert A, Richmond P, Prescott SL. Effects of probiotics on atopic dermatitis: a randomised controlled trial. Arch Dis Child. 2005;90:892–7. doi: 10.1136/adc.2004.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finegold SM, Molitoris D, Song Y, Liu C, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35:S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 27.Tuomilehto J, Lindstrom J, Hyyrynen J, Korpela R, et al. Effect of ingesting sour milk fermented using Lactobacillus helveticus bacteria producing tripeptides on blood pressure in subjects with mild hypertension. J Hum Hypertens. 2004;18:795–802. doi: 10.1038/sj.jhh.1001745. [DOI] [PubMed] [Google Scholar]
- 28.Han J, Datla R, Chan S, Borchers CH. Mass spectrometry-based technologies for high-throughput metabolomics. Bioanalysis. 2009;1:1665–84. doi: 10.4155/bio.09.158. [DOI] [PubMed] [Google Scholar]
- 29.Jansson J, Willing B, Lucio M, Fekete A, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antunes LC, Arena ET, Menendez A, Han J, et al. The impact of Salmonella infection on host hormone metabolism revealed by metabolomics. Infect Immun. 2011;79:1759–69. doi: 10.1128/IAI.01373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antunes LC, Han J, Ferreira RB, Lolic P, et al. The effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikoff WR, Anfora AT, Liu J, Schultz PG, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]