Abstract
Death-inducing signaling complex (DISC) formation is a critical step in Fas-mediated signaling for apoptosis. Previous experiments have demonstrated that the calmodulin (CaM) antagonist, trifluoperazine (TFP) regulates CaM-Fas binding and affects Fas-mediated DISC formation. In this study, we investigated the anti-cooperative characteristics of TFP binding to CaM and the effect of TFP on the CaM-Fas interaction from both structural and thermodynamic perspectives using combined molecular dynamics simulations and binding free energy analyses. We studied the interactions of different numbers of TFP molecules with CaM and explored the effects of the resulting conformational changes in CaM on CaM-Fas binding. Results from these analyses showed that the number of TFP molecules bound to CaM directly influenced α-helix formation and hydrogen bond occupancy within the α-helices of CaM, contributing to the conformational and motion changes in CaM. These changes affected CaM binding to Fas, resulting in secondary structural changes in Fas and conformational and motion changes of Fas in CaM-Fas complexes, potentially perturbing the recruitment of Fas-associated death domain (FADD) for DISC formation. The computational results from this study reveal the structural and molecular mechanisms that underlie the role of the CaM antagonist, TFP, in regulation of CaM-Fas binding and Fas-mediated DISC formation in a concentration-dependent manner.
Keywords: CaM antagonist TFP; CaM/Fas binding; DISC; molecular dynamics, binding free energy, conformational and motion analysis
INTRODUCTION
The molecular mechanisms mediated by the prototypical death receptor Fas have been well characterized (1). The Fas receptor is a member of the tumor necrosis factor receptor superfamily. Upon stimulation by Fas ligand or agonistic antibodies, Fas induces receptor clustering and formation of the death-inducing signaling complex (DISC). Abnormal expression of Fas has been reported in numerous types of cancer. Down-regulation of Fas expression has been observed in breast cancer, glioma, hepatocellular carcinoma, and pulmonary adenocarcinoma (2-5). Mutations in the Fas gene that cause alterations in the structure and/or function of Fas have also been detected in many cancers, including melanoma, T-cell leukemia, thyroid cancer, and lymphoma (6-9), and are also associated with lymphoproliferative and autoimmune disorders in both mice and humans (10-14). Therefore, regulation of Fas-mediated apoptosis represents a promising diagnostic and therapeutic avenue for treatment of multiple cancers, including cholangiocarcinoma (15-20).
Calmodulin (CaM) is a small intracellular protein that primarily functions as an intracellular mediator of Ca2+ signals (21). Elevated levels of Ca2+-bound CaM are associated with cancer (22), and CaM antagonists inhibit tumor cell invasion in vitro and metastasis in vivo (23), suggesting that CaM antagonists may prove to be promising chemotherapeutic agents. CaM antagonists induce apoptosis of cholangiocarcinoma cells, at least in part, through Fas-mediated apoptotic pathways (24). Recent experimental studies showed that CaM binds to Fas and regulates Fas-mediated DISC formation (25, 26). Furthermore, binding of CaM to Fas can be inhibited by the CaM antagonist, trifluoperazine (TFP), in a concentration-dependent manner (25, 26). The concentration-dependent effect of TFP on CaM-Fas binding could be directly related to the number of TFP molecules bound to CaM.
CaM is a ubiquitous Ca2+ binding protein of 148 residues that regulates a variety of physiological processes in a Ca2+-dependent manner (21, 27, 28). CaM consists of two globular domains similar in sequence and structure (the N-domain and the C-domain), linked by a central helix (29). Each globular domain consists of two helix-loop-helix Ca2+-binding regions referred to as EF-hand structures (30). In both the Ca2+-bound and Ca2+-free states, CaM adopts an ‘elongated’ structure in which the two globular domains are connected by a highly flexible linker (31-37). However, when bound to the CaM antagonist TFP, the structure of CaM is modified from a ‘dumb-bell’ shape to a compact globular shape (38). This change in conformation may then inhibit CaM binding to Fas, ultimately affecting Fas-induced signaling pathways. Fas has a six alpha-helix structure with a single hydrophobic site located within helices 5 and 6 (39). The Fas death receptor has the ability to self-associate or to bind other target proteins, including CaM and the Fas-associated death domain (FADD), to form the DISC (39, 40), which activates downstream signaling. Previous studies have shown that helices 1-2 of Fas associate with CaM (41, 42), and this interaction is tightly regulated during Fas-mediated apoptosis (25). Our recent study showed that the conformation of Fas could be altered by the Fas V254N mutation or by a C-terminal deletion mutation, and that these changes directly affect the degree of binding between Fas and CaM (6). Our study also showed that CaM binding to Fas resulted in conformational changes in both CaM and Fas, which likely further affects Fas recruitment of FADD to form the DISC (6). Furthermore, binding of the CaM antagonist, TFP to Ca2+-bound CaM results in conformational changes in CaM (38, 43-45) and affects CaM/Fas binding in a concentration-dependent manner (26), thereby inhibiting Fas-mediated DISC formation.
In this study, we investigated the anticooperative characteristics of TFP molecules that were bound to CaM and the effect of TFP on CaM-Fas binding from both structural and thermodynamic perspectives using the combined molecular dynamics simulations and binding free energy analyses. This study provides structural and thermodynamic evidence elucidating the role of the CaM antagonist TFP in regulating CaM-Fas binding and Fas-mediated DISC formation and apoptosis. The results provide the potential for identifying new drug candidates for regulation of the Fas-mediated signaling pathway of apoptosis for cancer chemotherapy.
MATERIALS AND METHODS
We performed molecular dynamics (MD) simulations and binding free energy analyses on CaM bound to different numbers of TFP molecules to characterize the anticooperative characteristics of TFP molecules that were bound to CaM and to evaluate the conformational and motion changes in CaM elicited by binding to the different numbers of TFP molecules. Using the equilibrated conformations of CaM bound to different numbers of TFP molecules, we constructed complexes of CaM and Fas. These complexes were then used to assess the effect of TFP molecules on CaM-Fas binding, providing structural insight into the effects of the CaM antagonist TFP on Fas-mediated recruitment of FADD for DISC formation.
Complex structure preparation
Complex structures of CaM bound to TFP
The initial complex structures of Ca2+-bound CaM to one TFP (PDBID: 1CTR) (43), two TFPs (PDBID: 1A29) (44), and four TFPs (PDBID: 1LIN) (45) were obtained from the Protein Data Bank. For the crystal structure of CaM bound to one TFP, residues 75-81 linking the N- and C- domains were missing (43). Using the structure of CaM bound to two TFPs as a template, we constructed the missing residues of CaM bound to one TFP with a homology modelling method using the MODELLER 9v2 package (46). We performed 30ns MD simulations for the three structures of CaM bound to different numbers of TFP molecules using the AMBER 9 MD package (47).
CaM-Fas complex construction
The initial protein structure of the Fas death domain (1ddf) (39) was obtained from the Protein Data Bank. The structures of CaM bound to different numbers of TFPs were obtained from equilibrated MD simulations. The initial complex of Fas with CaM (TFP was not included in the complex) was determined using the docking utilities, ZDOCK and RDOCK (48-50) using the experimentally known binding site of CaM in Fas as docking constraint. The performance of the combined ZDOCK/RDOCK programs has been demonstrated in the Critical Assessment of Prediction of Interactions (CAPRI) for protein docking (51, 52) and provided satisfactory results from MD simulations using the predicted protein complexes (6, 53-55). We also have successfully adopted ZDOCK and RDOCK program to construct the complex of CaM with no TFP bound with the Fas death domain and validated the calculated change of the binding free energy of CaM-Fas complex by Fas mutations with those from the experimental studies (6). CaM binding region in Fas is located within helices 1 and 2 (residues 231-254) of the Fas death domain (Fas DD) (27). Using the known binding site as a docking constraint, 2000 possible CaM-Fas complexes were generated using ZDOCK 2.3 docking software (56). Complexes were scored based on their geometry, desolvation, and electrostatics (48). The RDOCK program was used to perform the energy minimization for all of the 2000 predicted complexes through the CHARMM MD simulation package (57). The electrostatic, van der Waals, and contact energies associated with each of the proposed complexes were determined and used for ranking (49, 52). The binding energy results of the top three complexes of Fas with CaM bound to different number of TFP molecules were shown in Table 2S. The highest ranking structure was chosen to be the most probable CaM-Fas complex. 30 ns MD simulations were performed for the chosen complexes and the degree of changed binding free energy of the complexes by binding to different number of TFP molecules were calculated based on the MD trajectories and validated with experimental results to confirm the validity of the constructed CaM-Fas complexes.
Molecular dynamics simulation
To elucidate the structural basis and molecular mechanisms underlying the role of the CaM antagonist TFP in regulating CaM/Fas binding and Fas-mediated DISC formation, we performed a total of six 30 ns MD simulations using the AMBER 9 MD package (47) (Table 1). In our previous study, we have performed 30 ns MD simulation of CaM with no TFP bound and the complex of Fas with CaM not bound to TFP (6). We used the “parm99” parameters of the AMBER force field for the proteins in the simulated system. The initial TFP (Figure 1S) structure was obtained from the crystal structure of CaM-1TFP complex (PDBID: 1CTR) (43). The force field for TFP that is compatible with the general AMBER force field was determined using the ANTECHAMBER program (58) that is an accessory module in the AMBER 9 package. The RESP charges for TFP were obtained using the Gaussian 03 quantum mechanics program, using Hartree-Fock calculation with the 6-31G* basis set (58-61). The final charges of TFP used in the MD simulation were listed in Table 1S. While different local environment and conformational differences could cause small charge variations for TFP, using a fixed charge parameter set for TFP was necessary for the long molecular dynamics simulations. We expect that the new efficient polarizable force field that is actively developed by Ponder's group (62) will help overcome the need for the fixed charge in the future.
Table.1.
Six systems simulated in this study
| System | TFP - CaM complex | CaM - Fas complex |
|---|---|---|
| 1 | 1TFP – CaM | |
| 2 | 2TFP – CaM | |
| 3 | 4TFP – CaM | |
| 4 | 1TFP bound CaM – Fas | |
| 5 | 2TFP bound CaM – Fas | |
| 6 | 4TFP bound CaM – Fas |
A standard MD simulation protocol was performed for the six simulated systems. The TFP-CaM complex, or the CaM-Fas complex, was first minimized and solvated with TIP3P water molecules (63) in a periodic box with 150 mM physiological NaCl concentration. The periodic box size of the simulated systems were shown in Table 3S. The solvated system was first energy minimized with the protein complex and ions restrained, but with water unrestrained. The solvent was further equilibrated with the protein complex and ions restrained at a constant number-pressure-temperature (NpT) at 50 K and 1 atm for 20 ps. The simulated system was warmed via constant number-volume-temperature (NVT) MD simulations to 300 K by steps of 50 K lasting 10 ps each, with SHAKE constraints and a 2 fs time step. Production MD simulation of 30 ns at NpT of 300 K and 1 atm was performed for the simulated system. In the production simulations, SHAKE constraints with a relative tolerance of 1×10-5 were set on all hydrogen-heavy atom bonds to permit a dynamics time step of 2 fs. The particle-mesh Ewald method was used for electrostatic interaction calculations (64) with grid spacing of 0.12 nm and interpolation of order 4, and Lennard-Jones cutoffs set at 1.0 nm. All of the MD simulations were performed on a local DELL Infiniband Xeon cluster, a local BlueGene cluster, and on the AMD Opterons cluster in the Alabama Supercomputer Center.
Binding free energy analyses
We calculated and compared the binding free energy difference among the three TFP-CaM complexes and among the three CaM-Fas complexes to understand the effects of the number of TFP molecules bound to CaM on TFP-CaM binding and the effects of conformational changes in CaM caused by binding to TFP(s) on CaM-Fas binding. Binding free energy of the complex structure over the cumulative time was also used to verify the equilibration of the complex structure during the MD simulations. The binding free energy of the complex was calculated with the MM-PBSA method, as described in previous studies (6, 30, 65-67), which was implemented using the AMBER 9 Molecular Dynamics Software Package. The MM-PBSA method combines molecular mechanics, continuum electrostatics, solvent accessible surface area calculations, and normal mode analyses for entropy to calculate the binding free energy from a series of snapshots obtained from the trajectories of the MD simulations. The binding free energy was calculated by finding the energy difference between the complex and the receptor and ligand as shown in equation (1):
| (1) |
where Ḡ is the average value over the production trajectory
The binding energy is the sum of the molecular mechanics contribution to the energy (also known as the gas phase energy) and the solvation free energy of the system (Eq. 2). The solvation free energy is made up of both polar and nonpolar contributions (Eq. 3). The polar contribution to the solvation free energy is based on the electrostatics of the system, and calculated using the Poisson-Boltzman equation. In AMBER, this term is computed with the pbsa solver. The non-polar component is dependent upon the surface area of the molecule that is accessible to the solvent.
| (2) |
| (3) |
The gas phase contribution to the binding free energy is equal to the difference between the molecular mechanics energy and the entropy of the system (Eq. 4). The molecular mechanics portion of the binding energy calculation (Eq. 5) is equal to the sum of the van der Waals energy, electrostatics energy, and internal energy resulting from bond, angle, and torsion energies. T represents the temperature and S is the entropy of the system. To calculate the entropy of the system, the structure of the system was subjected to rigorous energy minimization, until the step change of energy < 10-5 kcal/mol. Then the mass-weighted Hessian matrix for each minimized structure was calculated and diagonalized by using normal mode analysis (68-70). The frequency of the normal mode was then used to calculate the conformational entropy (71), using MM-PBSA utility in AMBER 9.
| (4) |
| (5) |
The MM_PBSA program in the AMBER 9 MD package was used to calculate the binding free energy based on snapshots from the MD simulation trajectories. For the polar solvation energy calculation with the Poisson-Boltzmann equation using the PBSA solver in the AMBER 9 MD program, dielectrics values for the solute and solvent were chosen as 1 and 78.4 respectively (30). A 150 mM ionic strength and a temperature of 300 K were used for the energy calculation, identical to conditions for the MD simulations. A grid space of 0.5 Å was used for continuum electrostatics calculations. Calculation of the apolar solvation energy was based on the solvent-accessible surface area using the Molsurf solver in AMBER 9 MD program. Snapshots at a time interval of 20 ps from the MD simulation trajectories after initial equilibration were used to calculate all components of binding free energy except entropy. Due to the high computational cost for entropy calculations, snapshots at a time interval of 200 ps were used for the normal mode analysis to calculate the entropy of the system. The means and standard deviations of the binding free energy over the MD simulation trajectories were calculated after the system reached initial equilibration using the bootstrap statistical method (72). A Student's t-test with 95% confidence was used to determine whether the effect of the number of TFP molecules bound to CaM on TFP-CaM binding and the effect of conformational change of CaM caused by binding to TFP(s) on CaM-Fas binding were statistically significant.
Conformational and structural analyses
A series of analyses were carried out to better understand the conformational and motion character of CaM bound to different numbers of TFP molecules and the conformational and motion character of Fas binding to TFP-bound CaM. Root mean square deviations (RMSD) of protein backbone atoms were analyzed in conjunction with the binding free energy results to determine the systems’ equilibration tendencies and its convergence. Root mean square fluctuations (RMSF) of the proteins were calculated on a residue-by-residue basis, and averaged over the production simulation trajectories to observe the conformation fluctuation of protein domains. Dynamical cross-correlation maps (normalized covariance matrices) between residues provided insight into which residues have general movements correlating to the motions of other residues and the degree of this correlation (6, 65). Principal component analysis (PCA) was calculated to evaluate the essential modes of motion at the hydrophobic binding pocket of CaM induced by binding to TFP(s) and the distribution of the relative contribution of the first ten PCA modes of the Fas DD in CaM-Fas complexes. The primary motion of the system is usually represented by the relatively lower frequency modes obtained from PCA, and the first PCA mode is usually interpreted as the direction of the largest conformational fluctuation of the system during MD simulations (73-75). For PCA analyses, each PCA mode is associated with an eigenvalue that corresponds to the amplitude of fluctuations along that mode, and each eigenvalue normalized by the sum of all eigenvalues can be used to represent the relative contribution of a mode to the dynamic motion of the selected atoms observed in the MD simulation (6).
We calculated hydrogen bond formation within the α-helices, the occupancy of the residues of the α-helices for CaM and Fas, and the dihedral angle transitions of the residues in Fas to evaluate structural changes in CaM induced by binding to TFP molecule(s) and structural changes in Fas caused by binding to CaM. A hydrogen bond was assigned when the distance between the hydrogen and the acceptor was less than 4 Å and the angle of donor – hydrogen – acceptor was less than 30°. OH and NH groups were treated as donors, and oxygen and nitrogen atoms were defined as acceptors. The occupancy of each hydrogen bond was calculated based on the percentage of time that the hydrogen bond existed over the entire simulation time. The hydrogen bond occupancy of each α-helix in the protein was considered to be the average occupancy of the hydrogen bonds of each α-helix. An α-helix was defined to be at least five residues long in a coiled or spiral conformation. If the NH group of an amino acid formed a hydrogen bond with the C=O group of the amino acid found four residues earlier in the sequence, the amino acid was assigned to belong to the α-helix. The occupancy of each residue in an α-helix was determined based on the percentage of time that the residue existed in the α-helix over the simulation. Dihedral angle transitions for the residues in the protein, which are representative of backbone transformations, were also evaluated for Fas bound to CaM. These analyses were performed with the MD simulation trajectories obtained after the initial equilibration using the ptraj program of AMBER. Matlab (76) was used to generate the cross-correlation plots.
Statistical methods
To determine the binding characteristics of TFP molecules that were bound to CaM and to determine the effect of TFP on CaM-Fas binding, the means and standard deviations of the binding free energies were calculated. To overcome the tendency of adjacent snapshots from the MD trajectories to correlate with one another, we calculated the autocorrelation time τ (77, 78) for the calculated variables in order to re-sample the trajectories into statistically independent periods for calculation of the standard deviations for the calculated variables. To avoid the potential problem that the individual quantity could couple to the longest time-scale system property, we calculated the autocorrelation time with the system RMSD as performed in our previous studies (6, 78, 79). With a decorrelation time of 2τ, we used bootstrap analyses (72), with a similar protocol to that of Chen and Pappu (80) and our previous study (79), to calculate the mean and standard deviation of the binding free energy. Significant differences in the means and standard deviations for the interested variables were determined using the Student's t-test (81) with 95% confidence.
RESULTS & DISCUSSION
Equilibration of the simulated systems
TFP/CaM complexes
For the complexes of CaM bound to different numbers of TFP molecules, the RMSD of the backbone of CaM in the complexes over the 30 ns MD simulation indicated that the system reached initial equilibrium after 15 ns of simulation (Figure 2S). The binding free energies for the TFP-CaM complexes as a function of cumulative time over the 30 ns MD simulation also showed that the complexes converged after 15 ns of MD simulation (Figure 3S). The last 15 ns of MD trajectories were used for the analyses of the binding free energy and the conformational and motion changes of CaM induced by binding to TFP.
CaM/Fas complexes
RMSD of protein backbone atoms were analyzed in conjunction with the binding free energy results to determine the systems’ equilibration tendencies and its convergence. The binding free energies of the CaM-Fas complexes over the 30ns MD simulations, which was calculated as the ensemble-averaged value over the production trajectory, showed that the energy of the systems converged after the first 15 ns of the production simulation (Figure 4S). The RMSD of the Fas death domain protein core (res 225-318) over the 30 ns MD simulations also showed that the system reached initial equilibration after 15 ns of simulation (Figure 5S). Although the RMSD of CaM still have slight drifting after 15 ns of simulation (Figure 6S), it is mainly due to the fact that CaM is a highly flexible protein resulted from its flexible central linker region. The remaining 15 ns were then used for the analyses of Fas and CaM conformational changes and binding thermodynamics.
Binding thermodynamics analysis
Binding characteristics of TFP molecules with CaM
Experimental results from circular dichroism spectroscopy, together with X-ray diffraction for CaM bound to two TFPs (CaM-2TFP), suggest that the first TFP molecule binds to the C-terminal hydrophobic pocket and the second TFP binds to the interdomain site of CaM (44, 82). The crystal structure of CaM bound to four TFPs (CaM-4TFP) further shows that two TFPs bind at the hydrophobic pockets of the C-terminal and N-terminal halves, called ‘intradomain sites,’ and the other two TFPs bind to the central linker region, termed ‘interdomain sites’ (45). Furthermore, binding of these four TFP molecules changes the conformation of CaM from an elongated dumb-bell form to a compact globular form (45). High-performance liquid chromatography binding assay data suggests that binding of the four TFPs to CaM is not cooperative (83). The calculated binding free energy of the first TFP (TFP-1) with CaM in the three TFP-CaM complexes: CaM-1TFP, CaM-2TFP, and CaM-4TFP was shown in Table 2A. The mean of the binding free energy was obtained by averaging over trajectories, and the standard deviation was calculated with the independent periods of the MD simulation trajectories using the bootstrap statistics method described in the Methods section. The results showed that the second TFP (TFP-2) bound to CaM significantly decreased the binding affinity between TFP-1 and CaM, as the binding free energy increased from -14.3±3.3 kcal/mol to - 3.8±2.1 kcal/mol. However, the difference in decreased binding affinity between TFP-1 and CaM caused by the addition of the third and fourth TFP was not significantly different from that of the second TFP, as the binding free energy of TFP-1 with CaM was -5.3±2.7 kcal/mol in the CaM-4TFP complex (Table 2A). We also analyzed the binding free energy of TFP-2 with CaM in the CaM-2TFP and CaM-4TFP complexes (Table 2B). The results from these analyses showed that the third TFP and the fourth TFP bound to CaM significantly decreased the binding of TFP-2 to CaM, as the binding free energy of TFP-2 with CaM was increased from -13.4±5.7 kcal/mol to -4.1±3.8 kcal/mol (Table 2B). Analysis of the energy components demonstrated that electrostatic energy, van der Waals energy, polar solvation energy, and entropy all significantly contributed to the change in the binding free energy of TFP with CaM caused by additional TFP molecules bound to CaM (Table 2). These calculated results elucidated the binding characteristics of TFP molecules with CaM from the thermodynamics point of view. The calculated binding free energy results showed that binding of TFP molecules to CaM was not cooperative, consistent with experimental observations that binding of four TFPs to CaM is not cooperative (83).
Table 2 (A).
Binding free energy between TFP-1 and CaM
| CaM bound with one TFP | CaM bound with two TFPs | CaM bound with four TFPs | |
|---|---|---|---|
| Δ EvdW | -48.9 ± 3.5 | -37.7 ± 2.7 | -33.6 ± 3.2 |
| Δ Eelectrostatic | -4.9 ± 2.1 | -2.7 ± 1.8 | -8.7 ± 2.6 |
| Δ Gnonpolar–solvation | -6.7 ± 0.2 | -6.6 ± 0.3 | -5.8 ± 0.4 |
| Δ Gpolar–solvation | 23.1 ± 3.6 | 16.0 ± 2.9 | 20.4 ± 4.3 |
| Δ TS | -23.1 ± 6.5 | -27.2 ± 6.0 | -22.7 ± 5.5 |
| Δ Gbinding | -14.3 ± 3.3 | -3.8 ± 2.1 | -5.3 ± 2.7 |
| Δ Δ Gbinding | 10.5* ± 2.8 | 9.0* ± 3.0 |
All values in this table were expressed in terms of kcal/mol. ΔEvdW is the van der Waals energy, ΔEelectrostatic is the electrostatic energy, ΔGpolar–solvation is the polar solvation energy, ΔGnopolar–solvation is the nonpolar solvation energy, ΔTS is the energy contributed from solute entropy, ΔGbinding is the binding free energy for the complex, and ΔΔGbinding is the relative binding free energies between CaM and the first TFP for the systems of CaM bound to two TFPs or four TFPs with respect to the system of CaM bound to one TFP.
Differences are statistically significant (Student's t-test, p<0.05)
Table 2 (B).
Binding free energy between TFP-2 and CaM
| CaM bound with two TFPs | CaM bound with four TFPs | |
|---|---|---|
| Δ EvdW | -38.5 ± 3.0 | -28.8 ± 3.0 |
| Δ Eelectrostatic | -8.6 ± 2.0 | -5.6 ± 2.5 |
| Δ Gnonpolar–solvation | -6.3 ± 0.4 | -5.3 ± 0.4 |
| Δ Gpolar–solvation | 20.0 ± 2.8 | 17.3 ± 3.1 |
| Δ TS | -19.9 ± 5.7 | -18.3 ± 4.4 |
| Δ Gbinding | -13.4 ± 5.7 | -4.1 ± 3.8 |
| Δ Δ Gbinding | 9.3* ± 4.8 |
All values in this table were expressed in terms of kcal/mol. ΔEvdW is the van der Waals energy, ΔEelectrostatic is the electrostatic energy, ΔGpolar–solvation is the polar solvation energy, ΔGnopolar–solvation is the nonpolar solvation energy, ΔTS is the energy contributed from solute entropy, ΔGbinding is the binding free energy for the complex, and ΔΔGbinding is the relative binding free energies between CaM and the second TFP for the system of CaM bound to four TFPs with respect to the system of CaM bound to two TFPs.
Differences are statistically significant (Student's t-test, p<0.05)
Effect of TFP on CaM-Fas binding
The binding free energy of the CaM-Fas complexes, including the mean value and the standard deviation was shown in Table 3. The results showed that one TFP bound to CaM significantly reduced CaM/Fas binding, as the binding free energy of the CaM-Fas complex changed from -7.1±4.7 kcal/mol for the non-TFP-bound CaM/Fas complex (6) to -2.3±1.6 kcal/mol for the one TFP-bound CaM/Fas complex. However, as the number of TFP molecules bound to CaM was increased to two and four, the binding affinity of the CaM-Fas complex almost completely recovered. The binding free energies of the two TFP-bound CaM-Fas complex and the four TFP-bound CaM-Fas complex were equivalent to that of the non-TFP-bound CaM-Fas complex. Analysis of the changes in the energy components contributing to the binding free energy showed that the changes of electrostatic energy, van der Waals energy, polar solvation energy, and entropy resulting from the CaM-Fas interactions all significantly contributed to the change in the binding free energy (Table 3). The results showed that the effect of TFP on CaM-Fas binding is dependent on the number of TFP bound to CaM. The number of TFP molecules bound to CaM is likely directly related to the TFP concentration in the experimental systems. Therefore, the calculated changes in the binding free energy of CaM-Fas complexes containing CaM bound to different numbers of TFP molecules were consistent with the experimental observations that TFP treatment affected CaM/Fas binding in a concentration-dependent manner (26).
Table 3.
Free energy calculation for Fas binding to CaM bound to TFPs
| CaM (6) | CaM bound to one TFP | CaM bound to two TFPs | CaM bound to four TFPs | |
|---|---|---|---|---|
| Δ EvdW | 43.4 ± 4.2 | -33.9 ± 6.2 | -59.8 ± 5.7 | -64.6 ± 8.3 |
| Δ Eelectrostatic | -484.8 ± 53.5 | -364.5 ± 52.8 | -223.5 ± 39.2 | -448.4 ± 36.8 |
| Δ Gsolvation–electrostatic | 476.9 ± 49.9 | 373.9 ± 50.2 | 245.6 ± 24.0 | 473.8 ± 42.3 |
| Δ Gsolvation–nopolar | -8.4 ± 0.6 | -6.1 ± 0.7 | -10.9 ± 1.1 | -11.0 ± 0.6 |
| Δ TS | -52.5 ± 2.9 | -28.4 ± 2.6 | -41.5 ± 3.5 | -44.2 ± 4.2 |
| Δ Gbinding | -7.1 ± 4.7 | -2.3 ± 1.6 | -6.9 ± 3.7 | -6.1 ± 2.2 |
| Δ Δ Gbinding | 4.9* ± 3.5 | 0.2 ± 4.2 | 1.1 ± 3.6 |
All values in this table were expressed in terms of kcal/mol. ΔEvdW is the van der Waals energy, ΔEelectrostatic is the electrostatic energy, ΔGpolar–solvation is the polar solvation energy, ΔGnopolar–solvation is the nonpolar solvation energy, ΔTS is the energy contributed from solute entropy, ΔGbinding is the binding free energy for the complex, and ΔΔGbinding is the relative binding free energies with respect to the complex of Fas and CaM not bound to TFP.
Differences are statistically significant (Student's t-test, p<0.05)
Conformational and structural changes of CaM in CaM-TFP complexes
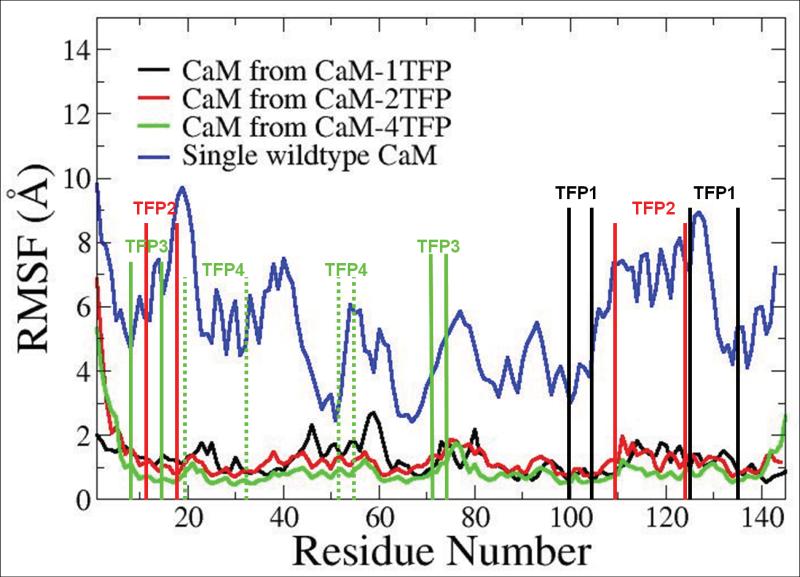
With the understanding of the thermodynamic basis of the binding characteristics of TFP molecules with CaM and the validation of the calculated results with experimental results regarding the anticooperative characteristics of TFP molecules binding to CaM, we further examined the conformational and motion changes in CaM induced by binding to TFP. The crystal structure of the Ca2+-bound CaM-4TFP complex shows that the first TFP (TFP1) binds to CaM in the region of res 92-144, the second TFP (TFP2) binds to CaM in the regions of res 11-18 and res 109-124, the third TFP (TFP3) binds to CaM in the regions of res 8-15 and res 72-147, and the fourth TFP (TFP4) binds to CaM in the regions of res 19-32 and res 51-71 (45). These data indicate that more than one region of the protein is involved in forming a particular TFP-binding site, and that the involved regions of the protein interact with more than one TFP (45). The calculated RMSF results for CaM in the CaM not bound to TFP, CaM-1TFP, CaM-2TFP, and CaM-4TFP complexes are shown in Figure 1. The overall magnitude of RMSF for CaM in CaM-TFP complexes was less than 3 Ǻ, which was significantly less than the RMSF of CaM not bound to TFP of more than 5 Ǻ overall (6). The more stable structure of CaM bound to TFP compared to CaM not bound to TFP was directly related to the structural change in CaM from an elongated dumb-bell form without TFP to a compact globular form when TFP was bound. The RMSF of CaM bound to different numbers of TFP molecules also exhibited differences, particularly in the binding regions for TFP molecules, and these changes were dependent on the number of TFP molecules bound to CaM (Figure 1). The conformational stability changes in CaM induced by binding to different numbers of TFP molecules could directly contribute to the experimental observations and the calculated results regarding the non-cooperative characteristics of TFP molecules binding to CaM, and could further affect binding of CaM to Fas.
Figure 1.
RMSF comparison of CaM in different CaM/TFP complexes.
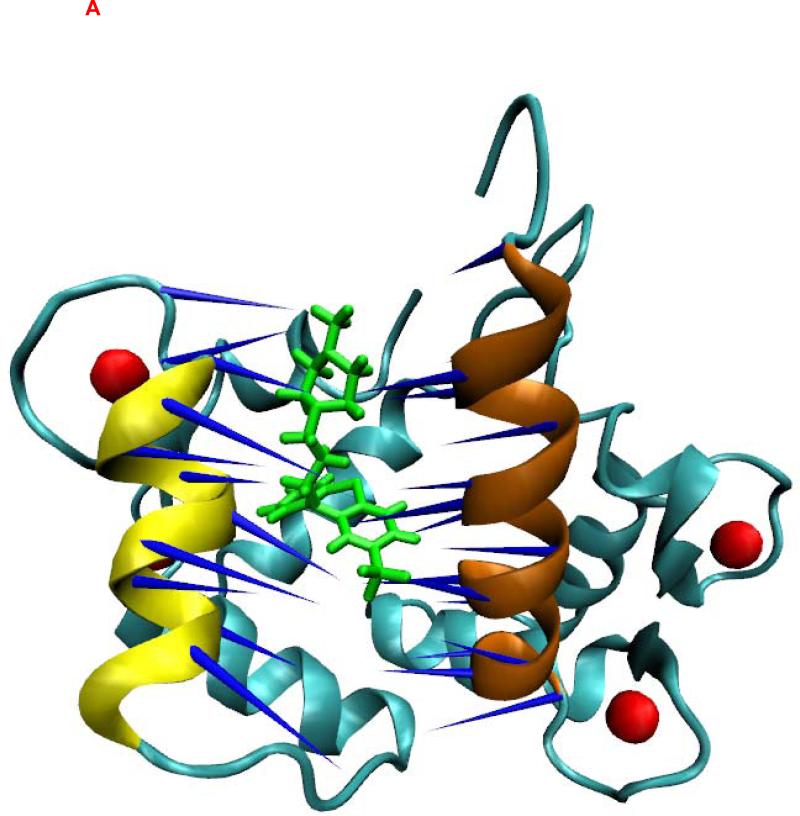
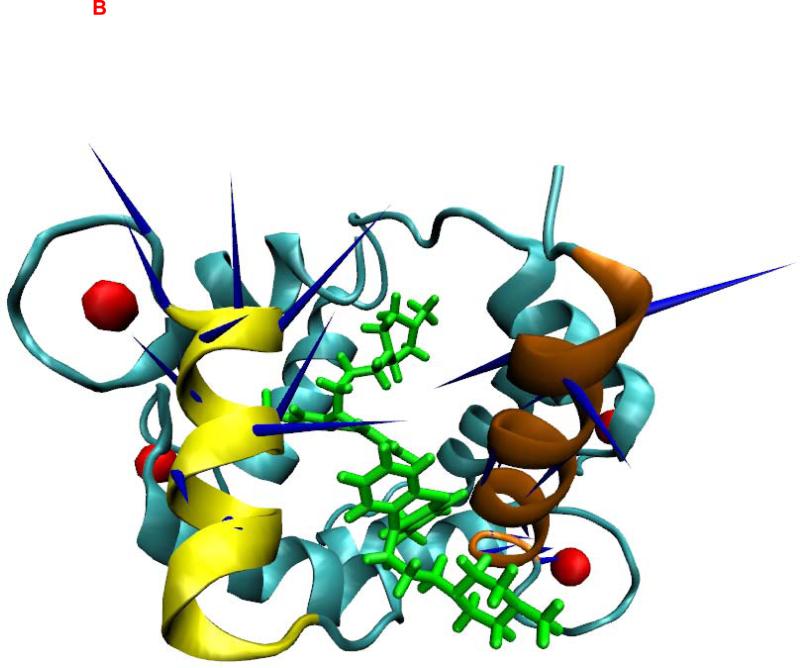
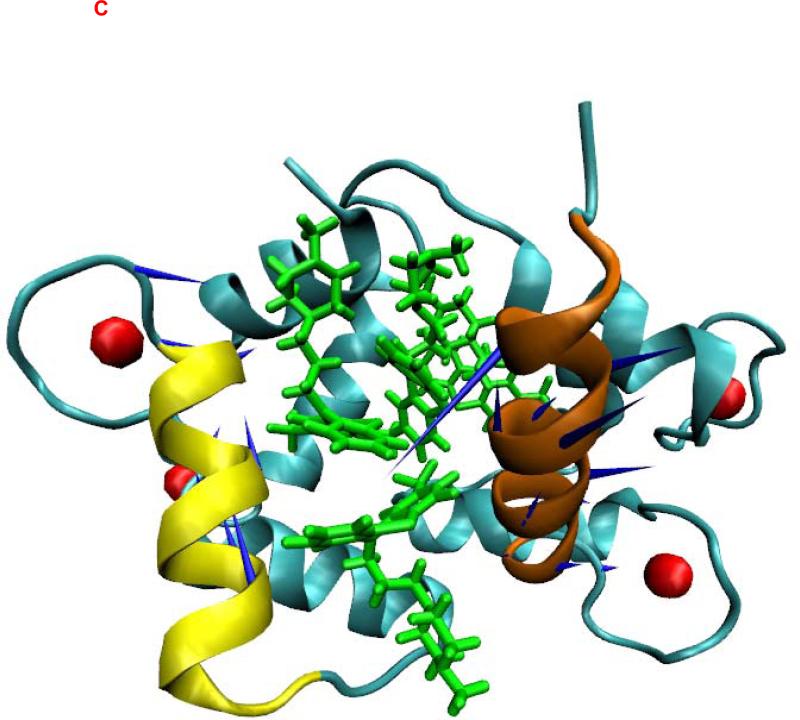
The crystal structures of CaM bound to TFP (43-45) showed that the hydrophobic pockets of the C-terminal and N-terminal halves of CaM and the central linker region of CaM form the binding regions for different TFP molecules. To understand the binding characteristics of TFPs bound to CaM, we focused on elucidating the PCA mode of the binding pocket formed by the C-terminal and N-terminal halves of CaM in the CaM-TFP complexes (Figure 2). The regions of res 5-20 (shown in red) and res 120-130 (shown in brown) correspond to the helixes in the C and N termini of CaM that form a binding pocket for TFP (Figure 2). The motion directions of the first PCA mode of the two helices that form the TFP binding pocket are shown on the atomic structures of the TFP-CaM complexes (Figure 2). Comparison of the direction and amplitude of the first PCA mode at the binding pocket of CaM in the TFP-CaM complexes showed that for the C-terminal helix (res 5-20) and the N-terminal helix (res 120-130), a stronger and more parallel anti-correlated motion tendency was observed for the CaM-1TFP complex compared to the CaM-2TFP and CaM-4TFP complexes. A dispersive motion direction was observed for the CaM-2TFP and CaM-4TFP complexes, further demonstrating the anticooperative characteristics of TFP molecules bound to CaM as observed in the experimental study (83).
Figure 2.
Principal dynamic modes for the residues 5-20 of CaM (brown) and the residues 120-130 of CaM (yellow) from CaM/TFP complexes. CaM was shown as new cartoon. TFPs were shown as licorice in green. Ca2+ was shown in red sphere. (A) CaM from CaM/1TFP complex. (B) CaM from CaM/2TFP complex. (C) CaM from CaM/4TFP complex. The images were made with VMD program.
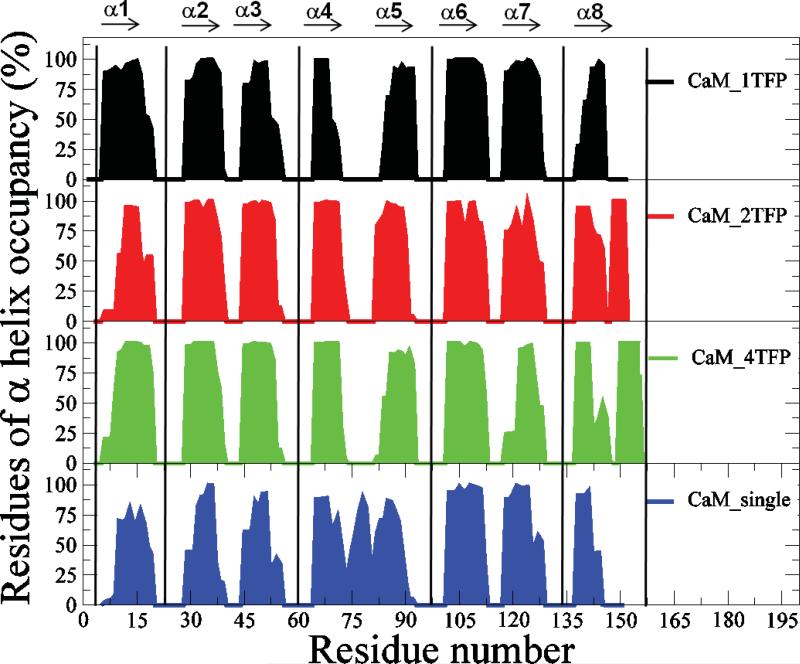
In the absence of TFP, CaM adopts an ‘elongated’ structure in which the two globular domains, composed of six α-helices, are connected by a highly flexible linker composed of one α-helix (31-37). With TFP molecules bound to CaM, the structure of CaM is modified from a “dumb-bell’ shape to a compact globular shape (38). We analyzed α-helix formation and hydrogen bond formation within the α-helices of CaM in CaM-TFP complexes over the last 15 ns of MD simulation trajectories. Results from this analysis revealed that TFP molecules bound to CaM caused the CaM linker region, which is composed of one α-helix, to split into two α-helices (Figure 3). Significantly different residue occupancies of the α1 and α8 helices in CaM were observed for the CaM-1TFP complex compared to that of the CaM-2TFP and CaM-4TFP complexes (Figure 3). Different numbers of TFP molecules bound to CaM resulted in different hydrogen bond occupancies for the α-helices, particularly for the α4 helix, which is located within the linker region of CaM that serves as a binding region for TFP molecules (Figure 8S). The altered α-helix length contributed to changes in the hydrogen bond occupancy within the α-helix. These changes could directly affect the conformational stability and motion of CaM in TFP-CaM complexes as observed in Figure 1 and Figure 2.
Figure 3.
Residues of α helix occupancy of CaM in TFP-CaM complexes
Conformational and structural changes of Fas in CaM-Fas complexes
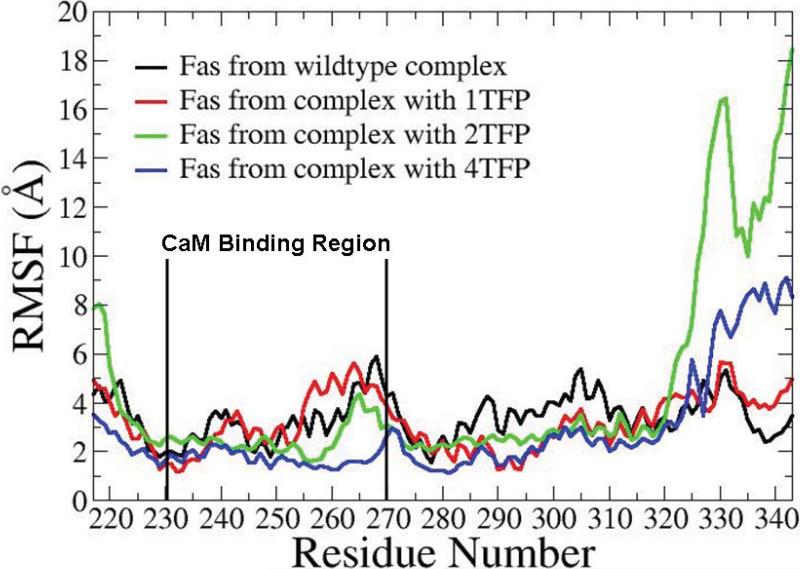
With the validation of the calculated results of the effect of TFP on CaM/Fas binding with the experimental results (26), we further investigated the conformational and motion changes of Fas by binding to TFP-bound CaM, providing structural insight about the effect of TFP on Fas-induced DISC formation. We examined the structural stability of Fas with RMSF analyses, and assessed the motion of Fas with dynamical cross correlation maps and PCA analyses. RMSF of the Fas death domain (Fas DD) showed that CaM bound to a different number of TFPs caused a different degree of fluctuation for the CaM binding site in Fas (res 231–270), including the residues around the key residue Val 254 that is critical for CaM-Fas binding (27) (Figure 4). Compared to the RMSF of Fas bound to non TFP-bound CaM (6), 1TFP-bound CaM resulted in an increased fluctuation at the CaM binding site in Fas, while either 2TFP-bound CaM or 4TFP-bound CaM resulted in a decreased fluctuation at the CaM binding site in Fas. The conformational stability changes in Fas induced by interactions with CaM bound to different number of TFP molecules could provide structural evidence to support previous experimental observations that TFP treatment affected CaM/Fas binding in concentration-dependent manners (26).
Figure 4.
RMSF comparison of the Fas Death Domain protein core (res 225-318) in the CaM/Fas complex for different simulation cases.
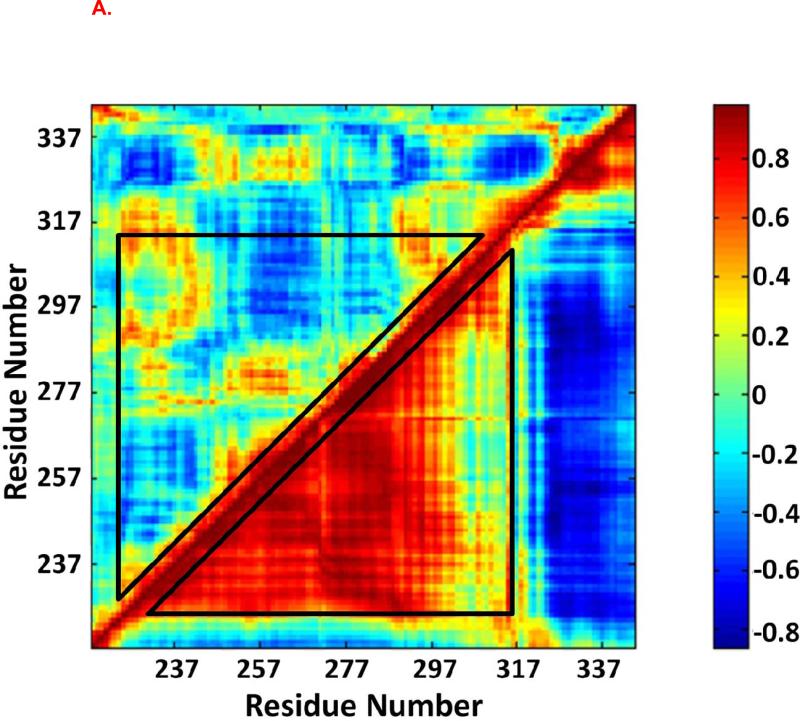
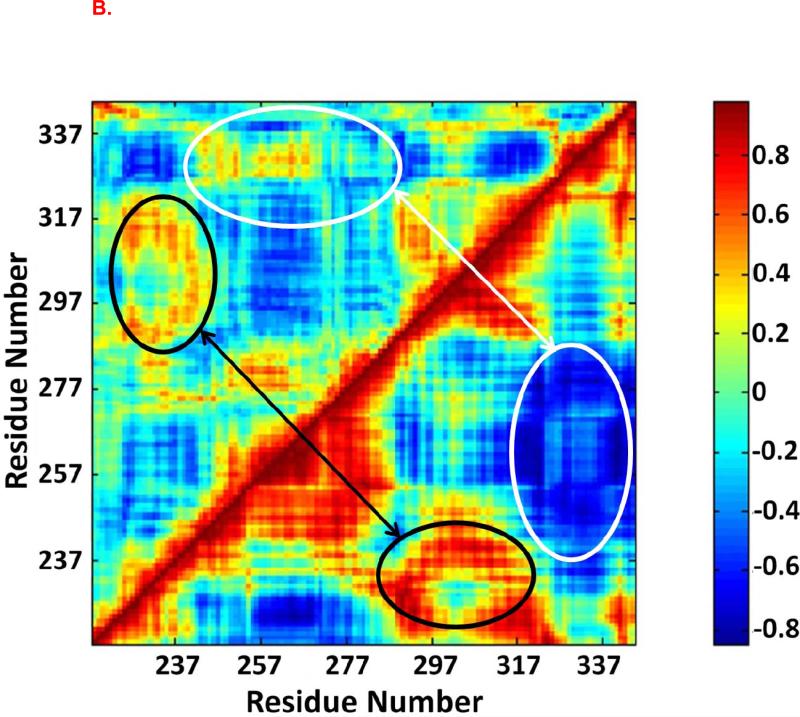
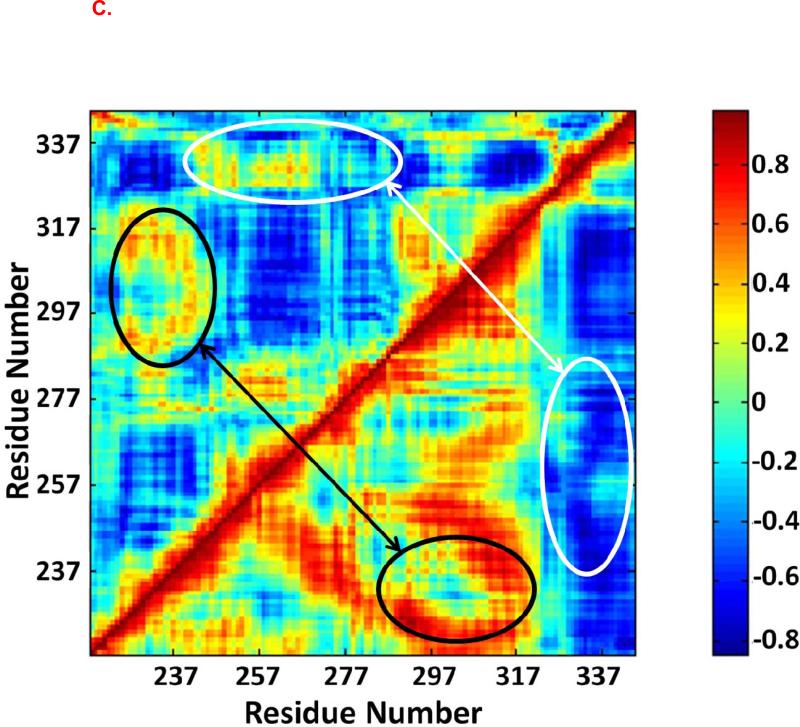
Dynamical cross-correlation maps were used to investigate the correlated motion between residues in the Fas DD (Figure 5). The top left halves of Figure 5 are the cross-correlation maps for the Fas DD in the non TFP-bound CaM-Fas complex (6), while the bottom right halves are the cross-correlation maps for the Fas DD in the 1TFP-bound CaM-Fas complex, 2TFP-bound CaM-Fas complex, and 4TFP-bound CaM-Fas complex. When compared to the dynamic cross-correlation map for the Fas DD in the CaM-Fas complex with CaM not bound to TFP (6), the Fas DD showed a significantly increased degree of correlated motion among residues in the binding sites of CaM in Fas (res 231-270) and a significantly increased degree of anti-correlated motion between C-terminal of Fas (res 320-340) and the binding site of CaM in Fas (res 231-270) when bound to 1TFP-bound CaM (Figure 5A). For the 2TFP-bound CaM-Fas complex, the Fas DD showed a significantly increased degree of correlated motion (red) between res 280-320 and the partial binding site of CaM in Fas (res 220-240) (black ellipse) and a significantly increased degree of anti-correlated motion (blue) between the binding site of Fas in CaM (res 231-270) and C-terminal of Fas (res 320-340) (white ellipse) compared to the Fas DD in the non-TFP-bound CaM/Fas complex (Figure 5B). For the 4TFP-bound CaM-Fas complex, the Fas DD showed an overall significant increase in correlated motion for res 237-317, including an increase in the degree of correlated motion between res 280-320 and the partial binding site of CaM in Fas (res 220-240) (black ellipse), and an overall significant increase in anti-correlated motion between C-terminal of Fas (res 320-340) and the binding site of CaM in Fas (res 231-270), including an increase in the degree of anti-correlated motion between the partial binding site of CaM in Fas (res 257-277) and the C-terminal of Fas (res 320-340) (white ellipse) when compared to the Fas DD in the non-TFP-bound CaM-Fas complex (Figure 5C). The significantly changed degree of correlated and anti-correlated motion for residues in the Fas DD in TFP-bound CaM-Fas complexes compared to those in the non-TFP-bound CaM-Fas complex could directly contribute to effects on CaM/Fas binding. Effects on formation of the CaM-Fas complex may then affect Fas-mediated recruitment of FADD for DISC formation to signal apoptosis, as observed in experiments showing that TFP treatment affects CaM/Fas binding and apoptosis (26).
Figure 5.
Dynamic cross-correlation maps for the degree of correlated motion of the residues in the Fas DD from CaM-Fas complexes. (A) Fas DD from wildtype CaM-Fas complex (6) (top left) compared to the Fas DD from CaM-1TFP-Fas complex (bottom right). (B) Fas DD from wildtype CaM-Fas complex (6) (top left) compared to the Fas DD from CaM-2TFPs-Fas complex (bottom right). (C) Fas DD from wildtype CaM-Fas complex (6) (top left) compared to the Fas DD from CaM-4TFPs-Fas complex (bottom right).
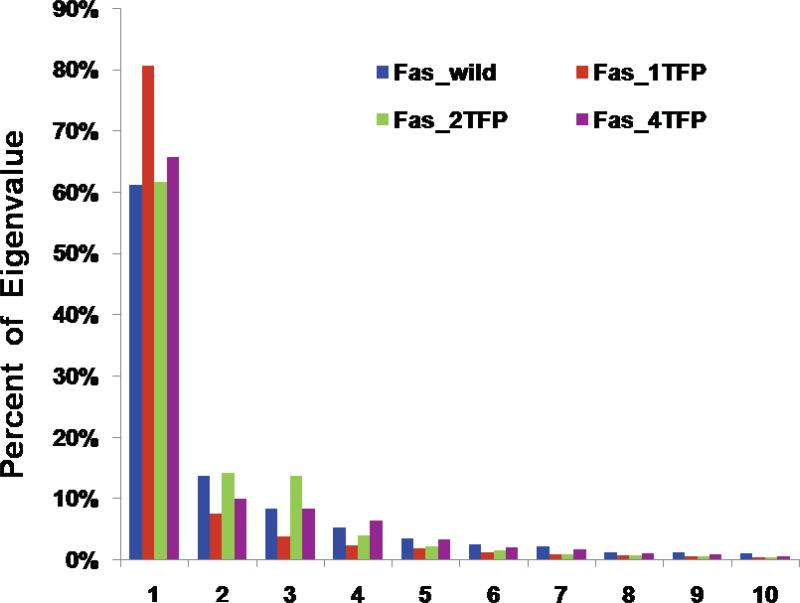
We also performed PCA analysis for the Fas in the CaM-Fas complexes to calculate the distribution of the relative contribution of the first ten PCA modes of the Fas in CaM-Fas complexes (Figure 6). The results showed that the contribution of the first mode to the dynamic motion of the Fas in the 1TFP-bound CaM-Fas complex was significantly larger than that of the Fas in the non-TFP-bound CaM-Fas complex. However, the contribution of the first mode to the dynamic motion of the Fas in the 2TFP-bound and 4TFP-bound CaM-Fas complexes were very similar to that of the non-TFP-bound CaM-Fas complex (Figure 6). These dynamic motion changes induced by one TFP bound to CaM could directly contribute to the binding changes in the CaM-Fas complex caused by TFP binding to CaM (Table 3).
Figure 6.
Distribution of percent of eigenvalues obtained from PCA analysis for Fas in the CaM-Fas complexes.
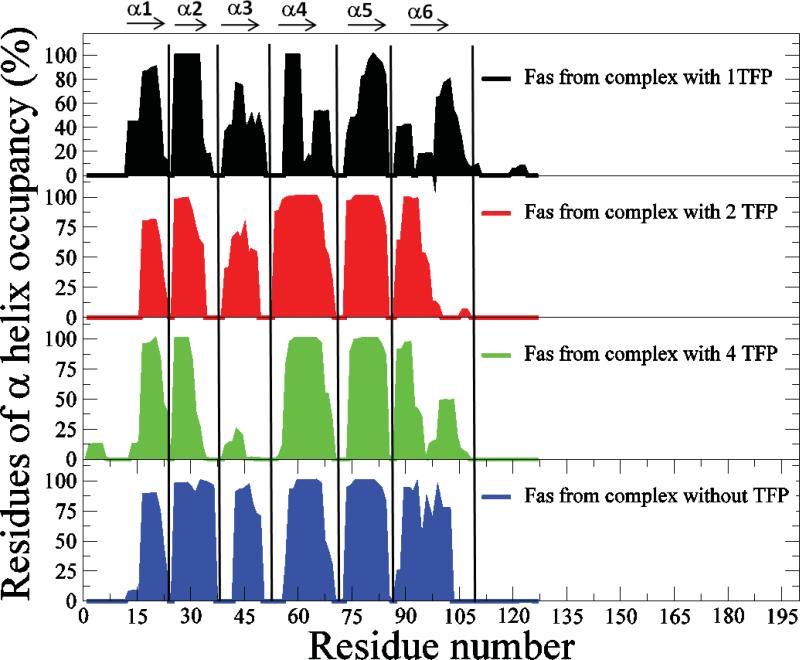
The structure of the Fas DD is primarily composed of six α-helices (39). The CaM binding region is located within the α1 and α2 helices of the Fas death domain, and the α5 and α6 helices of the Fas death domain are involved in FADD binding (27, 84). Therefore, we analyzed α-helix formation and hydrogen bond occupancy within the α-helices of Fas, as well as the number of dihedral angle transitions for all Fas residues in CaM-Fas complexes over the last 15 ns of MD simulation trajectories (Figure 7, Figure 9S, and Figure 10S). α-helix formation analysis revealed a significantly changed residue occupancy of the α-helices and changed lengths of the α2, α3, α4 and α6 helices in Fas for the 1TFP-bound, 2TFP-bound, and 4TFP-bound CaM-Fas complexes compared to the non-TFP-bound CaM-Fas complex (Figure 7).The α2 and α3 helices of Fas are either the binding site or are located near the binding site for CaM, and the α4 and α6 helices of Fas are either adjacent to or serve as the binding site in Fas for FADD. Altered α-helix formation could contribute to CaM-Fas binding as observed in Table 3, and could affect binding of Fas to FADD. Different numbers of TFP molecules bound to CaM resulted in different hydrogen bond occupancies for the α-helices of Fas, particularly for the α2, α3, α4 and α6 helices of Fas (Figure 9S). The number of dihedral angle transitions for all residues of Fas in CaM-Fas complexes over time is shown in Figure 10S. These results reveal a significantly decreased number of dihedral angle transitions for all Fas residues in the 1TFP-bound, 2TFP-bound, and 4TFP-bound CaM-Fas complexes compared to the non-TFP-bound CaM-Fas complex (Figure 10S A, B and C). However, changes in the number of TFP molecules bound to CaM also resulted in different numbers of dihedral angle transitions for all Fas residues in CaM-Fas complexes (Figure 9S D). The altered α-helix formation, hydrogen bond occupancy within the α-helices, and dihedral angle transitions for residues in Fas induced by binding to CaM bound to different numbers of TFP molecules could contribute to the conformational and motion changes of Fas in CaM-Fas complexes as observed in Figures 4, 5, and 6. These changes may also affect Fas recruitment of FADD to form the DISC complex and signal apoptosis.
Figure 7.
Residues of α helix occupancy of Fas in CaM-Fas complexes
CONCLUSIONS
Taken together, these data demonstrate that the number of TFP molecules bound to CaM directly affects the degree of conformational and motion changes of CaM in TFP-CaM complexes and Fas in CaM-Fas complexes. Binding of one TFP molecule to CaM resulted in strong and almost parallel anti-correlated motion between the C-terminal helix and the N-terminal helix, which form a binding pocket for TFP, while binding of two or four TFPs caused an increase in dispersive motion direction between the two helices. Binding of TFP to CaM is negatively affected by binding of additional TFP molecules, consistent with experimental observations that binding of four TFPs to CaM is not cooperative (83). The number of TFP molecules bound to CaM also directly influenced α-helix formation and hydrogen bond occupancy within the α-helices of CaM, contributing to the conformational and motion changes within the protein. These changes in CaM affected binding of CaM to Fas, resulting in changes in α-helix formation, hydrogen bond occupancy within the α-helices, and dihedral angle transitions for the residues of Fas, further influencing the conformational and motion changes of Fas in CaM-Fas complexes. These changes in Fas may interfere with the recruitment of FADD for DISC formation. The effect of TFP on the CaM-Fas interaction is dependent on the number of TFP molecules bound to CaM, consistent with experiments showing that CaM-Fas binding is affected by TFP in a concentration-dependent manner (26). These results provide important molecular evidence to understand the structural consequences induced by binding of CaM to different numbers of TFP molecules and to explain the effects on Fas caused by interactions with CaM bound to different numbers of TFP molecules. These results will further facilitate understanding of the concentration-dependent effects of TFP on the CaM-Fas interaction and Fas-mediated DISC formation, as observed in experimental studies (26). These studies also provide important insight into strategies for the identification of other CaM antagonists for use as as potential drug candidates to regulate Fas-mediated apoptosis for cancer chemotherapy.
Supplementary Material
ACKNOWLEDGMENT
Y.H.Song acknowledges NIH K25 award (5K25CA140791) and NSF sponsored UAB ADVANCE program support. J.M. McDonald acknowledges support from a Veterans Administration Merit Award.
Footnotes
SUPPORTING INFORMATION AVAILABLE
The structure and force field charge parameters of TFP (Fig. 1S and Table 1S) were included in the supplemental materials. Binding energy of the top three complexes of Fas with CaM bound to different number of TFP calculated from RDOCK program was shown in Table.2S. Size of the periodic box for the six systems simulated in this study was shown in Table.3S. RMSD for CaM in the TFP-CaM complexes over the 30ns MD simulation for different simulation cases (Fig. 2S) and binding free energy between CaM with TFP molecule(s) in the TFP-CaM complexes as a function of cumulative time over the 30ns MD simulation for different simulation cases to demonstrate the equilibration of the TFP/CaM complexes were shown in the supplemental materials (Fig. 3S). Binding free energy of Fas-CaM complexes as a function of cumulative time over the 30ns MD simulation for different simulation cases (Fig. 4S), RMSD for the Fas Death Domain protein core (res 225-318) and RMSD for CaM in the CaM/Fas complexes over the 30ns MD simulation for different simulation cases (Fig. 5S and Fig. 6S), to demonstrate the equilibration of the TFP-bound CaM/Fas complexes were shown in the supplemental materials. Root mean squared fluctuation (RMSF) comparison of CaM in different CaM/TFP complexes was included in the supplemental materials (Fig. 7S). Hydrogen bond occupancy of each α helix of CaM in TFP-CaM complexes (Fig. 8S), hydrogen bond occupancy of each α helix of Fas in CaM-Fas complexes (Fig. 9S) and the number of dihedral angle transition for all residues of Fas from CaM-Fas complexes over time (Fig. 10S) were included in the supplemental materials.
REFERENCES
- 1.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 2.Lee SH, Shin MS, Lee HS, Bae JH, Lee HK, Kim HS, Kim SY, Jang JJ, Joo M, Kang YK, Park WS, Park JY, Oh RR, Han SY, Lee JH, Kim SH, Lee JY, Yoo NJ. Expression of Fas and Fas-related molecules in human hepatocellular carcinoma. Human pathology. 2001;32:250–256. doi: 10.1053/hupa.2001.22769. [DOI] [PubMed] [Google Scholar]
- 3.Nambu Y, Hughes SJ, Rehemtulla A, Hamstra D, Orringer MB, Beer DG. Lack of cell surface Fas/APO-1 expression in pulmonary adenocarcinomas. The Journal of clinical investigation. 1998;101:1102–1110. doi: 10.1172/JCI1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keane MM, Ettenberg SA, Lowrey GA, Russell EK, Lipkowitz S. Fas expression and function in normal and malignant breast cell lines. Cancer research. 1996;56:4791–4798. [PubMed] [Google Scholar]
- 5.Weller M, Malipiero U, Rensing-Ehl A, Barr PJ, Fontana A. Fas/APO-1 gene transfer for human malignant glioma. Cancer research. 1995;55:2936–2944. [PubMed] [Google Scholar]
- 6.Suever JD, Chen Y, McDonald JM, Song Y. Conformation and free energy analyses of the complex of calcium-bound calmodulin and the Fas death domain. Biophysical journal. 2008;95:5913–5921. doi: 10.1529/biophysj.108.130542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takakuwa T, Dong Z, Takayama H, Matsuzuka F, Nagata S, Aozasa K. Frequent mutations of Fas gene in thyroid lymphoma. Cancer research. 2001;61:1382–1385. [PubMed] [Google Scholar]
- 8.Shin MS, Park WS, Kim SY, Kim HS, Kang SJ, Song KY, Park JY, Dong SM, Pi JH, Oh RR, Lee JY, Yoo NJ, Lee SH. Alterations of Fas (Apo-1/CD95) gene in cutaneous malignant melanoma. The American journal of pathology. 1999;154:1785–1791. doi: 10.1016/S0002-9440(10)65434-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda T, Yamada Y, Moriuchi R, Sugahara K, Tsuruda K, Joh T, Atogami S, Tsukasaki K, Tomonaga M, Kamihira S. Fas gene mutation in the progression of adult T cell leukemia. The Journal of experimental medicine. 1999;189:1063–1071. doi: 10.1084/jem.189.7.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 11.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 12.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts I, Debatin K, Fischer A, de Villartay J. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 13.Puck J, Straus S, Le Deist F, Rieux-Laucat F, Fischer A. In Primary Immunodeficiency Diseases: A Molecular and Genetic Approach. Oxford University Press; New York: 1999. Inherited Disorders with Autoimmune and Defective Lymphocyte Regulation. [Google Scholar]
- 14.Bi LL, Pan G, Atkinson TP, Zheng L, Dale JK, Makris C, Reddy V, McDonald JM, Siegel RM, Puck JM, Lenardo MJ, Straus SE. Dominant inhibition of Fas ligand-mediated apoptosis due to a heterozygous mutation associated with autoimmune lymphoproliferative syndrome (ALPS) Type Ib. BMC medical genetics. 2007;8:41. doi: 10.1186/1471-2350-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jhala NC, Vickers SM, Argani P, McDonald JM. Regulators of apoptosis in cholangiocarcinoma. Archives of pathology & laboratory medicine. 2005;129:481–486. doi: 10.5858/2005-129-481-ROAIC. [DOI] [PubMed] [Google Scholar]
- 16.Ahn EY, Pan G, Vickers SM, McDonald JM. IFN-gammaupregulates apoptosis-related molecules and enhances Fas-mediated apoptosis in human cholangiocarcinoma. International journal of cancer. 2002;100:445–451. doi: 10.1002/ijc.10516. [DOI] [PubMed] [Google Scholar]
- 17.Li ZY, Zou SQ. Fas counterattack in cholangiocarcinoma: a mechanism for immune evasion in human hilar cholangiocarcinomas. World J Gastroenterol. 2001;7:860–863. doi: 10.3748/wjg.v7.i6.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimonishi T, Isse K, Shibata F, Aburatani I, Tsuneyama K, Sabit H, Harada K, Miyazaki K, Nakanuma Y. Up-regulation of fas ligand at early stages and down-regulation of Fas at progressed stages of intrahepatic cholangiocarcinoma reflect evasion from immune surveillance. Hepatology (Baltimore, Md. 2000;32:761–769. doi: 10.1053/jhep.2000.18192. [DOI] [PubMed] [Google Scholar]
- 19.Que FG, Phan VA, Phan VH, Celli A, Batts K, LaRusso NF, Gores GJ. Cholangiocarcinomas express Fas ligand and disable the Fas receptor. Hepatology (Baltimore, Md. 1999;30:1398–1404. doi: 10.1002/hep.510300618. [DOI] [PubMed] [Google Scholar]
- 20.Pan G, Vickers SM, Pickens A, Phillips JO, Ying W, Thompson JA, Siegal GP, McDonald JM. Apoptosis and tumorigenesis in human cholangiocarcinoma cells. Involvement of Fas/APO-1 (CD95) and calmodulin. The American journal of pathology. 1999;155:193–203. doi: 10.1016/S0002-9440(10)65113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moser MJ, Lee SY, Klevit RE, Davis TN. Ca2+ binding to calmodulin and its role in Schizosaccharomyces pombe as revealed by mutagenesis and NMR spectroscopy. The Journal of biological chemistry. 1995;270:20643–20652. doi: 10.1074/jbc.270.35.20643. [DOI] [PubMed] [Google Scholar]
- 22.Hait WN, Lazo JS. Calmodulin: a potential target for cancer chemotherapeutic agents. J Clin Oncol. 1986;4:994–1012. doi: 10.1200/JCO.1986.4.6.994. [DOI] [PubMed] [Google Scholar]
- 23.Ito H, Wang JZ, Shimura K. Inhibition of lung metastasis by a calmodulin antagonist, N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W-7), in mice bearing Lewis lung carcinoma. Anticancer research. 1991;11:249–252. [PubMed] [Google Scholar]
- 24.Ahn EY, Pan G, Oh JH, Tytler EM, McDonald JM. The combination of calmodulin antagonists and interferon-gamma induces apoptosis through caspase-dependent and -independent pathways in cholangiocarcinoma cells. The American journal of pathology. 2003;163:2053–2063. doi: 10.1016/s0002-9440(10)63563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Pawar P, Pan G, Ma L, Liu H, McDonald JM. Calmodulin binding to the Fas-mediated death-inducing signaling complex in cholangiocarcinoma cells. Journal of cellular biochemistry. 2008;103:788–799. doi: 10.1002/jcb.21447. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Ahn EY, McKenna MA, Yeo H, McDonald JM. Fas binding to calmodulin regulates apoptosis in osteoclasts. The Journal of biological chemistry. 2005;280:29964–29970. doi: 10.1074/jbc.M500710200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn EY, Lim ST, Cook WJ, McDonald JM. Calmodulin binding to the Fas death domain. Regulation by Fas activation. J Biol Chem. 2004;279:5661–5666. doi: 10.1074/jbc.M311040200. [DOI] [PubMed] [Google Scholar]
- 28.Eldik L, Watterson D. Calmodulin and Calcium Signal Transduction. Academic Press; San Diego: 1998. [Google Scholar]
- 29.Ikura M. CALMODULIN TARGET DATABASE. [DOI] [PubMed]
- 30.Ganoth A, Friedman R, Nachliel E, Gutman M. A molecular dynamics study and free energy analysis of complexes between the Mlc1p protein and two IQ motif peptides. Biophysical journal. 2006;91:2436–2450. doi: 10.1529/biophysj.106.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn BE, Evenas J, Drakenberg T, Waltho JP, Thulin E, Forsen S. Calcium-induced structural changes and domain autonomy in calmodulin. Nat Struct Biol. 1995;2:777–783. doi: 10.1038/nsb0995-777. [DOI] [PubMed] [Google Scholar]
- 32.Kretsinger RH, Rudnick SE, Weissman LJ. Crystal structure of calmodulin. J Inorg Biochem. 1986;28:289–302. doi: 10.1016/0162-0134(86)80093-9. [DOI] [PubMed] [Google Scholar]
- 33.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 35.Seaton BA, Head JF, Engelman DM, Richards FM. Calcium-induced increase in the radius of gyration and maximum dimension of calmodulin measured by small-angle X-ray scattering. Biochemistry. 1985;24:6740–6743. doi: 10.1021/bi00345a002. [DOI] [PubMed] [Google Scholar]
- 36.Heidorn DB, Trewhella J. Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry. 1988;27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- 37.Persechini A, Kretsinger RH. The central helix of calmodulin functions as a flexible tether. J Biol Chem. 1988;263:12175–12178. [PubMed] [Google Scholar]
- 38.Matsushima N, Hayashi N, Jinbo Y, Izumi Y. Ca2+-bound calmodulin forms a compact globular structure on binding four trifluoperazine molecules in solution. Biochem J. 2000;347(Pt 1):211–215. [PMC free article] [PubMed] [Google Scholar]
- 39.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 40.Imtiyaz HZ, Zhang Y, Zhang J. Structural requirements for signal-induced target binding of FADD determined by functional reconstitution of FADD deficiency. The Journal of biological chemistry. 2005;280:31360–31367. doi: 10.1074/jbc.M504138200. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, McKenna MA, Feng X, Nagy TR, McDonald JM. Osteoclast apoptosis: the role of Fas in vivo and in vitro. Endocrinology. 2003;144:5545–5555. doi: 10.1210/en.2003-0296. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Xu J, Jhala N, Pawar P, Zhu ZB, Ma L, Byon CH, McDonald JM. Fas-mediated apoptosis in cholangiocarcinoma cells is enhanced by 3,3'-diindolylmethane through inhibition of AKT signaling and FLICE-like inhibitory protein. The American journal of pathology. 2006;169:1833–1842. doi: 10.2353/ajpath.2006.060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cook WJ, Walter LJ, Walter MR. Drug binding by calmodulin: crystal structure of a calmodulin-trifluoperazine complex. Biochemistry. 1994;33:15259–15265. doi: 10.1021/bi00255a006. [DOI] [PubMed] [Google Scholar]
- 44.Vertessy BG, Harmat V, Bocskei Z, Naray-Szabo G, Orosz F, Ovadi J. Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: crystallographic and spectroscopic studies. Biochemistry. 1998;37:15300–15310. doi: 10.1021/bi980795a. [DOI] [PubMed] [Google Scholar]
- 45.Vandonselaar M, Hickie RA, Quail JW, Delbaere LT. Trifluoperazine-induced conformational change in Ca(2+)-calmodulin. Nature structural biology. 1994;1:795–801. doi: 10.1038/nsb1194-795. [DOI] [PubMed] [Google Scholar]
- 46.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 47.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen R, Weng Z. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 50.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 51.Chen R, Tong W, Mintseris J, Li L, Weng Z. ZDOCK predictions for the CAPRI challenge. Proteins. 2003;52:68–73. doi: 10.1002/prot.10388. [DOI] [PubMed] [Google Scholar]
- 52.Wiehe K, Pierce B, Mintseris J, Tong WW, Anderson R, Chen R, Weng Z. ZDOCK and RDOCK performance in CAPRI rounds 3, 4, and 5. Proteins. 2005;60:207–213. doi: 10.1002/prot.20559. [DOI] [PubMed] [Google Scholar]
- 53.Yi H, Qiu S, Cao Z, Wu Y, Li W. Molecular basis of inhibitory peptide maurotoxin recognizing Kv1.2 channel explored by ZDOCK and molecular dynamic simulations. Proteins. 2008;70:844–854. doi: 10.1002/prot.21706. [DOI] [PubMed] [Google Scholar]
- 54.Jin L, Wu Y. Molecular mechanism of the sea anemone toxin ShK recognizing the Kv1.3 channel explored by docking and molecular dynamic simulations. Journal of chemical information and modeling. 2007;47:1967–1972. doi: 10.1021/ci700178w. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y, Cao Z, Yi H, Jiang D, Mao X, Liu H, Li W. Simulation of the interaction between ScyTx and small conductance calcium-activated potassium channel by docking and MM-PBSA. Biophysical journal. 2004;87:105–112. doi: 10.1529/biophysj.103.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 57.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Journal of computational chemistry. 1983;4:187–217. [Google Scholar]
- 58.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. Journal of molecular graphics & modelling. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the multimolecule and multiconformational RESP methodology to biopolymers: Charge derivation for DNA, RNA, and proteins. Computational Chemistry. 2004;16:1357–1377. [Google Scholar]
- 60.Zhang W, Hou T, Qiao X, Xu X. Parameters for the Generalized Born Model Consistent with RESP Atomic Partial Charge Assignment Protocol. J. Phys. Chem B. 2003;107:9071–9078. [Google Scholar]
- 61.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery J, J. A., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Gaussian, Inc; Wallingford, CT: 2004. [Google Scholar]
- 62.Ponder JW, Case DA. Force fields for protein simulations. Adv Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 63.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 64.Darden T, York D, Pedersen LG. Particle mesh Ewald: An Nâlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 65.Li L, Uversky VN, Dunker AK, Meroueh SO. A computational investigation of allostery in the catabolite activator protein. Journal of the American Chemical Society. 2007;129:15668–15676. doi: 10.1021/ja076046a. [DOI] [PubMed] [Google Scholar]
- 66.Wang W, Lim WA, Jakalian A, Wang J, Wang J, Luo R, Bayly CI, Kollman PA. An analysis of the interactions between the Sem-5 SH3 domain and its ligands using molecular dynamics, free energy calculations, and sequence analysis. Journal of the American Chemical Society. 2001;123:3986–3994. doi: 10.1021/ja003164o. [DOI] [PubMed] [Google Scholar]
- 67.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Accounts of chemical research. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 68.Brooks B, Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc Natl Acad Sci U S A. 1983;80:6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Go N, Noguti T, Nishikawa T. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc Natl Acad Sci U S A. 1983;80:3696–3700. doi: 10.1073/pnas.80.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levitt M, Sander C, Stern PS. The normal modes of a protein: Native bovine pancreatic trypsin inhibitor. Int J Quantum Chem. Quantum Biol. Symp. 1983;24:181–199. [Google Scholar]
- 71.McQuarrie DA. Statistical Thermodynamics. Harper & Row; New York: 1973. [Google Scholar]
- 72.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman & Hall; New York: 1998. [Google Scholar]
- 73.Hayward S, Kitao A, Go N. Harmonicity and anharmonicity in protein dynamics: a normal mode analysis and principal component analysis. Proteins. 1995;23:177–186. doi: 10.1002/prot.340230207. [DOI] [PubMed] [Google Scholar]
- 74.Hayward S, Kitao A, Hirata F, Go N. Effect of solvent on collective motions in globular protein. Journal of molecular biology. 1993;234:1207–1217. doi: 10.1006/jmbi.1993.1671. [DOI] [PubMed] [Google Scholar]
- 75.Berendsen HJ, Hayward S. Collective protein dynamics in relation to function. Current opinion in structural biology. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 76.MathWorks T. MATLAB.
- 77.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University Press; New York: 1987. [Google Scholar]
- 78.Lee SJ, Song Y, Baker NA. Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+-lipid interactions and finite size effects. Biophysical journal. 2008;94:3565–3576. doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y, Pan D, Bellis SL, Song Y. Effect of altered glycosylation on the structure of the I-like domain of beta1 integrin: A molecular dynamics study. Proteins. 2008;73:989–1000. doi: 10.1002/prot.22126. [DOI] [PubMed] [Google Scholar]
- 80.Chen AA, Pappu RV. Quantitative characterization of ion pairing and cluster formation in strong 1:1 electrolytes. The journal of physical chemistry. 2007;111:6469–6478. doi: 10.1021/jp0708547. [DOI] [PubMed] [Google Scholar]
- 81.Bailey NTJ. Statistical methods in biology. Cambridge University Press; New York, NY: 1995. [Google Scholar]
- 82.Harmat V, Bocskei Z, Naray-Szabo G, Bata I, Csutor AS, Hermecz I, Aranyi P, Szabo B, Liliom K, Vertessy BG, Ovadi J. A new potent calmodulin antagonist with arylalkylamine structure: crystallographic, spectroscopic and functional studies. Journal of molecular biology. 2000;297:747–755. doi: 10.1006/jmbi.2000.3607. [DOI] [PubMed] [Google Scholar]
- 83.Massom L, Lee H, Jarrett HW. Trifluoperazine binding to porcine brain calmodulin and skeletal muscle troponin C. Biochemistry. 1990;29:671–681. doi: 10.1021/bi00455a012. [DOI] [PubMed] [Google Scholar]
- 84.Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.