Abstract
AIM: To quantitatively investigate the effect of p16 hypermethylation on hepatocellular carcinoma (HCC) and hepatocirrhosis using a meta-analysis of available case-control studies.
METHODS: Previous studies have primarily evaluated the incidence of p16 hypermethylation in HCC and corresponding control groups, and compared the incidence of p16 hypermethylation in tumor tissues, pericancer liver tissues, normal liver tissues and non-tumor liver tissues with that in other diseases. Data regarding publication information, study characteristics, and incidence of p16 hypermethylation in both groups were collected from these studies and summarized.
RESULTS: Fifteen studies, including 744 cases of HCC and 645 non-tumor cases, were identified for meta-analysis. Statistically significant odds ratios (ORs) of p16 hypermethylation were obtained from tumor tissues and non-tumorous liver tissues of HCC patients (OR 7.04, 95% CI: 3.87%-12.78%, P < 0.0001), tumor tissues of HCC patients and healthy liver tissues of patients with other diseases (OR 12.17, 95% CI: 6.64%-22.31%, P < 0.0001), tumor tissues of HCC patients and liver tissues of patients with non-tumorous liver diseases (OR 6.82, 95% CI: 4.31%-10.79%, P < 0.0001), and cirrhotic liver tissues and non-cirrhotic liver tissues (OR 4.96, 95% CI: 1.45%-16.96%, P = 0.01). The pooled analysis showed significantly increased ORs of p16 hypermethylation (OR 6.98, 95% CI: 4.64%-10.49%, P < 0.001) from HCC tissues and cirrhotic tissues.
CONCLUSION: P16 hypermethylation induces the inactivation of p16 gene, plays an important role in hepatocarcinogenesis, and is associated with an increased risk of HCC and liver cirrhosis.
Keywords: P16 hypermethylation, Hepatocellular carcinoma, Liver cirrhosis, Meta-analysis, Odds ratio
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the major causes of cancer death worldwide[1]. The HCC incidence is still increasing in developed countries although considerable progress has been made in diagnostic and therapeutic modalities[2]. The molecular genetics of HCC have recently been extensively characterized[3]. Among these molecular genetics, aberrant DNA cytosine methylation is one of the most consistent epigenetic changes in human cancers. Generally, the overall DNA methylation level is lower in cancer cells than in normal cells. However, some loci tend to show increased DNA methylation in cancer cells[4].
The p16INK4A gene is located on chromosome 9p21 and is one of the most frequently altered genes observed in various human neoplasms[4,5]. It is a cell cycle-related gene encoding a p16 protein that binds competitively to cyclin-dependent kinase 4 protein (Cdk4), thereby inhibiting the interaction of Cdk4 and cyclin D1 to stimulate passage through the G1 phase of the cell cycle[6]. The disruption of p16-mediated cell cycle control seems to play a role in hepatocarcinogenesis because inactivation of the p16INK4A gene resulting from methylation of the p16INK4A gene, has been reported in HCC[7].
Although previous reports indicated that inactivation of the p16INK4A gene is mainly induced by the methylation of the p16 gene, and it is one of the important genetic alterations in HCCs, the reported rates of p16 methylation in HCCs were remarkably diverse. Moreover, whether it is associated with the incidence of hepatocirrhosis is still unclear. The various results of these studies underpin the need for assessing the evidence of the relationship between p16 inactivation and HCC. Hence, we conducted a systematic review and meta-analysis to quantitatively evaluate the effects of p16 hypermethylation on the incidence of HCC.
MATERIALS AND METHODS
The meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses[8] and the recommendations of the Cochrane Collaboration[9].
Data source and search
To avoid publication bias, both published and unpublished studies, with an English or Chinese language restriction, were included, and several methods were used to identify all relevant studies. The databases screened were PubMed (1976 onward), EMBASE (1966 onward), Cochrane Library (no date restriction), Biological Abstracts (no date restriction), Science Citation Index (no date restriction), China National Knowledge Infrastructure (no date restriction), and the Chinese BioMedical Literature Database (no date restriction). Medical Subject Headings were used in the searching in both Chinese and English languages. The keywords used were p16 methylation, HCC and hepatocirrhosis. Relevant reviews and meta-analysis of the role of p16 methylation in the incidence of HCC and hepatocirrhosis were examined for potential inclusive studies. We also searched http://www.jamas.gr.jp and http://www.cdc.gov websites for studies completed but not yet published.
Study selection
The following studies were included in this meta-analysis: studies primarily evaluating the incidence of p16 hypermethylation in HCC and corresponding control groups, and comparing the incidence of p16 hypermethylation in tumor tissues, pericancer liver tissues, normal liver tissues, and non-tumor liver tissues with other identified diseases. The bibliographies of the search results were manually scanned and independently reviewed by two authors (Xie F and Zang JJ) to identify relevant studies that met the inclusion criteria (full text or abstract). If there was any disagreement between the two authors, it was settled by discussion with a third author (He J) until a consensus was reached. One author (Xu JF) contacted the authors of the article for missing data if necessary.
Data extraction
Data extraction was independently conducted by two reviewers (Xu JF and Qin YY) using a standardized approach. Data for publication information (year of publication and name of first author), study characteristics (sample size and distributions of age and sex), and rates of p16 hypermethylation were collected using standard data extraction forms. Point estimates for selected variables were extracted and checked by the other two reviewers (Xie F and Qin YY). Disagreement was adjudicated by a third reviewer (He J) after referring back to original articles.
Statistical analysis
Odds ratios (ORs) were used as a measure of the relationship between p16 hypermethylation and the risk of HCC for case-control studies and the corresponding 95% CIs. The pooled ORs were combined by the Mantel-Haenszel methods. When there were trials with no events in one or both arms, the Peto method was used[6,10].
An OR > 1 indicated a higher incidence of p16 methylation in HCC tissues than in corresponding controls. The percentage of variability across studies attributable to heterogeneity beyond chance was assessed by χ2 test (P < 0.1) and I2 statistics[11]. When there was no statistically significant heterogeneity, a pooled effect was calculated with a fixed-effects model; otherwise, a random-effects model was employed. We also assessed the probability of publication bias with funnel plots[12] and Egger’s test[13]. A P value of less than 0.1 indicated statistically significant publication bias. In addition, we conducted a sensitivity analysis to evaluate whether the results were statistically affected. Statistical significance was defined as a two-tailed P value of 0.05. All statistical analyses were conducted with RevMan version 5 from the Cochrane Collaboration.
RESULTS
Search results
Fifteen[14-28] articles met the inclusion criteria according to the aforementioned search strategies and provided data regarding p16 hypermethylation in 744 cases of HCC tumor tissues and 645 cases of non-tumor tissues. Hypermethylation profile of tumorous and paired non-tumorous liver tissue samples from nine studies, HCC tumor tissues and normal tissues (normal liver tissues or blood samples) from five studies, and HCC tumor tissues and abnormal and non-tumorous tissues (dysplastic nodule, liver cirrhosis, and chronic hepatitis) from four studies was compared, respectively. Twelve eligible trials were conducted in Asia from 1999 to 2010, and the other three were conducted in the United States and Germany. Three of them were published in Chinese, and the others were published in English. The median sample size was 79 patients (range, 22-176). The median age of the study participants ranged from 48.5 to 66.2 years. All of the specimens in the 15 studies were surgically obtained from HCC patients or non-HCC patients who underwent liver surgery. The characteristics of the included studies are shown in Table 1.
Table 1.
Demographic data of studies included in meta-analysis
| Study | HCC tissue/control | No. of patients | Country or area | Median age(Yr) | Sex (M/F) | Year of publication |
| Formeister et al[14] | Tumor/non-tumor tissues | 43/45 | America | 66.28 ± 8.1 | 37/12 | 2010 |
| Zhu et al[15] | Tumor/non-tumor tissues | 88/88 | China | 52.7 ± 10.62 | 78/10 | 2010 |
| Zhang et al[16] | Tumor/liver cirrhosis/normal liver tissue | 120/120/10 | China | 52.8 ± 10.2 | 106/14 | 2008 |
| Xu et al[28] | Tumor/non-tumor tissues from other patients | 30/5 | China | NR | NR | 2006 |
| Liu et al[17] | Tumor/pericancer tissues | 50/50 | China | 48.5 | 46/4 | 2006 |
| Qin et al[18] | Tumor/pericancer/non-tumor tissues | 20/20/20 | China | NR | NR | 2004 |
| Lee et al[19] | Tumor/dysplastic nodule/liver cirrhosis/chronic hepatitis tissues | 60/22/30/34 | Korea | 53.8 | 47/13 | 2003 |
| Schagdarsurengin et al[20] | Tumor/non-tumor/liver cirrhosis/normal liver tissues | 14/14/6/8 | Germany | NR | NR | 2003 |
| Zhang et al[21] | Tumor/pericancer/normal tissues | 83/10/12 | China | NR | NR | 2002 |
| Yu et al[22] | Tumor/pericancer tissues | 29/29 | China | NR | NR | 2003 |
| Saito et al[24] | Tumor/non-tumor tissues | 59/48 | Japan | 61 ± 12 | 42/7 | 2001 |
| Zhang et al[27] | Tumor/pericancer tissues | 35/35 | China | NR | NR | 2002 |
| Kondo et al[23] | Tumor/non-tumor tissues | 40/40 | Japan | 20-77 | 32/8 | 2000 |
| Wong et al[25] | Tumor/non-tumor tissues from other patients | 25/35 | Hong Kong | NR | NR | 2000 |
| Liew et al[26] | Tumor/non-tumor tissues | 48/30 | Hong Kong | NR | NR | 1999 |
P16 hypermethylation in tumorous liver tissues and non-tumorous liver tissues of HCC patients
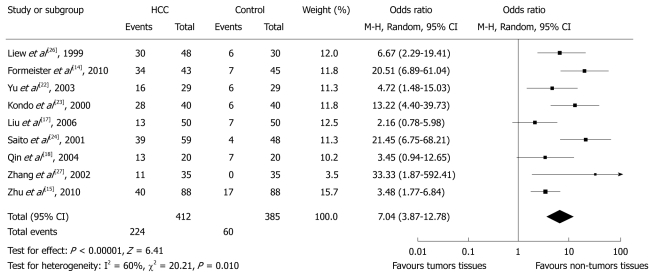
Data for this comparison were available in nine studies which included 412 and specimens of HCC tissues and 385 non-tumorous pericancer tissues. Overall, 224 (54.5%) and 60 (15.6%) cases of p16 hypermethylation were observed in tumorous and non-tumorous tissues of HCC patients, respectively. The pooled analysis showed significantly increased ORs of HCC for p16 hypermethylation compared with controls (OR 7.04, 95% CI: 3.87%-12.78%, P < 0.0001). There was, however, evidence of heterogeneity across the studies (P for heterogeneity = 0.01, I2 = 60%, Figure 1). The heterogeneity was incorporated into the random-effects model. Funnel plots did not show any evidence of publication bias.
Figure 1.
Pooled analysis of p16 hypermethylation in tumorous liver tissues and non-tumorous liver tissues of hepatocellular carcinoma patients. HCC: Hepatocellular carcinoma.
P16 hypermethylation in tumorous liver tissues of HCC patients and healthy liver tissues of patients with other diseases
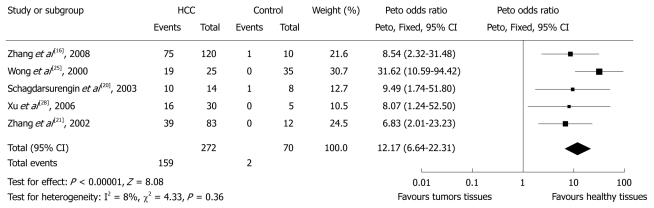
Five studies calculated the OR of p16 hypermethylation in HCC patients and non-HCC healthy patients (Figure 2). There were 159 cases of methylated p16 genes among 272 (58.5%) HCC patients and 2 in 70 (2.9%) non-HCC patients, indicating an OR for p16 hypermethylation of 12.17 (95% CI: 6.64%-22.31%, P < 0.0001). There was no evidence of heterogeneity across the studies (P for heterogeneity = 0.36; I2 = 8%). Funnel plots did not show any evidence of publication bias.
Figure 2.
Pooled analysis of p16 hypermethylation in tumorous liver tissues of hepatocellular carcinoma patients and healthy liver tissues of patients with other diseases. HCC: Hepatocellular carcinoma.
P16 hypermethylation in tumorous liver tissues of HCC patients and liver tissues of patients with non-tumorous liver diseases
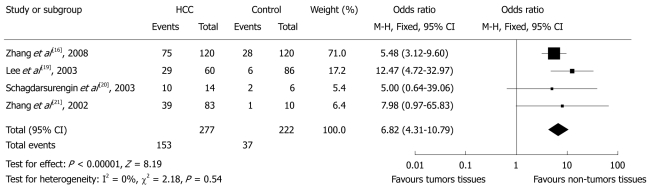
Four studies calculated the OR of p16 hypermethylation in liver tissues of HCC patients and those of patients with liver diseases (Figure 3). There were 153 cases of hypermethylated p16 genes in 277 (55.2%) HCC patients and 37 in 222 patients (16.7%) with liver diseases, indicating an OR for p16 hypermethylation of 6.82 (95% CI: 4.31%-10.79%, P < 0.0001). There was no evidence of heterogeneity across the studies (P for heterogeneity = 0.54; I2 = 0%). There was no evidence of publication bias in the funnel plots.
Figure 3.
Pooled analysis of p16 hypermethylation in tumorous liver tissues of hepatocellular carcinoma patients and liver tissues of patients with non-tumorous liver diseases. HCC: Hepatocellular carcinoma.
Among these studies, data on the comparison of p16 hypermethylation in HCC tissues and cirrhotic tissues were also extracted. Overall, 133 (60.7%) and 30 (15.9%) cases of p16 hypermethylation were observed in 219 HCC tissues and 189 cirrhotic tissues, respectively. The pooled analysis showed significantly increased OR (6.98, 95% CI: 4.64%-10.49%, P < 0.001, data not shown).
P16 hypermethylation in cirrhotic liver tissue and non-cirrhotic liver tissue
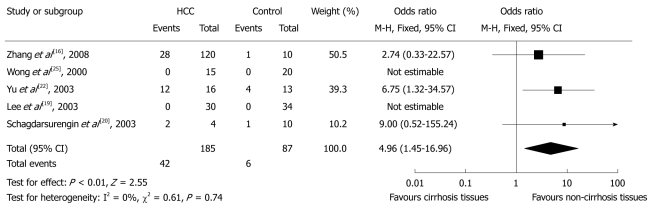
Five studies did this comparison, which included 185 specimens of cirrhotic tissues and 87 specimens of non-cirrhotic tissues. Overall, 42 (22.7%) and 8 (9.2%) cases of p16 hypermethylation were observed in cirrhotic tissues and non-cirrhotic tissues, respectively. The pooled analysis showed significantly increased OR of liver cirrhosis for p16 hypermethylation compared with matched controls (OR 4.96, 95% CI: 1.45%-16.96%, P = 0.01, Figure 4). There was no evidence of heterogeneity across the studies (P for heterogeneity = 0.74; I2 = 0%). There was no evidence of publication bias in the funnel plots.
Figure 4.
Pooled analysis of p16 hypermethylation in cirrhotic liver tissues and non-cirrhotic liver tissues. HCC: Hepatocellular carcinoma.
DISCUSSION
Gene-specific promoter alterations are common epigenetic aberrations found in human liver tumors; however, the epigenetic changes of p16 gene hypermethylation specific to the underlying disease etiology remains elusive. Based on 15 studies and a total of 744 cases of HCC tumor tissues and 645 cases of non-tumor tissues, this pooled analysis comprehensively assessed the relationship between p16 gene hypermethylation and the incidence of HCC or liver cirrhosis. Using the pooled crude ORs from the included studies, we found that p16 gene hypermethylation was associated with 6.16-, 12.17-, and 6.82-fold increased risks of HCC compared with non-tumorous tissues of HCC patients, healthy liver tissues of patients with other diseases, and liver tissues of patients with non-tumorous liver diseases, respectively. Moreover, a 4.96-fold increased risk of liver cirrhosis was also found when compared with non-cirrhotic tissues.
The relationship between p16 gene hypermethylation and the incidence of HCC has been verified by other studies that assessed p16 mRNA expression and its promoter CpG island methylation. Kaneto et al[29], using methylation-specific PCR and immunohistochemistry, detected methylation of the p16 promoter in HCC (72.6%, 16/22) and loss of expression in all methylation-positive HCCs. Roncalli et al[30] reported that methylation of the p16 promoter with complete loss of immunoreactivity occurred in 27 of 33 HCCs (82%). Our results, which were consistent with those of other reports, suggested that p16 gene methylation might play an important role in hepatocarcinogenesis and it might be the major mechanism of p16 gene inactivation.
This review quantitatively assessed the relationship of p16 gene methylation between HCC tissues and non-HCC tissues using well-designed case control studies. To our knowledge, this has not been presented in other meta-analyses or reviews[31-33]. Consistent results were shown in sensitivity analyses, and no evidence of publication bias was found.
This study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders cannot be completely excluded because all of the included studies were observational. Second, the searching strategy was restricted to articles published in English or Chinese. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. Third, most studies included in this meta-analysis were conducted in Eastern Asia, where HCC more frequently occurs. Fourth, comparisons of p16 hypermethylation in cirrhotic non-tumorous liver tissues and normal tissues, chronic hepatitis tissues, or non-cirrhotic HCC tissues were involved in five of the included studies. However, there was no distinction between cirrhotic liver tissues with or without HCC. Thus, we could not perform comparisons under these circumstances. Hence, cautions should be taken when our findings are interpretated among the general populations.
In conclusion, we found that p16 hypermethylation was associated with an increased risk of HCC and liver cirrhosis. P16 hypermethylation, which induced the inactivation of the p16 gene, plays an important role in hepatocarcinogenesis.
COMMENTS
Background
The inactivation of the p16INK4A gene is one of the important genetic alterations in hepatocellular carcinoma (HCC), which is mainly induced by the hypermethylation of p16 gene. However, the role of p16 hypermethylation in HCC or hepatocirrhosis is unclear. Hence, the authors performed a systematic review and meta-analysis to quantitatively evaluate the effects of p16 hypermethylation in the incidence of HCC and hepatocirrhosis.
Research frontiers
Inconsistent results have been reported on the effect of p16 hypermethylation on HCC or hepatocirrhosis and its corresponding controls, and the incidence of p16 hypermethylation.
Innovations and breakthroughs
This is the first systematic review and meta-analysis to investigate quantitatively the effect of p16 hypermethylation in HCC or hepatocirrhosis.
Applications
P16 hypermethylation induces the inactivation of p16 gene and plays an important role in hepatocarcinogenesis, and it is associated with an increased risk of HCC and liver cirrhosis. Detection of p16 hypermethylation using a methylation-specific PCR is favorable for the differential diagnosis of HCC from liver cirrhosis.
Peer review
The authors aimed to quantitatively evaluate the effects of p16 hypermethylation on the incidence of HCC and hepatocirrhosis by systematic review and meta-analysis. The authors found that p16 hypermethylation was associated with an increased risk of HCC and liver cirrhosis. The article is well organized. The methods utilized were appropriate and they presented convincing evidence.
Footnotes
Supported by The Ministry of Science and Technology of China, No. 2009ZX09312-025 and No. 2008ZX10002-018
Peer reviewer: Yoshiaki Iwasaki, MD, PhD, Associate Professor, Health Service Center, Okayama University, 2-1-1, Tsushima-Naka, Kita-ku, Okayama 700-8530, Japan
S- Editor Sun H L- Editor Ma JY E- Editor Zheng XM
References
- 1.Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271–285. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Sheu JC. Molecular mechanism of hepatocarcinogenesis. J Gastroenterol Hepatol. 1997;12:S309–S313. doi: 10.1111/j.1440-1746.1997.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 4.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto A, Demetrick DJ, Spillare EA, Hagiwara K, Hussain SP, Bennett WP, Forrester K, Gerwin B, Serrano M, Beach DH. Mutations and altered expression of p16INK4 in human cancer. Proc Natl Acad Sci U S A. 1994;91:11045–11049. doi: 10.1073/pnas.91.23.11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 7.Biden K, Young J, Buttenshaw R, Searle J, Cooksley G, Xu DB, Leggett B. Frequency of mutation and deletion of the tumor suppressor gene CDKN2A (MTS1/p16) in hepatocellular carcinoma from an Australian population. Hepatology. 1997;25:593–597. doi: 10.1002/hep.510250317. [DOI] [PubMed] [Google Scholar]
- 8.Moher D, Liberati A, Tetzlaff J, Altman DG. Reprint--preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Phys Ther. 2009;89:873–880. [PubMed] [Google Scholar]
- 9.Bero L, Rennie D. The Cochrane Collaboration. Preparing, maintaining, and disseminating systematic reviews of the effects of health care. JAMA. 1995;274:1935–1938. doi: 10.1001/jama.274.24.1935. [DOI] [PubMed] [Google Scholar]
- 10.Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions version 5.0.2 (updated September 2009). The Cochrane Collaboration, 2009. Accessed March 1, 2010 [Google Scholar]
- 11.Woodward M. Epidemiology: design and data analysis. 2nd ed. Boca Raton: Chapman and Hall/CRC Press; 2005. [Google Scholar]
- 12.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 13.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Formeister EJ, Tsuchiya M, Fujii H, Shpyleva S, Pogribny IP, Rusyn I. Comparative analysis of promoter methylation and gene expression endpoints between tumorous and non-tumorous tissues from HCV-positive patients with hepatocellular carcinoma. Mutat Res. 2010;692:26–33. doi: 10.1016/j.mrfmmm.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu YZ, Zhu R, Fan J, Pan Q, Li H, Chen Q, Zhu HG. Hepatitis B virus X protein induces hypermethylation of p16(INK4A) promoter via DNA methyltransferases in the early stage of HBV-associated hepatocarcinogenesis. J Viral Hepat. 2010;17:98–107. doi: 10.1111/j.1365-2893.2009.01156.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Guo X, Jiang G, Zhang L, Yang Y, Shen F, Wu M, Wei L. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer. 2008;123:998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 17.Liu WJ, Wang L, Wang JP, Li JQ, Zhang CQ, Zheng L, Yuan YF. [Correlations of CpG island methylator phenotype and OPCML gene methylation to carcinogenesis of hepatocellular carcinoma] Ai Zheng. 2006;25:696–700. [PubMed] [Google Scholar]
- 18.Qin Y, Liu JY, Li B, Sun ZL, Sun ZF. Association of low p16INK4a and p15INK4b mRNAs expression with their CpG islands methylation with human hepatocellular carcinogenesis. World J Gastroenterol. 2004;10:1276–1280. doi: 10.3748/wjg.v10.i9.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schagdarsurengin U, Wilkens L, Steinemann D, Flemming P, Kreipe HH, Pfeifer GP, Schlegelberger B, Dammann R. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22:1866–1871. doi: 10.1038/sj.onc.1206338. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YJ, Ahsan H, Chen Y, Lunn RM, Wang LY, Chen SY, Lee PH, Chen CJ, Santella RM. High frequency of promoter hypermethylation of RASSF1A and p16 and its relationship to aflatoxin B1-DNA adduct levels in human hepatocellular carcinoma. Mol Carcinog. 2002;35:85–92. doi: 10.1002/mc.10076. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Zhang HY, Ma ZZ, Lu W, Wang YF, Zhu JD. Methylation profiling of twenty four genes and the concordant methylation behaviours of nineteen genes that may contribute to hepatocellular carcinogenesis. Cell Res. 2003;13:319–333. doi: 10.1038/sj.cr.7290177. [DOI] [PubMed] [Google Scholar]
- 23.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 24.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 25.Wong IH, Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res. 2000;6:3516–3521. [PubMed] [Google Scholar]
- 26.Liew CT, Li HM, Lo KW, Leow CK, Chan JY, Hin LY, Lau WY, Lai PB, Lim BK, Huang J, et al. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene. 1999;18:789–795. doi: 10.1038/sj.onc.1202359. [DOI] [PubMed] [Google Scholar]
- 27.Zhang YL, Xiao WH, Zhang YM, Liang HJ. Detection and significance of p16 and its methylation in primary hepatocellular carcinoma. Disan Junyi Daxue Xuebao. 2002;24:1182–1184. [Google Scholar]
- 28.Xu J, Li X, Gao RT, Xu XC, Zhai ZM, Ma JL, Ye SL. Detection of methylation of p16 and p15 gene promoter in hepatocellular carcinoma. Anhui Yike Daxue Xuebao. 2006;41:365–368. [Google Scholar]
- 29.Kaneto H, Sasaki S, Yamamoto H, Itoh F, Toyota M, Suzuki H, Ozeki I, Iwata N, Ohmura T, Satoh T, et al. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut. 2001;48:372–377. doi: 10.1136/gut.48.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roncalli M, Bianchi P, Bruni B, Laghi L, Destro A, Di Gioia S, Gennari L, Tommasini M, Malesci A, Coggi G. Methylation framework of cell cycle gene inhibitors in cirrhosis and associated hepatocellular carcinoma. Hepatology. 2002;36:427–432. doi: 10.1053/jhep.2002.34852. [DOI] [PubMed] [Google Scholar]
- 31.Fang JY, Xiao SD. Alteration of DNA methylation in gastrointestinal carcinogenesis. J Gastroenterol Hepatol. 2001;16:960–968. doi: 10.1046/j.1440-1746.2001.02554.x. [DOI] [PubMed] [Google Scholar]
- 32.Chu HJ, Heo J, Seo SB, Kim GH, Kang DH, Song GA, Cho M, Yang US. Detection of aberrant p16INK4A methylation in sera of patients with liver cirrhosis and hepatocellular carcinoma. J Korean Med Sci. 2004;19:83–86. doi: 10.3346/jkms.2004.19.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda Y. Molecular mechanism underlying the functional loss of cyclindependent kinase inhibitors p16 and p27 in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1734–1740. doi: 10.3748/wjg.14.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]