Abstract
Background
Current biomarkers for breast cancer have little potential for detection. We determined if breast cancer subtypes influence circulating protein biomarkers.
Methods
A sandwich-ELISA microarray platform was used to evaluate 23 candidate biomarkers in plasma samples that were obtained from subjects with either benign breast disease or invasive breast cancer. All plasma samples were collected at the time of biopsy, after a referral due to a suspicious screen (e.g., mammography). Cancer samples were evaluated based on breast cancer subtypes, as defined by the HER2 and estrogen receptor statuses.
Results
Ten proteins were statistically altered in at least one breast cancer subtype, including four epidermal growth factor receptor ligands, two matrix metalloproteases, two cytokines, and two angiogenic factors. Only one cytokine, RANTES, was significantly increased (P<0.01 for each analysis) in all four subtypes, with areas under receiver operating characteristic curves (AUC) that ranged from 0.76 to 0.82, depending on cancer subtype. The best AUC values were observed for analyses that combined data from multiple biomarkers, with values ranging from 0.70 to 0.99, depending on the cancer subtype. Although the results for RANTES are consistent with previous publications, the multi-assay results need to be validated in independent sample sets.
Conclusions
Different breast cancer subtypes produce distinct biomarker profiles, and circulating protein biomarkers have potential to differentiate between true and false positive screens for breast cancer. Impact: Subtype-specific biomarker panels may be useful for detecting breast cancer or as an adjunct assay to improve the accuracy of current screening methods.
Introduction
Breast cancer is responsible for more than 40,000 deaths per year in the United States. Early detection appears to be the best method to increase survival rates (1,2), and considerable effort has been directed towards improving mammography and other imaging tools to improve detection of small tumors (3,4). Even so, early detection of breast cancer remains problematic. In the case of mammography, the area under the curve (AUC) values for receiver operating characteristic (ROC) curves has been estimated to range from 0.76 to 0.82 (5,6). The limitations of existing screening methods suggest a need for alternative tests to assist in the early detection of breast cancer. This need has emphasized the potential value of circulating biomarkers for early detection. However, established breast cancer biomarkers have not proven useful for this purpose (7).
One reason for the lack of good markers for early detection may be the phenotypic diversity of breast cancer, which would be expected to complicate biomarker identification. Five major subtypes of breast cancer have been identified based on gene-expression or protein profiles in tumor tissue (8-12). These subtype expression patterns closely align with traditional histological classifiers related to the overexpression of estrogen receptor (ER+) and/or the HER2 receptor (HER2+) (10,13). Results from the current study suggest that patterns of circulating biomarkers also show breast-cancer subtype dependence.
Materials and Methods
Samples
All samples were obtained and analyzed in accordance with the human subjects institutional review boards at Duke University Medical Center, Durham, NC, and the Pacific Northwest National Laboratory, Richland, WA. All blood samples were collected after a positive mammogram or breast exam that led to referral for image-guided biopsy. All samples were collected at the time of biopsy, such that the benign controls and cancer patients were not defined until later, after pathology. It is unknown if the samples were collected before or after the biopsy, but all samples were collected during the same visit. Therefore, there was concurrent collection of both benign and malignant cases in the same clinical venue using identical methods, with both the patient and the phlebotomist ignorant of the presence or absence of breast cancer. This sample-collection design eliminates the possibility of some potentially confounding factors, including systematic differences in sample acquisition, handling, and storage, and patient-specific confounders, such as pre-surgery fasting and elevated stress levels that could result from knowing that cancer is present. Benign controls were patients with a positive screen but without breast cancer, including ductal carcinoma in situ, as determined by pathology evaluation of the biopsy.
The human plasma samples (both benign and invasive) were collected at Duke University Medical Center from 2000-2005. 125 samples were selected such that the different groups were age-matched and each group contained equal numbers of women younger than 50 and older than 50 years. The cancer groups were also selected based on receptor status and analyzed in two sets.
The first group contained 20 estrogen receptor positive/HER2 negative (ER+/HER2-) tumors, 19 HER2 positive/estrogen receptor negative tumors (ER-/HER2+), and 20 benign controls. The second group contained 22 estrogen receptor positive/HER2 positive (ER+/HER2+) tumors, 24 estrogen receptor negative/HER2 negative (ER-/HER2-) tumors, and 21 benign controls. The patients for both tumor sets were between the ages of 19 and 78, with a median age of 56 years. After collection, samples were aliquoted and stored at -80 °C until analyzed.
ELISA microarray assay
The protocols and reagents used in the ELISA microarray have been previously described in detail (14). The sandwich ELISA microarray platform, when employing the same set of reagents and assays as used in this study, has been shown to lack any assay cross-reactivity and to be able to quantitatively measure purified antigens spiked into human serum (15). All reagents have been previously described in detail, including commercial sources and concentrations used (15). Capture antibodies were printed on aminosilanated glass slides (Erie Scientific, Portsmouth, NH) that were stamped by the manufacturer with a hydrophobic barrier to create 16 identical wells (1 chip per well) on each slide. A sandwich ELISA for green fluorescent protein, which was spiked into the samples and standards at 100 pg/ml, was used for quality evaluation and data normalization across chips, as previously described (16). Each reagent was printed four times per chip, once in each of four identical quadrants. Successful printing of the capture antibodies was confirmed using the Red Reflect option on a ScanArray ExpressHT microarray laser scanner (PerkinElmer, Santa Clara, CA). Standard curves for the ELISAs were generated as described previously (14). Fifteen microliters of each diluted sample or standard mixture were placed on each of three replicate chips. Thus, each sample was analyzed on three chips, and each chip contained quadruplicates spots for each assay, for a total of 12 replicates per sample for each ELISA analysis. Sample positioning was blocked by study group, and each of the three sample replicates was analyzed on a different slide. The data from a single dilution of the plasma was used for each of the 23 ELISAs (Table S1), with the selected dilution being the one that produced the most signal intensities for the samples that were within the optimal range of the standard curve.
Data, Statistics and ROC analyses
Standard curves from the ELISA microarray analyses were generated using the Protein Microarray Analysis Tool (ProMAT), a custom software program that we developed specifically for this use (17). This software is freely available at (18). Mann-Whitney nonparametric tests (P ≤0.05) were used to compare the protein concentrations in the human plasma samples across groups (19). The Mann-Whitney rank sum and Spearman correlation P-values were calculated using SigmaPlot (SYSTAT; Chicago, IL). Cancer/no cancer evaluations were quantified and visualized using receiver operator characteristic (ROC) curves (20). These curves were generated using R (21). The empirical “stepped” ROC curves, and the AUC values that were derived from these curves, were estimated with the ROCR library (22). Smooth ROC curves and approximate 90% confidence bounds were estimated using Monte Carlo simulation. For these analyses, the data was log transformed to produce normally distributed values. The success of the log transformation was evaluated with Kolomogorov-Smirnov and Shapiro-Wilk Normality tests (23). Twenty samples were simulated with draws from an estimated normal distribution representing each of the cancer/no cancer classes, and an empirical ROC curve was constructed and stored. This was repeated 500 times. The 5%, 50%, and 95% quantiles of the point-wise distribution of the 500 ROC curves were calculated and then smoothed with a moving average to estimate the mid-ROC curve with approximate 90% confidence interval bounds. Smooth ROC curves complementing their empirical ROC counterparts were accepted for assays with admissible Normality tests (P < 0.10) and empirical ROC curves contained within the ROC confidence envelope.
A linear combination of assay scores was used to estimate a composite assay score from multiple assays. A composite multivariate assay score was calculated using a linear discriminant algorithm. The ability of the multivariate assay test to discriminate between cancer and no cancer was evaluated using the previously described ROC method on the composite multivariate assay scores
Results
EGFR ligands produce a unique signature in plasma from women whose tumors are either singly positive for the ER or for HER2
We compared plasma protein profiles between women with breast cancer and age-matched controls with benign disease. This initial sample set was comprised of individual plasma samples from 20 women with ER+/HER2-, 18 with ER-/HER2+ breast cancer, and 18 with benign breast disease. The individual biomarkers that were significantly increased in plasma from the ER+/HER2- cases compared to the benign controls were AmR (P= 0.015), HB-EGF (P= 0.03), RANTES (P= 0.002), and TGFα (P= 0.01) (Fig. 1; see Table S1 for assay abbreviations). For Her2+/ER- group: EGF (P= 0.02), HB-EGF (P= 0.001), RANTES (P= 0.002), and TGFα (P= 0.001) were significantly increased compared to subjects with benign breast disease. Only one analyte, MMP9, was significantly decreased (P=0.009), and this decrease was only observed in patients with ER+/HER2- tumors.
Figure 1.
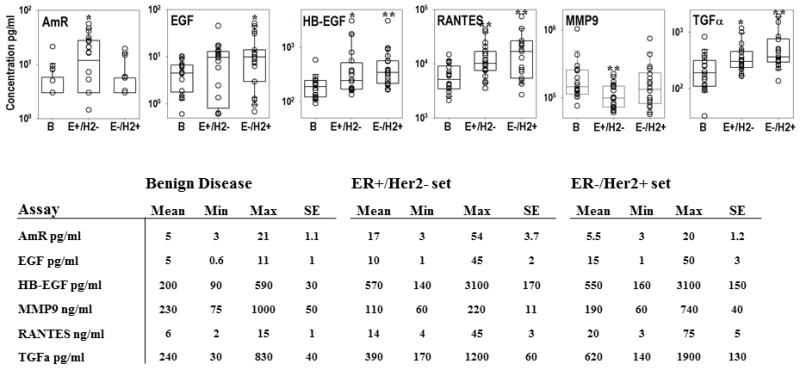
Protein concentrations in plasma, as determined by ELISA microarray analysis of 18 benign (B), 19 estrogen receptor positive/HER2 receptor negative (ER+/HER2-), and 20 estrogen receptor negative/HER2 receptor positive (ER-/HER2+) patients. Plasma concentrations are graphed for all assays that showed a statistically significant difference between groups, including amphiregulin (AmR), epidermal growth factor (EGF), heparin binding-epidermal growth factor (HB-EGF), matrix metalloproteinase 9 (MMP9), RANTES and transforming growth factor alpha (TGFα). The center line, box and crossbars indicate the group median and 75th and 90th percentiles, respectively. *Significantly different at p<0.05. **Significantly different at p<0.01. Values shown in the table are the mean, minimal (Min) and maximal (Max) concentration values and the standard error (SE) for each assay.
ROC curves were generated for the biomarkers that were statistically different (see above) between the singly receptor-positive cases and the benign controls. The RANTES ROC curves were similar (Fig. 2) for ER-/HER2+ and ER+/HER2- groups, each having an AUC value of 0.82 (Table S2). AmR concentrations, which were significantly altered for the ER+/HER2- group but not the ER-/HER2+, produced AUC values of 0.73 and 0.53, respectively (Figs. 1 and 2). Differences were also observed for EGF (AUC values of 0.67, 0.75, respectively), HB-EGF (0.73, 0.85), TGFα (0.84, 0.77) and MMP9 (0.79, 0.63). We evaluated the effect of combining multiple assays using Linear Discriminant Analysis (Fig. 3). Based on the AUC values, the multi-assay tests (Fig. 3) generally had higher sensitivity and specificity than the single-analyte tests (Fig.2). Of the paired biomarker analyses, MMP9 and TGFα yielded the best AUC value of 0.96 for the ER+/Her2- versus benign control comparison (Table S2).
Figure 2.
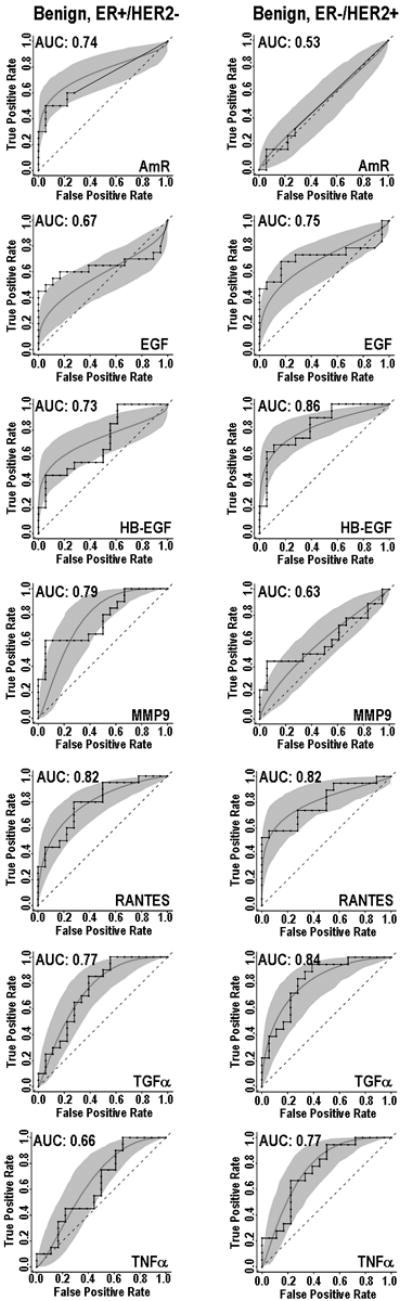
Receiver operator characteristic (ROC) curves from single assays for amphiregulin (AmR), epidermal growth factor (EGF), heparin binding epidermal growth factor (HB-EGF), matrix metalloprotease 9 (MMP9), and RANTES, all of which are significantly altered in patients with breast cancer in subjects with either ER+/HER2- tumors (left column) or ER-/HER2+ tumors (right column). Each graph contains step (empirical) and smooth (fitted, parametric) ROC curves, and a grey shaded area that presents the 90% confidence intervals of the fitted curve, as generated by Monte Carlo analysis.
Figure 3.
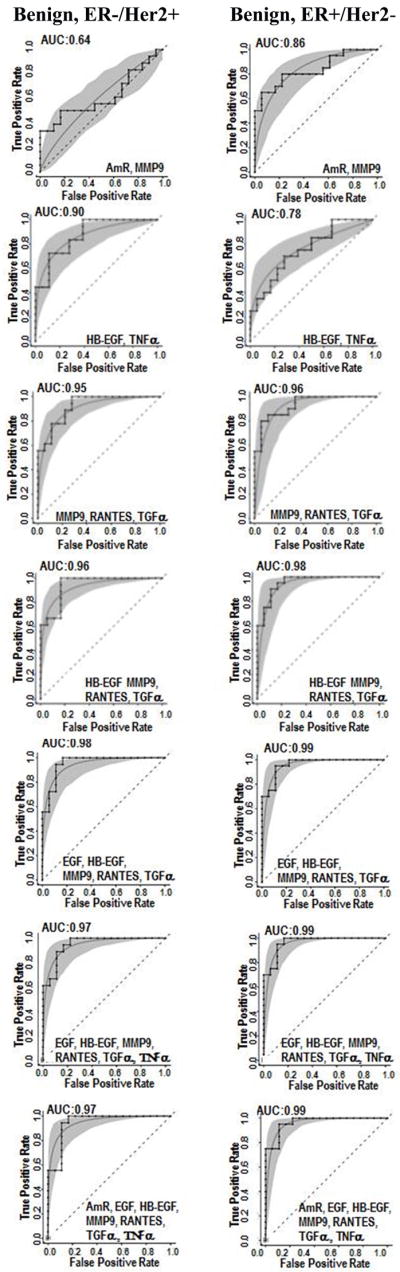
Multiple-assay ROC curves for patients with either ER+/HER2- tumors (left column) or ER-/HER2+ tumors (right column) versus controls with benign breast disease. Each graph contains step (empirical) and smooth (fitted, parametric) ROC curves, and a grey shaded area that presents the 90% confidence intervals of the fitted curve, as generated by Monte Carlo analysis.
The clinical utility of combinations of different candidate biomarkers might be greater if the individual biomarkers were independent indicators of the presence of cancer. We therefore compared biomarker concentrations across groups using the Spearman correlation test for those biomarkers that showed significant differences between study groups. We also determined if the concentrations of these biomarkers correlated with age. These tests demonstrated that plasma concentrations of the four EGFR ligands, AmR, EGF, HB-EGF, and TGFα were generally significantly correlated, with the sole exception of AmR and TGFα (Table S3). In addition, RANTES significantly correlated with EGF (P ≤ 0.001), HB-EGF (P = 0.022) and TGFα (P ≤ 0.001). Age did not correlate with the levels of the different biomarkers. Overall, subsets of the biomarkers are correlated, but not all of the biomarkers are. Thus, these results suggest that multiple biological processes may be regulating concentrations of these circulating proteins.
RANTES and angiogenic factors PDGF and VEGF are increased in ER-/HER2- and/or ER+/HER2+ tumors
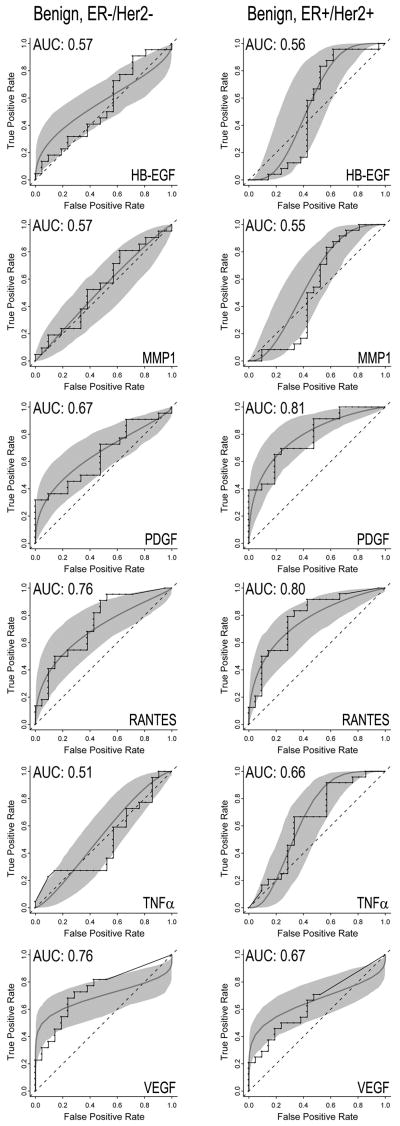
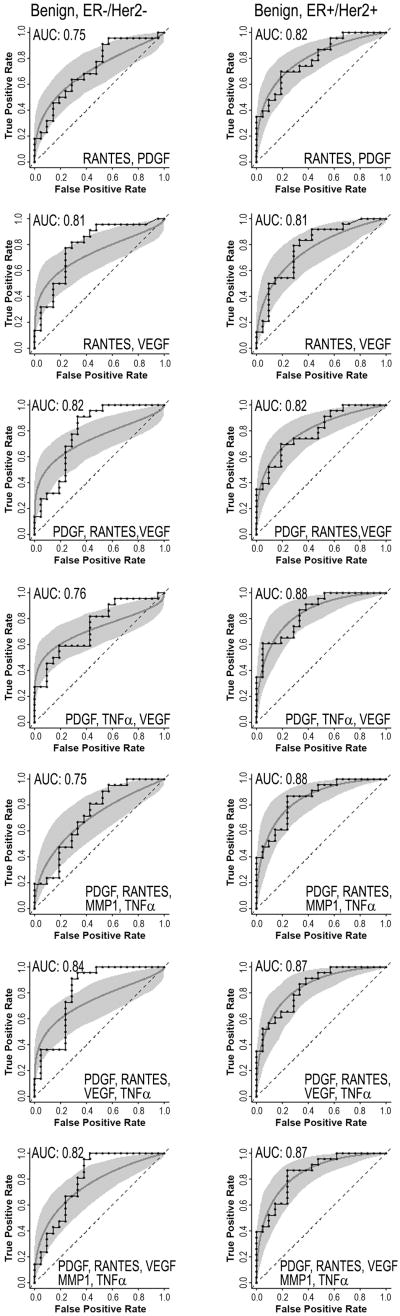
In order to better determine the effects of breast cancer subtypes on biomarker profiles, we analyzed a second set of plasma samples that were obtained from 22 ER-/HER2- cases, 24 ER+/HER2+ cases, and 21 aged-matched benign controls. Plasma from ER-/HER2- patients had significantly increased concentrations of VEGF (P = 0.006) and RANTES (P = 0.009) relative to the benign controls (Fig. 4). In plasma samples from patients with ER+/HER2+ tumors, PDGF (P = 0.001) and RANTES (P = 0.002) were significantly increased relative to plasma samples from benign controls (Fig. 4). In contrast to the first set of data, the EGFR ligand HB-EGF was decreased (P = 0.044) in the plasma from subjects with ER+/HER2+ tumors compared to the benign disease. In addition, TNFα (P = 0.006) and MMP1 (P = 0.048) were significantly decreased in plasma samples from ER+/HER2+ (Fig.4). None of the proteins we assayed were significantly decreased in the ER-/HER2- patients. Single-assay ROC curves for either PDGF, RANTES or VEGF produced AUC values of 0.67, 0.76, and 0.76, respectively, for ER-/HER2- and 0.81, 0.80 and 0.67, respectively, for ER+/HER2+ (Fig. 5). When results from PDGF, RANTES, and VEGF assays were combined, the AUC values for each of the breast-cancer subtypes improved to 0.82 (Fig. 6). The highest AUC value was 0.88, and was observed for the ER+/HER2+ plasma set using RANTES, PDGF, VEGF and TNFα (Table S4).
Figure 4.
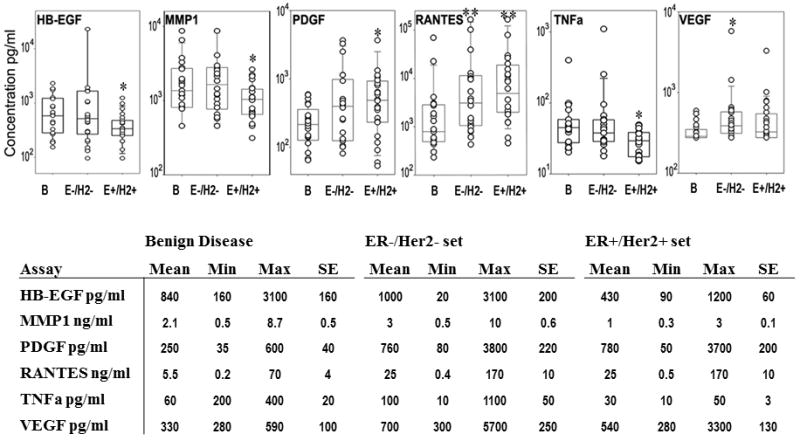
Plasma protein concentration determined by ELISA microarray analysis for 20 benign (B), 22 estrogen receptor negative/HER2 receptor negative (E-/H2-), and 24 estrogen receptor positive/HER2 receptor positive (E+/H2+) patients. The center line, box and crossbar represent the group median and 75th and 90th percentiles, respectively. Values in the table are the mean, minimal (Min), and maximum (Max) concentration values, and the standard error (SE) for each assay. *significantly different at p<0.05. **significantly different at p<0.001.
Figure 5.
Receiver operating characteristic (ROC) curves from single assays for heparin binding epidermal growth factor (HB-EGF), matrix metalloprotease 1 (MMP1), platelet derived growth factor (PDGF), RANTES, tumor necrosis factor alpha (TNFα), or vascular endothelial growth factor (VEGF) that are significantly altered in either double-negative (ER-/Her2-, left column) or double-positive (ER+/Her2+) breast cancer cases. Each graph contains step (empirical) and smooth (fitted, parametric) ROC curves, and a grey shaded area that presents the 90% confidence intervals of the fitted curve, as generated by Monte Carlo analysis.
Figure 6.
Multi-assay receiver operating characteristic (ROC) curves for ER-/HER2- (left column) and the ER+/HER2+ (right column) breast cancer groups. Each graph contains step (empirical) and smooth (fitted, parametric) ROC curves, and a grey shaded area that presents the 90% confidence intervals of the fitted curve, as generated by Monte Carlo analysis.
We determined if these biomarkers in this second sample set correlated with each other or age (Table S5). Plasma concentrations of RANTES were significantly correlated with PDGF and VEGF (P ≤ 0.001); however, PDGF and VEGF levels were not significantly correlated with each other. Of the three proteins that decreased significantly in this sample set, only HB-EGF and MMP1 levels were correlated (P < 0.001). Age did not correlate with any of the ELISA data except for a negative correlation with MMP1 (P < 0.001).
Discussion
We used a custom ELISA microarray platform to evaluate 23 candidate biomarkers in plasma samples from women with newly diagnosed breast cancer. Data from these analyses were compared to data from plasma samples from women with benign breast disease who were also undergoing image-guided biopsy over the same time period and using the same collection and processing methods as the breast cancer cases. We found that ten analytes (AmR, EGF, HB-EGF, MMP1, MMP9, PDGF, RANTES, TGFα, TNFα, and VEGF) were significantly altered in at least one of the four breast cancer subtypes, which were defined by ER and HER2 tumor expression levels. With the exception of RANTES, significant changes in biomarker expression were subtype dependent. This observation is consistent with gene and protein expression studies that show unique expression patterns in tumors based on cancer subtypes (8-13). All ten proteins that we found to be altered in the circulation have previously been reported to have altered expression in breast cancer tissue at the mRNA or protein level (12,24-28), suggesting that altered protein secretion by the breast tumors may be responsible for the changes we observe in the circulating proteins. However, when levels of a biomarker are decreased, it is unlikely that this effect is due to tumor secretion of the biomarker. Rather, indirect effects of the tumor on biomarker levels are likely to be important. For example, tumor-dependent secretion of proteolytic enzymes could be a factor.
RANTES has been previously reported to be increased in the plasma of women with breast cancer (29,30). Our results not only verify these prior studies, but we extend these results by demonstrating that circulating levels of RANTES are increased in at least four subtypes of breast cancer. Notably, single-assay analysis of RANTES produced AUC values that ranged from 0.76 to 0.82, and RANTES consistently improved accuracy when included in subtype-specific panels of biomarkers. Plasma levels of RANTES correlated with levels of three EGFR ligands, EGF, HB-EGF, and TGFα, in the first sample set (Table S3). We have observed that RANTES secretion is increased upon EGFR activation in cultured human mammary epithelial cells (31), suggesting that the correlation of RANTES and the EGFR ligand levels in the plasma may also reflect EGFR regulation of RANTES secretion in humans. RANTES is an inflammatory cytokine and circulating levels of this protein are altered in a variety of diseases besides breast cancer, suggesting that RANTES alone will not be a specific marker for early detection of breast cancer. Even so, our data suggest that RANTES may be useful in discerning between mammographic true and false positives, and that RANTES may provide other useful data when included in a panel of biomarkers. The combination of biomarkers may potentially prove more specific for breast cancer than any individual marker.
Previous studies have shown that overexpression of individual HER receptors or their ligands in breast tumors promotes survival and resistance to chemotherapy (32-36). AmR, EGF, and TGFα also have important roles in normal mouse mammary gland development, and each of these EGFR ligands apparently has a different role in this process (37). In the current study, these EGFR ligands were commonly increased in plasma from women with breast cancer. In addition, we observed subtype-dependent differences in the EGFR ligands, suggesting that, as in mammary development, expression of the individual ligands may reflect differential tumor characteristics. Furthermore, it has been demonstrated that normal serum levels of EGFR ligands are sufficient to stimulate EGFR activity in cultured cells (38), further supporting the concept that an increase in circulating EGFR ligands may directly affect tumor biology. That is, a further increase in circulating levels of EGFR ligands in breast cancer patients could further activate the EGFR and thereby facilitate mammary cell growth and tumor development. Thus, these results suggest that circulating levels of EGFR ligands may be an important etiological factor in the development of breast cancer.
MMP9 is synthesized as a “pro” form that is proteolytically cleaved to produce a smaller, but catalytically active, protease (reviewed in (39)). The ELISA analysis we use only detects the activated (i.e., proteolytically truncated) form of MMP9, which was decreased in ER+/HER2- breast cancer. A variety of proteases can activate MMP9 in artificial systems employing purified proteins, but it is unclear what protease(s) activates MMP9 in vivo (39). Our data are consistent with a previous report that circulating levels of truncated MMP9 are decreased in breast cancer patients (40). Even so, this prior study also found that the proMMP9 form is increased in the same subjects. Given the opposite effects of breast cancer on the precursor and processed forms of MMP9 (40), we speculate that the activity of other protease(s) that process MMP9 may be more important than MMP9 secretion in altering the circulating levels of MMP9 in breast cancer subjects. This concept is supported by evidence that cancer cells secrete a variety of proteases (41).
In conclusion, identifying useful circulating markers for the detection of breast cancer has remained problematic. Our results suggest that the discovery of biomarker panels for the early detection of breast cancer may be complicated by the heterogeneity of this disease. However, results from two independent sample sets confirm that plasma levels of RANTES are commonly increased in breast cancer. Furthermore, the use of non-specific biomarkers such as RANTES in combination with breast-cancer or breast-cancer-subtype specific biomarkers may produce a superior assay that may prove useful as an adjuvant to current screening methods. We also identify two panels of biomarkers that are able to discriminate between benign breast disease and the most common receptor statuses found in breast cancer, which are ER+ (about 60 to 67% of all breast cancers) and HER2+ (about 20%) (42). These biomarker panels are very promising, but the data are novel and need to be replicated in an independent sample set. Intriguingly, these panels contain several EGFR ligands. Given the central role of the EGFR in regulating mammary cell growth and migration, these data suggest that circulating levels of these ligands could be a factor in breast cancer development and/or progression.
Supplementary Material
Acknowledgments
This work was funded by NCI Early Detection Research Network grants CA117378 and CA084955.
Reference List
- 1.Dillman RO, McClure SE. Improving survival for patients with breast cancer compared with intramural and extramural benchmarks. Clin Breast Cancer. 2007;7:480–5. doi: 10.3816/CBC.2007.n.005. [DOI] [PubMed] [Google Scholar]
- 2.Siegelmann-Danieli N, Khandelwal V, Wood GC, Mainali R, Prichard J, Murphy TJ, et al. Breast cancer in elderly women: outcome as affected by age, tumor features, comorbidities, and treatment approach. Clin Breast Cancer. 2006;7:59–66. doi: 10.3816/CBC.2006.n.014. [DOI] [PubMed] [Google Scholar]
- 3.Mobley LR, Kuo TM, Driscoll D, Clayton L, Anselin L. Heterogeneity in mammography use across the nation: separating evidence of disparities from the disproportionate effects of geography. Int J Health Geogr. 2008;7:32. doi: 10.1186/1476-072X-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corsetti V, Houssami N, Ferrari A, Ghirardi M, Bellarosa S, Angelini O, et al. Breast screening with ultrasound in women with mammography-negative dense breasts: evidence on incremental cancer detection and false positives, and associated cost. Eur J Cancer. 2008;44:539–44. doi: 10.1016/j.ejca.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Fenton JJ, Taplin SH, Carney PA, Abraham L, Sickles EA, D’Orsi C, et al. Influence of computer-aided detection on performance of screening mammography. N Engl J Med. 2007;356:1399–409. doi: 10.1056/NEJMoa066099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrick RE, Cole EB, Pisano ED, Acharyya S, Marques H, Cohen MA, et al. Accuracy of soft-copy digital mammography versus that of screen-film mammography according to digital manufacturer: ACRIN DMIST retrospective multireader study. Radiology. 2008;247:38–48. doi: 10.1148/radiol.2471070418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen VN, Sorlie T, Geisler J, Langerod A, Yoshimura N, Karesen R, et al. Gene expression profiling of breast cancer in relation to estrogen receptor status and estrogen-metabolizing enzymes: clinical implications. Clin Cancer Res. 2005;11:878s–83s. [PubMed] [Google Scholar]
- 9.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–65. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 11.Gast MC, Schellens JH, Beijnen JH. Clinical proteomics in breast cancer: a review. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0263-3. [DOI] [PubMed] [Google Scholar]
- 12.Goncalves A, Charafe-Jauffret E, Bertucci F, Audebert S, Toiron Y, Esterni B, et al. Protein profiling of human breast tumor cells identifies novel biomarkers associated with molecular subtypes. Mol Cell Proteomics. 2008;7:1420–33. doi: 10.1074/mcp.M700487-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez R, Varnum S, Zangar R. Sandwich ELISA microarrays. Generating reliable and reproducible assays for high-throughput screens. In: Wang F, editor. Biomarker Methods in Drug Discovery and Development. Totowa, NJ: Humana Press; 2008. pp. 273–90. [Google Scholar]
- 15.Gonzalez RM, Seurynck-Servoss SL, Crowley SA, Brown M, Omenn GS, Hayes DF, et al. Development and validation of sandwich ELISA microarrays with minimal assay interference. J Proteome Res. 2008;7:2406–14. doi: 10.1021/pr700822t. [DOI] [PubMed] [Google Scholar]
- 16.Zangar RC, Daly DS, White AM, Servoss SS, Tan RM, Collett JR. ProMAT Calibrator: a tool for reducing experimental bias in antibody microarrays. J Proteome Res. 2009;8:3937–43. doi: 10.1021/pr900247n. [DOI] [PubMed] [Google Scholar]
- 17.White AM, Daly DS, Varnum SM, Anderson KK, Bollinger N, Zangar RC. ProMAT: protein microarray analysis tool. Bioinformatics. 2006;22:1278–9. doi: 10.1093/bioinformatics/btl093. [DOI] [PubMed] [Google Scholar]
- 18.www.pnl.gov/statistics/ProMAT/
- 19.Conover W. Practical Nonparametric Statistics. Third ed. New York: John Wiley and Sons; 1999. [Google Scholar]
- 20.Pepe M. The Statistical Evaluation of Medical Tests for Classification and Prediction. New York: Oxford University Press; 2003. [Google Scholar]
- 21.http://www.r-project.org
- 22.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 23.Panik MJ. Advanced statistics from an elementary point of view. San Diego, CA: Elsevier Academic Press; 2005. [Google Scholar]
- 24.Drukier AK, Grigoriev I, Brown LR, Tomaszewski JE, Sainsbury R, Godovac-Zimmermann J. Looking for Thom’s biomarkers with proteomics. J Proteome Res. 2006;5:2046–8. doi: 10.1021/pr060231q. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs JM, Waters KM, Kathmann LE, Camp DG, Wiley HS, Smith RD, et al. The mammary epithelial cell secretome and its regulation by signal transduction pathways. J Proteome Res. 2008;7:558–69. doi: 10.1021/pr0704377. [DOI] [PubMed] [Google Scholar]
- 26.Rogers S, Girolami M, Kolch W, Waters KM, Liu T, Thrall B, et al. Investigating the correspondence between transcriptomic and proteomic expression profiles using coupled cluster models. Bioinformatics. 2008;24:2894–900. doi: 10.1093/bioinformatics/btn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson KS, Roberts H, Leek R, Harris AL, Geradts J. Differential gene expression patterns in HER2/neu-positive and -negative breast cancer cell lines and tissues. Am J Pathol. 2002;161:1171–85. doi: 10.1016/S0002-9440(10)64394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, Yu J, Cordero KE, Johnson MD, Ghosh D, Rae JM, et al. A transcriptional fingerprint of estrogen in human breast cancer predicts patient survival. Neoplasia. 2008;10:79–88. doi: 10.1593/neo.07859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dehqanzada ZA, Storrer CE, Hueman MT, Foley RJ, Harris KA, Jama YH, et al. Assessing serum cytokine profiles in breast cancer patients receiving a HER2/neu vaccine using Luminex technology. Oncol Rep. 2007;17:687–94. [PubMed] [Google Scholar]
- 30.Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267:271–85. doi: 10.1016/j.canlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Gonzalez RM, Zangar RC. Protein secretion in human mammary epithelial cells following HER1 receptor activation: influence of HER2 and HER3 expression. BMC Cancer. 2011;11:69. doi: 10.1186/1471-2407-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boerner JL, Gibson MA, Fox EM, Posner ED, Parsons SJ, Silva CM, et al. Estrogen negatively regulates epidermal growth factor (EGF)-mediated signal transducer and activator of transcription 5 signaling in human EGF family receptor-overexpressing breast cancer cells. Mol Endocrinol. 2005;19:2660–70. doi: 10.1210/me.2004-0439. [DOI] [PubMed] [Google Scholar]
- 33.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. ErbB/HER receptor family in breast cancer--the more we search the more we learn. Ann Oncol. 2008;19:1020–1. doi: 10.1093/annonc/mdn061. [DOI] [PubMed] [Google Scholar]
- 34.Revillion F, Lhotellier V, Hornez L, Bonneterre J, Peyrat JP. ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis. Ann Oncol. 2008;19:73–80. doi: 10.1093/annonc/mdm431. [DOI] [PubMed] [Google Scholar]
- 35.Sundaresan S, Roberts PE, King KL, Sliwkowski MX, Mather JP. Biological response to ErbB ligands in nontransformed cell lines correlates with a specific pattern of receptor expression. Endocrinology. 1998;139:4756–64. doi: 10.1210/endo.139.12.6378. [DOI] [PubMed] [Google Scholar]
- 36.Zaczek A, Brandt B, Bielawski KP. The diverse signaling network of EGFR, HER2, HER3 and HER4 tyrosine kinase receptors and the consequences for therapeutic approaches. Histol Histopathol. 2005;20:1005–15. doi: 10.14670/HH-20.1005. [DOI] [PubMed] [Google Scholar]
- 37.Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–50. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 38.Koellensperger E, von HD, Markowicz M, Pallua N. Human serum from platelet-poor plasma for the culture of primary human preadipocytes. Stem Cells. 2006;24:1218–25. doi: 10.1634/stemcells.2005-0020. [DOI] [PubMed] [Google Scholar]
- 39.Murphy G, Gelatinase B, Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Second. London, UK: Elsevier Academic Press; 2004. pp. 503–511. [Google Scholar]
- 40.Somiari SB, Shriver CD, Heckman C, Olsen C, Hu H, Jordan R, et al. Plasma concentration and activity of matrix metalloproteinase 2 and 9 in patients with breast disease, breast cancer and at risk of developing breast cancer. Cancer Lett. 2006;233:98–107. doi: 10.1016/j.canlet.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Duffy MJ. Proteases as prognostic markers in cancer. Clin Cancer Res. 1996;2:613–8. [PubMed] [Google Scholar]
- 42.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.