SUMMARY
Sequences rich in glutamine (Q) and asparagine (N) residues often fail to fold at the monomer level. This, coupled to their unusual hydrogen-bonding abilities, provides the driving force to switch between disordered monomers and amyloids. Such transitions govern processes as diverse as human protein-folding diseases, bacterial biofilm assembly, and the inheritance of yeast prions (protein-based genetic elements). A systematic survey of prion-forming domains suggested that Q and N residues have distinct effects on amyloid formation. Here we use cell biological, biochemical, and computational techniques to compare Q/N-rich protein variants, replacing Ns with Qs and Qs with Ns. We find that the two residues have strong and opposing effects: N-richness promotes assembly of benign self-templating amyloids; Q-richness promotes formation of toxic non-amyloid conformers. Molecular simulations focusing on intrinsic folding differences between Qs and Ns suggest that their different behaviors are due to the enhanced turn-forming propensity of Ns over Qs.
Protein regions with little structure – intrinsically disordered regions (IDRs) – are abundant in eukaryotic proteomes (Radivojac et al., 2007). Such regions play critical roles in gene regulation, signaling circuitries, and intracellular transport and they are often centrally located in protein interaction networks (Fuxreiter et al., 2008; Turoverov et al., 2010). Many of these disordered regions undergo disorder-to-order transitions upon binding their interaction partners. But IDRs can also form promiscuous interactions that pose a burden for cellular protein homeostasis (Gsponer et al., 2008; Vavouri et al., 2009).
Some archetypal IDRs have low complexity amino acid sequences that are depleted of order-promoting residues (Romero et al., 2001). Here, our interest is in a subset of such sequences that are enriched in the polar uncharged residues glutamine (Q) and asparagine (N). Despite their general tendency toward disorder at the monomer level (Weathers et al., 2004; Pierce et al., 2005; Toombs et al., 2010), Q/N-rich sequences can, on occasion, self-assemble into some of the most ordered structures in biology – amyloids (Perutz et al., 2002; Uversky, 2008; Alberti et al., 2009).
Amyloids are pseudocrystalline fibrils stabilized by extensive intermolecular interactions between individual polypeptide monomers (Nelson and Eisenberg, 2006). Thus, during amyloid formation, Q/N-rich proteins transition from one extreme of conformational space to the other. This transition involves the association of disordered monomers into molten oligomers. Disorder-to-order transitions of individual polypeptides within these oligomers can be facilitated by intermolecular interactions, leading to an amyloid-nucleating species (Serio et al., 2000; Krishnan and Lindquist, 2005; Mukhopadhyay et al., 2007; Walters and Murphy, 2009; Williamson et al., 2010).
Several protein misfolding diseases, including Huntington’s disease and multiple spinocerebellar ataxias, are associated with the aggregation of polyQ sequences. Both the severity of the disease and the aggregation tendency of the protein are correlated with the length of the polyQ tract (Perutz and Windle, 2001). Q-rich and N-rich proteins can also undergo conformational switches to amyloid under non-pathological conditions, and in some cases these amyloids have important biological roles. Functional amyloids include extracellular adhesins of bacteria, an ancient and broadly distributed class of proteins that act as structural scaffolds for biofilm formation (Larsen et al., 2007; Hammer et al., 2008; Dueholm et al., 2010). Other Q/N-rich proteins can serve as “protein only” elements of inheritance when their IDRs switch to the amyloid state. The latter, comprising the majority of known prion proteins, are united only by the ability of their amyloid conformations to perpetuate through a protein folding reaction that is self-templating and heritable (Glover et al., 1997; Alberti et al., 2009).
In the baker’s yeast Saccharomyces cerevisiae, in which Q/N-rich prions have been characterized most extensively, the self-templating conformational conversions of prion proteins can produce robust new biological traits that derive either from sequestration of the protein’s globular domain, or from novel functions conferred by the prion itself (Halfmann and Lindquist, 2010). Unlike the well-known mammalian prion protein, PrP, which causes a deadly disease, yeast prions are not overtly toxic. In fact, the phenotypic diversity generated by yeast prion switching can be beneficial under many conditions. Hence prions might facilitate survival and adaptation in the rapidly changing natural environments of microbial cells (True and Lindquist, 2000; Halfmann et al., 2010). Whether yeast prions have evolved to this purpose remains to be established.
A recent genome-wide survey found that prion-forming proteins were more likely to be N-rich than Q-rich (Alberti et al., 2009). This observation was unexpected, as it challenged the common assumption that Ns and Qs are equivalent for prion formation (see for example Michelitsch and Weissman, 2000; Osherovich et al., 2004; Ross et al., 2005a). It was subsequently suggested (Toombs et al., 2010) that the observed bias was better explained by the presence of structure-breaking proline residues, which were more common in the Q-rich sequences studied. Here, we provide an in-depth analysis of the contributions of N and Q residues to prion formation. Our assessments are performed in identical sequence contexts, i.e., for each protein we compare the wild-type sequence, which has an admixture of Qs and Ns, to variants where all the Qs have been replaced by Ns or vice versa. This allowed us to make clear conclusions about intrinsic differences in the effects of these residues.
RESULTS
Qs and Ns have disparate effects on prion formation by Sup35
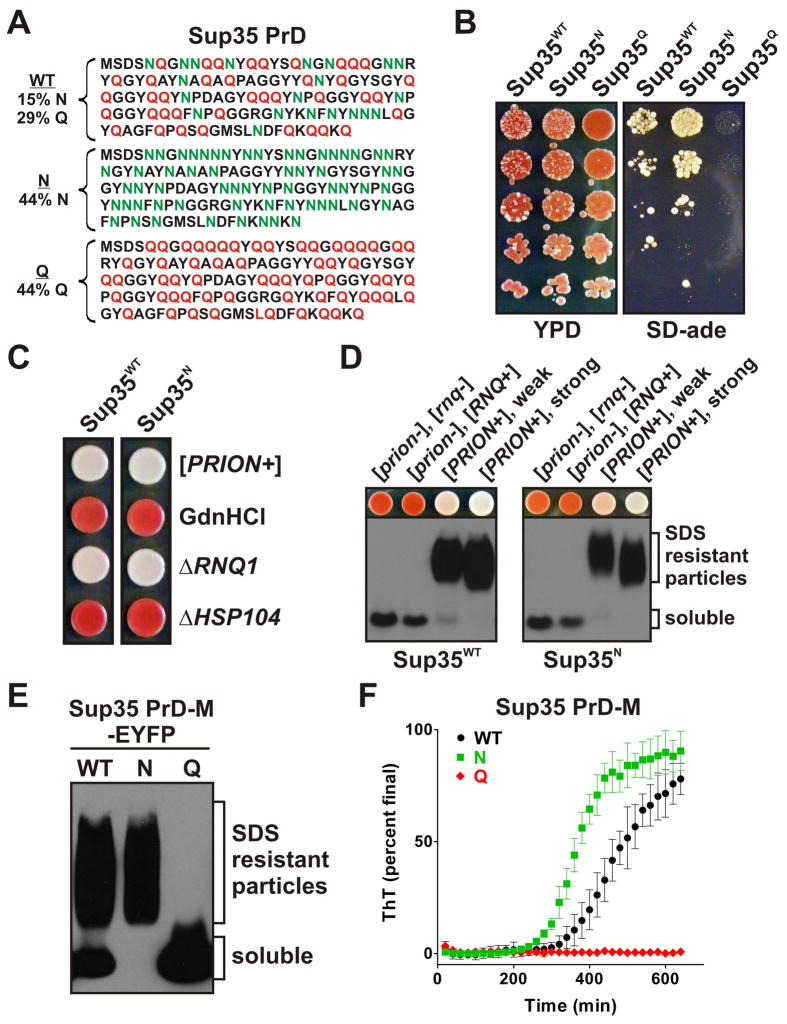
Our computational analyses indicated that N-rich proteins were more likely than Q-rich proteins to form prions, even when prolines were accounted for (Figure S1A-B). We therefore set out to compare the effects of Ns and Qs directly. We generated two variants of the amyloidogenic prion domain (PrD) of the yeast protein Sup35. Normally, 15% of the residues in this PrD are Ns and 29% are Qs (Sup35WT). In the two modified variants, either all Q residues were replaced with Ns (Sup35N), or all N residues were replaced with Qs (Sup35Q) (Figure 1A). The sequences were otherwise identical.
Figure 1. Prion formation by Sup35 is promoted by Ns, inhibited by Qs.
(A) WT sequence of the Sup35 PrD (top), and Q and N replacement variants. (B) Yeast strains expressing Sup35 variants spotted as 5-fold serial dilutions onto YPD (nonselective) or SD-ade (prion-selective) plates. Prion states were induced by the over-expression of PrD-M-EYFP fusions for 24 hours prior to plating. (C) The N-substituted variant of Sup35 can form a prion state that is equivalent to that of WT. White Ade+ Sup35N cells were isolated and passaged on plates containing 5 mM GdnHCl (“GdnHCl”) or transformed with gene-specific knockout cassettes to delete RNQ1 (“ΔRNQ1”) or HSP104 (“ΔHSP104”). All presumptive prion strains were curable and lost the prion state upon deletion of HSP104. A representative [PRION+] strain of Sup35N (right) is compared to a strong [PRION+] strain of Sup35WT (left). (D) Sup35N can form different conformational variants that are equivalent to those of Sup35WT. Colonies with weak and strong Ade+ phenotypes were isolated (Figure S1G). SDS-resistant aggregates were detected by SDD-AGE and immunoblotting with a Sup35C-specific antibody. (E) Variant Sup35 PrD-M-EYFP fusions were expressed for 24 hours in [RNQ+] cells prior to SDD-AGE analysis. PrD-M-EYFP was detected with a GFP-specific antibody. (F) Sup35 PrD-M-His7 variants were purified under denaturing conditions and then diluted to 5 μM in assembly buffer. Reactions were agitated for 10 sec every 2 min in the presence of non-binding plastic beads. Amyloid formation was monitored by ThT fluorescence. Data were normalized by the final values achieved for each variant after extended incubations. Data represent means +/− SEM. See also Figure S1.
We first analyzed the ability of the proteins to maintain normal Sup35 function and their propensity to form alternative self-propagating prion conformations in vivo. To do so, we used a simple phenotypic assay (Alberti et al., 2009). Sup35 is a translation termination factor. When the prion domain switches to the assembled amyloid state, it sequesters Sup35 from ribosomes, causing them to read through stop codons at an increased frequency. In cells carrying a premature stop codon in the ADE1 gene, prion-mediated read through changes colony color from red to white and allows cells to grow without adenine in the medium.
Each Sup35 variant (Sup35WT, Sup35N, and Sup35Q) was constitutively expressed in strains lacking endogenous Sup35. They accumulated to similar levels (Figure S1D). They also produced colonies with comparable red colors. That is, all possessed normal Sup35 activity and could stably maintain a soluble non-prion state (Figure S1E).
The spontaneous rate of Sup35 prion formation is quite low (~ one in 106 cells per generation; Chernoff et al., 1999; Allen et al., 2007; Lancaster et al., 2010). To allow better quantitative comparisons, we increased the likelihood of prion conversion by over-expressing each PrD variant (Derkatch et al., 1996; Alberti et al., 2009) as an EYFP fusion from an inducible promoter (GAL1, Figure S1C). This caused the expected increase in white Ade+ colonies with Sup35WT. Ade+ colonies were more frequent with Sup35N. They were essentially absent with Sup35Q (Figure 1B).
To confirm that the white Ade+ colonies of Sup35N cells were due to prion formation and not a genetic mutation, we asked if they depended on Hsp104, a AAA+ ATPase whose amyloid-fragmenting activity is critical for prion propagation (Chernoff et al., 1995). We passed presumptive prion colonies on media containing a low concentration of guanidine hydrochloride (GdnHCl), which selectively inhibits Hsp104 (Grimminger et al., 2004). This restored Sup35WT and Sup35N cells to their original red phenotypes (Figure 1C). Genetic ablation of HSP104 had the same effect (Figure 1C).
Second, we asked if white Ade+ colonies of Sup35N cells contained the SDS-resistant amyloids that constitute the prion template. Semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE) confirmed that they did. Moreover, Sup35N cells gave rise to a range of phenotypically distinct prion states (white and pink color variants; Derkatch et al., 1996) typical of the wild-type prion, and these were associated with the expected differences in amyloid size on SDD-AGE gels (Kryndushkin et al., 2003) (Figure 1D, S1G).
Having confirmed that the white Ade+ phenotypes of Sup35N cells were due to prions, we examined their dependence on the prion-inducing factor [RNQ+]. This factor is itself a prion conformer of the Rnq1 protein, hence the brackets (denoting cytoplasmic inheritance) and capital letters (denoting genetic dominance). The Sup35WT protein does not require [RNQ+] for prion maintenance, but it does require [RNQ+] to convert to its prion state de novo (Derkatch et al., 2001). We eliminated [RNQ+] by deleting the RNQ1 gene. Sup35WT and Sup35N cells that had already acquired prion phenotypes were not affected (Figure 1C). As expected the RNQ1 deletion eliminated de novo induction of prions in Sup35WT cells. However, while it reduced the appearance of prions in Sup35N cells, it did not eliminate them (Figure S1F). Thus, shifting the Sup35 prion sequence to an N-rich form increased its tendency to form amyloid to such an extent that it bypassed the normal requirement for the prion-inducing factor [RNQ+].
Using these same criteria, the rare white colonies that appeared in Sup35Q cells proved not to be due to the formation of prions. However, rare colonies with all of the hallmarks of prions could form in Sup35Q cells when the protein was expressed at extremely high levels (supplemental text and Figure S1I). These prion states were not toxic. But they were very unstable when expression of Sup35Q was returned to normal levels (not shown). Thus, shifting the Sup35 prion sequence to a Q-rich form virtually eliminated its ability to form prions.
Q and N have disparate effects on amyloid formation by Sup35
Next we asked if the Q and N variants had intrinsically different propensities to form amyloid de novo. We induced Sup35 PrD-EYFP variants for 24 hrs in prion-minus cells. Despite similar expression levels (Figure S1C), the variants showed very different behaviors (Figure 1E). The WT protein partitioned between SDS-soluble and amyloid states. All Sup35N coalesced into SDS-resistant polymers. All Sup35Q remained SDS-soluble.
We next asked if these differences depended upon the cellular environment, or reflected inherently different biochemical properties. We purified the variants from bacteria under fully denaturing conditions. They were then diluted into a physiological assembly buffer containing Thioflavin-T (ThT), a dye that binds amyloid (LeVine, 1993). Sup35WT and Sup35N formed ThT-binding species after a short lag phase, as is characteristic for prion proteins. Sup35N achieved this state more rapidly than Sup35WT. Sup35Q did not form amyloid in the time frame examined (Figure 1F, S1H).
Ns and Qs influence other proteins in similar ways
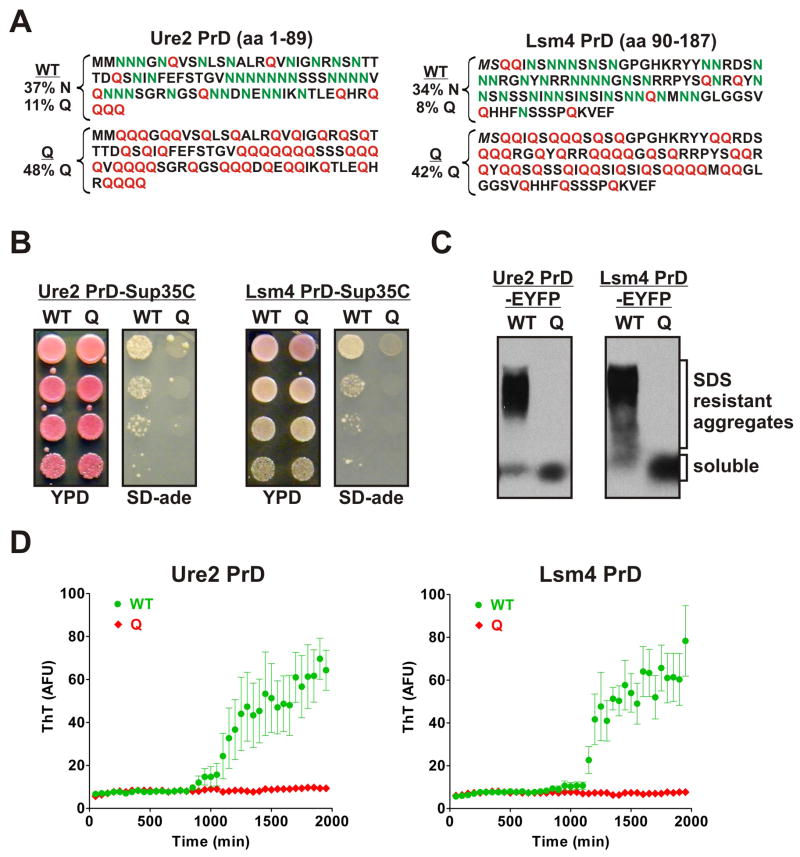
To determine if the effects of Ns and Qs were generalizable, we created N→Q variants of two PrDs that are already very N-rich, one from Ure2 and the other from Lsm4 (Ure2Q and Lsm4Q, Figure 2A). We subjected them to the same tests used for Sup35. The Ure2WT and Lsm4WT Sup35C-fusions drove prion formation at high frequencies. The corresponding Q-rich versions did not (Figure 2B, S2A-C). The Q-rich PrDs were also severely impaired for amyloid formation in vivo, when over-expressed as EYFP fusions (Figure 2C), and in vitro, following their purification and dilution into physiological buffer (Figure 2D).
Figure 2. Replacing Ns with Qs eliminates prion-formation by N-rich PrDs.
(A) The sequences of the Ure2 and Lsm4 PrDs (top), along with the Q variants. (B) Yeast strains containing variant Ure2 and Lsm4 PrDs fused to Sup35C were spotted to YPD and SD-ade plates as in Figure 1B. Prion states were induced by over-expression of PrD-EYFP fusions for 24 hours prior to plating. Representative Ade+ colonies for Ure2WT and Lsm4WT (but not the few Ade+ colonies observed for Ure2Q) showed SDS-resistant aggregates by SDD-AGE and were eliminated by growth on GdnHCl (not shown). (C) Variant Ure2 and Lsm4 PrD-EYFP fusions were expressed for 24 hrs in [RNQ+] cells prior to SDD-AGE analysis as in Figure 1E. (D) Purified denatured variants of Ure2 and Lsm4 PrD-His7 were diluted to 20 μM or 5uM, respectively, in assembly buffer. Reactions were agitated for 10 sec every 2 min in the absence of beads. Amyloid formation was monitored by ThT fluorescence. Data represent means +/− SEM. See also Figure S2.
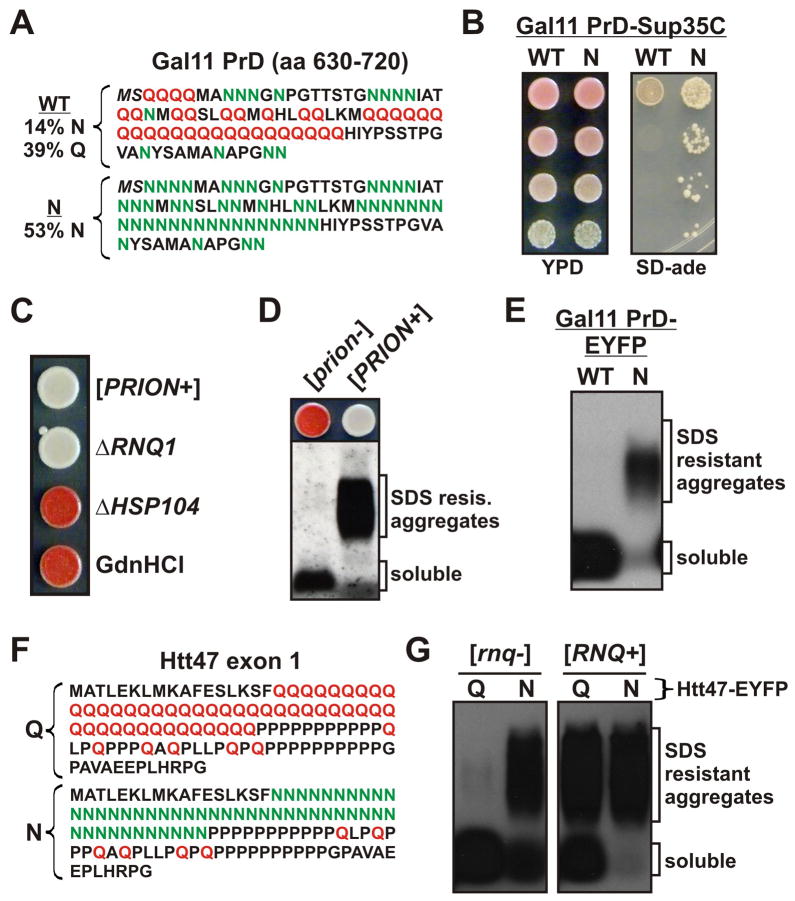
To determine if N-richness can drive prion formation in a protein that does not normally form them, we generated a Q→N variant of a fragment of the Gal11 transcription factor (Gal11N, Figure 3A). As reported previously (Alberti et al., 2009), the WT sequence lacks prion activity (Figure 3B, S3A-C). Gal11N variant readily produced Ade+ colonies with prion properties: the phenotype was reversed by Hsp104 inactivation (Figure 3C), did not require the continued presence of [RNQ+] (Figure 3C), and was associated with SDS-insoluble amyloids (Figure 3D). The Gal11 variants were too poorly expressed in E. coli for purification and in vitro analysis. However, the prion propensities of the Gal11 PrD variants corresponded to their amyloid-forming propensities when they were expressed de novo in yeast cells (Figure 3E).
Figure 3. Replacing Qs with Ns increases amyloid and prion formation by Q-rich proteins.
(A) WT and N variants of the putative PrD of Gal11, residues 630–720. (B) Yeast strains containing variants of the Gal11 PrD fused to Sup35C were spotted to YPD and SD-ade plates as in Figure 1B. Prion states were induced by over-expression of PrD-EYFP fusions for 24 hours prior to plating. (C–D) Gal11N PrD-Sup35C-expressing cells can convert to a prion state. Representative Ade+ cells were isolated and analyzed as in Figure 1C-D. (E) Variant PrD-M-EYFP fusions were expressed for 24 hrs in [RNQ+] cells, followed by SDD-AGE analysis as in Figure 1D. (F) The sequence of Huntingtin exon 1 with a homopolymeric expansion of 47 Qs (top), and the N variant (bottom). (G) HttQ47 and HttN47 fused to EYFP were expressed for 24 hrs in [rnq−] or [RNQ+] cells, followed by SDD-AGE analysis as in Figure 1E. See also Figure S3.
Many proteins associated with amyloid diseases contain long glutamine tracts (polyQ) with a propensity to aggregate in both the human brain (DiFiglia et al., 1997) and when heterologously expressed in yeast (Krobitsch and Lindquist, 2000; Meriin et al., 2002; Duennwald et al., 2006). To explore the distinction between Qs and Ns in such a polypeptide, we compared a disease-associated version of Htt exon 1 (HttQ47), with a Q→N variant of the same protein (HttN47, Figure 3F). When fused to EYFP and expressed for 24 hrs, both variants formed SDS-resistant aggregates that were strongly promoted by [RNQ+] (Figure 3G). However, regardless of [RNQ+] status, HttN47 partitioned much more completely to the SDS-resistant fraction than HttQ47.
N-richness reduces proteotoxicity of Q/N-rich proteins
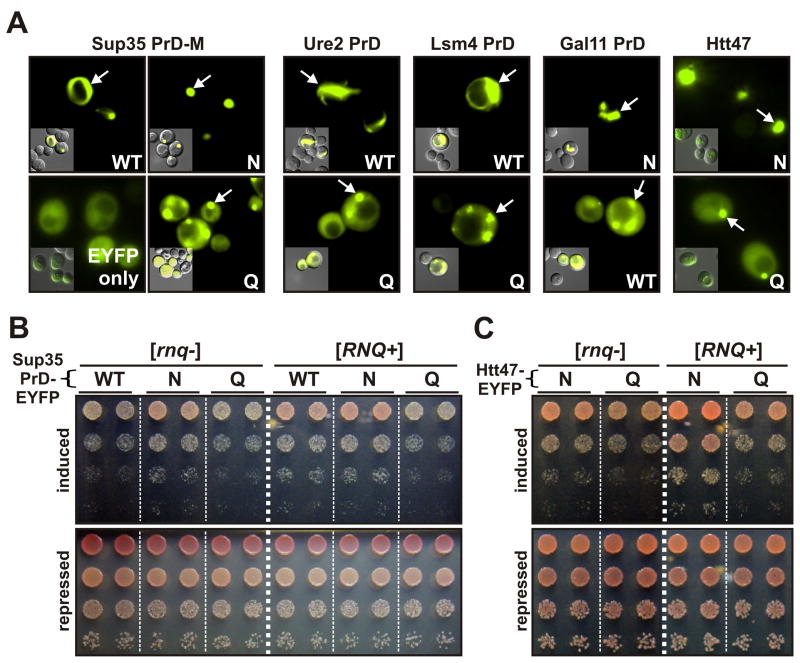
Over-expressed fluorescently-tagged prions typically form bright puncti or ribbon- like foci, indicative of bundled amyloid filaments (Alberti et al., 2009; Kawai-Noma et al., 2010; Tyedmers et al., 2010). Non prion-forming proteins remain diffuse or coalesce weakly into amorphous foci (Alberti et al., 2009). After 48 hrs of expression from a galactose-inducible promoter, all of the N-rich, prion-proficient proteins studied here formed intense foci, often with an elongated filament-like morphology (Figure 4A, top). The Q-rich proteins also formed foci, but these were less intense and surrounded by diffuse fluorescence (Figure 4A bottom and Figure S4G). Foci with filament-like morphology were not observed. To characterize the proteins more rigorously we employed SDD-AGE. The N-rich proteins had acquired an SDS-resistant aggregated state. The Q-rich proteins were largely SDS-sensitive (Figure S4A-D).
Figure 4. N-richness reduces proteotoxicity of Q/N-rich proteins.
(A) Single-copy plasmids coding for PrD-EYFP fusions were introduced into [RNQ+] cells. Expression was induced by addition of galactose for 48 hours and protein localization was determined by fluorescence microscopy. (B) Isogenic [rnq−] or [RNQ+] yeast bearing the indicated Sup35 PrD-EYFP variants were spotted as 5-fold serial dilutions to plates that either induced (galactose) or repressed (glucose). Growth on glucose established that equal cell densities were plated for each variant. Differences in growth on galactose indicate toxicity resulting from expression of the indicated protein. Duplicate transformants are shown. White dashed lines are provided only for clarity; comparisons are made between cells growing on the same plate. (C) As in (B), but with HttQ47- and HttN47-EYFP. See also Figure S4.
Disordered proteins tend to be toxic when over-expressed. Analyzing previously published data (Vavouri et al., 2009) we find that toxic proteins more commonly have strong enrichments for Qs than Ns (Figure S4E). To examine the toxicity of our variants, we transformed galactose-inducible constructs into cells carrying a chromosomal deletion of Sup35’s prion domain. This made the cells immune to a form of toxicity resulting from Sup35 sequestration. Cells were then spotted onto media that either induced or repressed expression of the variants. The Ure2, Lsm4, and Gal11 variants were not toxic (not shown). Of the Sup35 variants, Sup35Q and Sup35WT were mildly toxic, whereas Sup35N was benign (Figure 4B, left). Similarly, HttQ47 was toxic relative to HttN47 (Figures 4C and S4F).
To determine whether toxicity was enhanced or reduced by amyloid formation, we examined isogenic strains containing [RNQ+]. This amyloid-promoting factor decreased the toxicity of Sup35WT (Figure 4B) as well as both variants of Htt (Figures 4C and S4F), consistent with the hypothesis that the toxicity of non-amyloid conformers is suppressed by amyloid conversion. Notably, [RNQ+] did not affect the toxicity of Sup35Q (Figure 4B), a variant whose conversion to amyloid is not promoted by [RNQ+] (Figure S1F and data not shown).
Q-rich proteins preferentially form toxic non-amyloid conformers
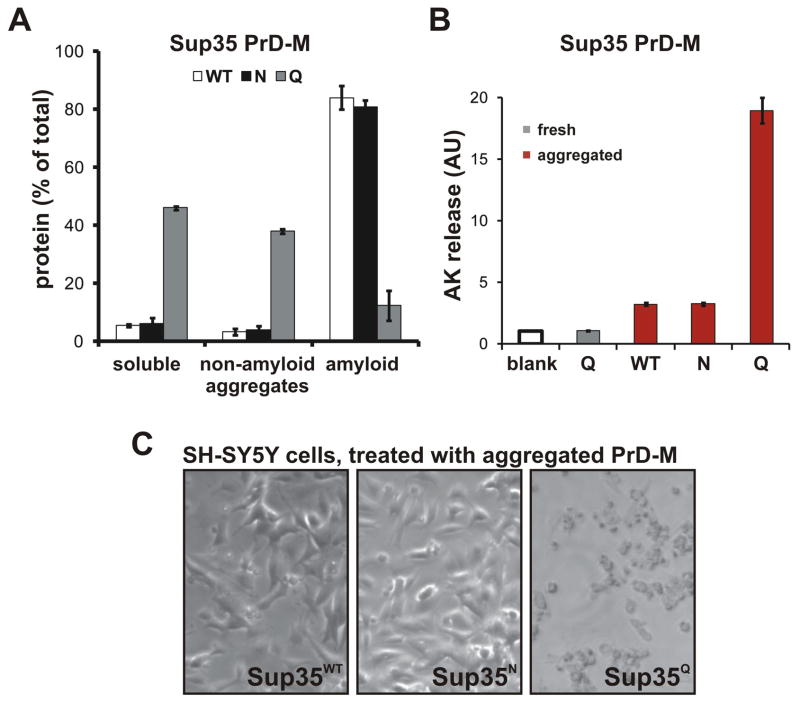
To investigate the inherent tendency of the Sup35 variants to partition between amyloid and non-amyloid states, we incubated purified proteins in assembly buffer for 24 hrs, with end-over-end agitation. After centrifugation to collect all aggregates, SDS was added, followed by a second centrifugation step to specifically collect amyoid. Sup35WT and Sup35N converted almost entirely to amyloids (Figure 5A). A large fraction of Sup35Q remained soluble and the fraction that did aggregate was mostly SDS-soluble.
Figure 5. Q-rich proteins preferentially form non-amyloid conformers.
(A) Quantitation of soluble, amyloid, and non-amyloid aggregated protein in assemblies of Sup35 PrD-M-His7 variants. Freshly diluted 5 μM solutions were induced to assemble with end-over-end agitation for 24 hrs. Soluble and aggregated fractions were partitioned by centrifugation at 39,000 rcf for 30 min. The aggregate fraction was further resuspended in 1 % SDS and allowed to incubate at 25°C for 30 min, followed by a second centrifugation step. Protein concentrations are shown (+/− SEM) for the original supernatant (“soluble”), post-SDS supernatant (“non-amyloid aggregation”) and post-SDS pellet (“amyloid aggregation”). (B–C) Toxicity of variant Sup35 PrD-M-His7 assemblies to human neuroblastoma cells. SH-SY5Y cells incubated for 15 hrs with 2.5 μM of either freshly diluted or pre-aggregated protein, as indicated, were visually inspected for cell detachment (B) or assayed for membrane disruption by adenylate kinase release (C). See also Figure S5.
To determine if conformers formed by the Sup35 variants are inherently toxic, we applied purified protein preparations to human neuroblastoma cells in culture. None were toxic when freshly diluted from denaturant (as shown for Sup35Q; Fig. 5B). When the proteins were allowed to aggregate for 24 hrs, Sup35Q became severely toxic, causing membrane permeabilization (quantified by adenylate kinase release; Figure 5B) and cell detachment (Figure 5C). Sup35N and Sup35WT became only mildly toxic. The extreme distinctions between Sup35 variants in this assay prompted us to examine the Ure2 PrD variants as well. After 24 hrs of aggregation, Ure2WT was only mildly toxic whereas Ure2Q was severely toxic (Figure S5).
Q-rich proteins have specific defects in amyloid conversion
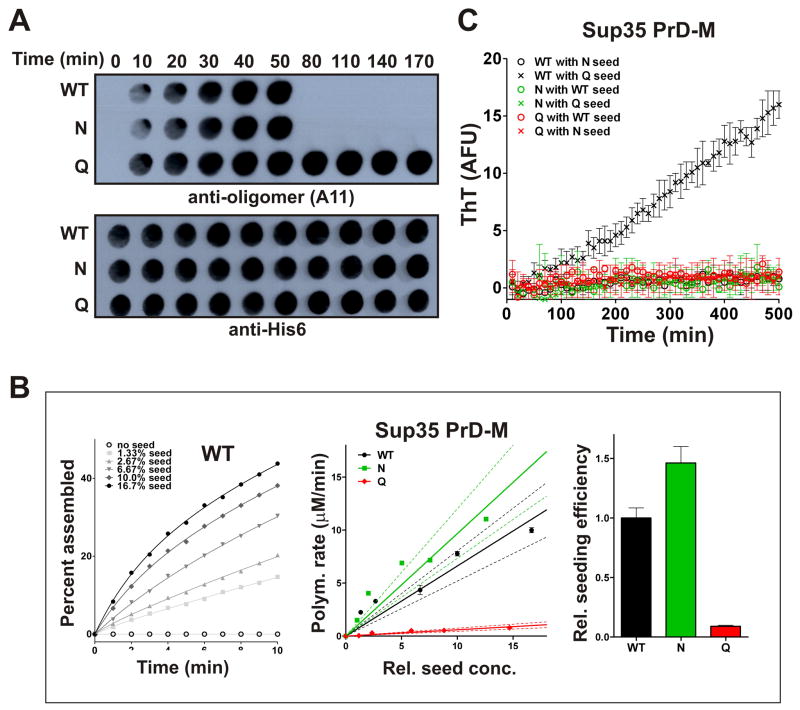
What aspects of amyloid formation govern the distinct behaviors of these proteins? Amyloid formation is a multistep process involving the formation of collapsed oligomeric intermediates, their conversion to amyloidogenic nuclei, and the polymerization of soluble protein onto those nuclei (Serio et al., 2000; Shorter and Lindquist, 2004; Kodali and Wetzel, 2007). The conformation-specific antibody (A11; Kayed et al., 2003) detects a toxic oligomeric species common to the polymeriztion of Aβ, α-Synuclein, and other disease-associated amyloidogenic proteins. It also recognizes an oligomer that is an obligate on-pathway species in Sup35 polymerization (Shorter and Lindquist, 2004). All three variants accumulated A11-reactive species (Figure 6A). Sup35Q formed these species more rapidly than Sup35WT or Sup35N and remained in this form much longer. Thus, Sup35Q is defective in the conversion of oligomeric intermediates into amyloids, providing a link between its defect in prion formation and its toxicity.
Figure 6. Q-rich proteins have reduced rates of conformational conversion to amyloid.
(A) Sup35 PrD-M-His7 variants were diluted to 2.5 μM in assembly buffer and incubated for the indicated times prior to the removal of 50 μl to a nitrocellulose membrane. Pre-amyloid oligomers (top) or total protein (bottom) were detected with A11 or anti-His6 antibodies respectively. (B) Sup35 PrD-M-His7 variants were diluted to 7.5 μM in assembly buffer containing ThT, followed immediately by the addition of various concentrations (% m/m) of the respective preformed sonicated amyloid fibers. Reactions were incubated without agitation and monitored for amyloid polymerization by ThT fluorescence. Nonlinear regression (as shown on left for WT, fit to one-phase association curves) was used to determine initial rates of amyloid elongation (as shown in middle, plotted against normalized seed concentrations). Dotted lines denote the 95% CI of the best fit line. Slopes of the best fit lines show the seeding efficiencies of each variant amyloid preparation, relative to WT (right). (C) The ability of individual variants to polymerize onto heterologous pre-assembled amyloids. 5 μM soluble protein was seeded with 10% (m/m) preformed aggregates in each case. Data show means +/− SEM. See also Figure S6.
Next we asked if Ns and Qs influence polymerization of soluble proteins onto their own preformed amyloid seeds. Each variant was incubated for one week in assembly buffer with agitation, which drove even Sup35Q into amyloid (Figure S1H). Amyloid fibers were then sonicated into similar sized fragments and normalized to contain approximately the same number of fiber ends (Figure S6A-B). Nonlinear regression of ThT fluorescence was used to determine initial polymerization rates of soluble protein across a range of added seed concentrations (Figure 6B, S6C). The rates of seeded polymerization differed dramatically between variants. Sup35N converted more rapidly than Sup35WT; Sup35Q converted much more slowly. The polymerization of Ure2 was altered in the same manner by Q substitutions (Figure S6A-D).
If seeded polymerization is driven by oligomers, however, these differences might simply reflect the inherently different oligomerization tendencies of the proteins. To investigate, we performed a complementary experiment: soluble proteins at different concentrations were seeded with a single concentration of pre-formed amyloid seeds. A linear relationship was observed in all cases. Because oligomer formation is concentration dependent, seeded amyloids must be assembling predominantly by monomer addition (Figure S6F-G). Thus, in addition to having different oligomerization properties, the variants differ in the rates at which their monomers polymerize onto preformed seeds.
Next, pre-formed sonicated amyloids of each of the Sup35 and Ure2 PrD variants were used to cross-seed amyloid formation by each of the other variants. In all but one case, cross-seeding was not observed, indicating that Ns and Qs generally create incompatible templates (Figure 6C, S6E). The single exception occurred between the pair of proteins with the greatest sequence identity: Sup35WT and Sup35Q. This relationship was asymmetric in a meaningful way. Sup35Q did not polymerize on Sup35WT seeds, but Sup35WT did polymerize on Sup35Q seeds. This indicates that Sup35Q amyloid seeds are competent for templating. The defect in seeded Sup35Q polymerization, therefore, appears to reflect a defect in the conformational conversion of soluble Sup35Q.
Towards a mechanistic distinction between Qs and Ns
Why does a subtle chemical distinction between N and Q side chains, namely, one methylene group, so strongly influence amyloid propensity? The conformational fluctuations that lead a disordered protein to convert to amyloid are difficult to dissect experimentally. Molecular simulations provide a tool for investigating the free energy landscapes and thermodynamics of β-sheet formation in such sequences (Wang et al., 2006; Vitalis et al., 2007; Pappu et al., 2008; Vitalis et al., 2008; Vitalis et al., 2009). To understand the intrinsic differences between Q- and N-rich sequences, without the confounding complexities imposed by different sequence contexts, we performed molecular simulations with polyQ and polyN molecules. For practical reasons we limited the simulations to molecules containing 30 glutamines (Q30) or 30 asparagines (N30). As in previous work (Vitalis et al. 2009), we performed two sets of simulations. In one set, we interrogated the unbiased free energy landscapes. In the second set, we imposed local conformational restraints to generate non-specific biases of the backbone dihedral angles in the β-basin of conformational space. The latter allowed us to observe rare conformations that might be sampled on-pathway to amyloid formation.
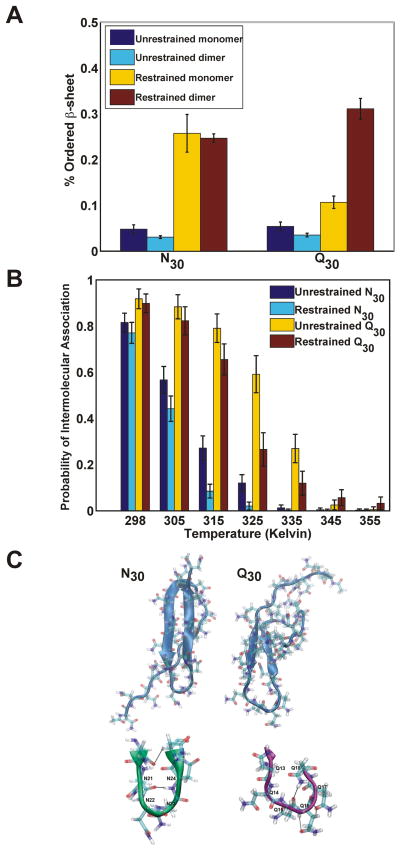
Conformational restraints allow us to design simulations where the entropic penalty is pre-paid equivalently for both Q30 and N30. Our analysis focused on two quantities, namely the degree of ordered intramolecular β-sheet formation, and the probability that a pair of conformationally biased molecules would self-associate. Figure 7A compares the degree of ordered β-sheet formation in N30 and Q30 in the presence and absence of conformational restraints. The extent of β-sheet formation was low for both N30 and Q30 in the absence of conformational restraints. However, when the entropic penalties for sampling the appropriate conformations were pre-paid, N30 monomers showed greatly increased ordered β-sheet formation; Q30 monomers were much less affected.
Figure 7. Molecular simulations of polyN (N30) and polyQ (Q30).
(A) Percentage of ordered β-sheet formed by N30 and Q30. Single N30 and Q30 molecules were simulated in the absence (dark blue) or presence (yellow) of local conformational restraints that restrict conformational sampling to dihedral angles drawn from the β-basin in conformational space. Pairs of N30 and Q30 molecules simulated with (cyan) or without (dark brown) local conformational restraints show the effects of homotypic intermolecular interactions on ordered βsheet content. Shown are means +/− SD from five simulations. (B) Temperature-dependent probabilities of realizing homotypic intermolecular associations, quantified as the probability that the intermolecular (center-of-mass to center-of-mass) distance between the pair of restrained / unrestrained N30 or Q30 molecules is ≤ 25Å (corresponding to less than 0.025% of the total volume available to the molecules in the simulation setup). Simulations were performed for pairs of N30 and Q30 molecules without (dark blue and yellow) and with (cyan and dark brown) local conformational restraints. Shown are means +/− SD from five simulations. (C) Visual comparison of ordered β-sheet structures formed by N30 (left) and Q30 (right) molecules in the presence of local conformational restraints. Note the tight type I β-turn formed by N30 relative to Q30, and the resulting differences in the lengths of intramolecular antiparallel β-sheets. When the entropic penalty is pre-paid using conformational restraints, we find a greater frequency of sampling intramolecular β-sheet structures with N30 because asparagine tracts can form canonical β-turns through backbone and sidechain hydrogen bonds, a representative of which is shown in the enlarged picture in green for N30. Conversely, Q-rich tracts form longer loops that lack any of the hallmarks of canonical turns and this increases the barrier for strand nucleation and propagation (Finkelstein, 1991).
Next, to simulate the effects of homotypic intermolecular interactions we used two N30 or two Q30 molecules. In simulations with two restrained Q30 molecules, there was positive coupling, and the overall β-sheet content of both Q30 molecules increased through intermolecular interactions (Figure 7A). This suggests that ordered β-sheet formation in such short Q-rich systems requires at least two interacting molecules (Zhang and Muthukumar, 2009) that have been appropriately biased to sample conformations drawn from the β-basin.
Next, we quantified the thermodynamics of bimolecular associations. The probability of intermolecular associations was smaller for N30 than for Q30 (Figure 7B). The intermolecular associations in such simulations are largely non-specific (Vitalis et al., 2008; Vitalis et al., 2009), i.e. spontaneous fluctuations lead disordered monomers to form disordered dimers. The presence of conformational restraints decreased this disorder and, in turn, systematically diminished intermolecular associations, an observation borne out by the temperature-dependence of these probabilities. The lower disorder of N30 and its increased ability to form ordered β-sheet structures (Figure 7A) led to weaker non-specific intermolecular associations. Q30 molecules showed a preference for increased conformational heterogeneity and hence an increased preference for non-specific associations.
These different association tendencies of Q30 and N30 appeared to result, at least in part, from a difference in turn formation between the two systems (Figure 7C). No more than four Ns were needed to form a tight turn. These were often canonical β-hairpin turns (Figure 7C) with characteristic intra-turn distances, backbone dihedral angles, and hydrogen bonding patterns. The bulkier side chain of Q in Q-rich tracts did not form tight turns but instead formed wider bulges and loops that required at least five (often more) residues to promote the reversal of chain direction.
DISCUSSION
A single methylene distinguishes Q from N. We find that this distinction profoundly alters one prominent activity of Q/N-rich proteins, prion formation, and influences another, toxicity. Changing Ns to Qs decreased prion formation and increased the accumulation of non-amyloid aggregates. These were toxic in the yeast cytoplasm and even more toxic when ectopically applied to cell lines of neuronal origin. In contrast, changing Qs to Ns enhanced prion formation and reduced toxicity. These observations were surprising, as the notion that Qs and Ns are equivalent for prion formation is pervasive (Si et al., 2003; Ross et al., 2005b; Decker et al., 2007; Patel et al., 2009; Salazar et al., 2010). Further, algorithms for identifying amyloidogenic sequences – TANGO (Fernandez-Escamilla et al., 2004) and Zyggregator (Tartaglia and Vendruscolo, 2008) – do not predict clear differences in the effects of our Q and N replacements (Figure S7), and do not predict amyloid formation by Q/N-rich PrDs (Alberti et al., 2009) or the PrD variants we analyzed here (Figure S7). Hopefully, the training sets created by our variants will lead to improved sequence-based predictions of amyloid formation.
Seeking a mechanistic explanation for our biological results, we asked how Qs and Ns affected various steps in both spontaneous amyloid assembly and templated assembly. The residues affected the specificity of templated and strongly altered the intrinsic efficiencies with which both the monomers and oligomers converted from the soluble disordered state to the amyloid. Molecular simulations suggest a possible rationale: the shorter N side chain enhances hydrogen bonding to the polypeptide backbone, increasing the formation of turns and β-sheets. We propose that this distinction is amplified as multiple monomers come together, allowing N-rich molecules to more effectively form ordered self-assemblies. Moreover, Ns reduced nonspecific interactions, which inhibit polymerization through off-pathway aggregation.
The disparity between our Q and N variants is likely the culmination of small contributions from many residues in the sequence, but local, contextual effects might also matter. For example, Ns (but not Qs) form hydrogen-bonded spines, or “asparagine-ladders”, in β-helix proteins (Jenkins and Pickersgill, 2001; Lenore Cowen, personal communication). The β-helix is a model structure for functional amyloids (Shewmaker et al., 2009) and fungal prions (Krishnan and Lindquist, 2005; Wasmer et al., 2008; Tessier and Lindquist, 2009; Dong et al., 2010). Moreover, Q stretches of sufficient length can overcome their intrinsically lower amyloid propensities, as occurs when a short segment of Sup35’s PrD, containing 13 Qs and 9 Ns, is replaced with a stretch of 62 Qs (Osherovich et al., 2004).
Recently, it was reported that many Q/N-rich proteins form coiled coils which might govern their aggregation (Fiumara et al., 2010). We note that Ns have a much lower coiled coil propensity than Qs – 0.25 vs. 0.99 using the Coils algorithm (Lupas et al., 1991) and 0.29 vs. 0.90 using Paircoil2 (McDonnell et al., 2006). A testable, unifying explanation for both our data and that of Fiumara arises: might a too-strong propensity to form coiled coils inhibit conversion to amyloid and favor the formation of toxic aggregates?
Many proteomes, including those of roundworms, insects, slime molds, and malarial parasites, contain an abundance of N-rich proteins (Michelitsch and Weissman, 2000; Harrison and Gerstein, 2003; Singh et al., 2004). It will be of great interest to determine whether these organisms take advantage of the functional, self-assembling properties of amyloids in their biology.
Other functions of Q/N rich proteins derive from the opposite conformational extreme they populate – disorder. Our Q and N variants are predicted to be highly disordered (9 of 10 disorder prediction web-servers reviewed in He et al., 2009). Yet, IDRs are typically enriched for Qs and depleted of Ns (Radivojac et al., 2007). Q-richness may be integral to the functions of dynamic protein assemblies: transcriptional regulatory complexes, RNA processing bodies and endocytic complexes (Xiao and Jeang, 1998; Titz et al., 2006; Decker et al., 2007; Meriin et al., 2007; Buchan et al., 2008; Fuxreiter et al., 2008; Alberti et al., 2009). The conformational heterogeneity of Q-rich polypeptides might expedite the assembly and remodeling of such complexes, and grant freedom to explore new binding partners, accelerating the functional diversification of network hubs and the evolution of novel circuitries.
These desirable properties come at a price, however. Conformational disorder is a burden for protein homeostasis, in part due to mass action-driven interaction promiscuity (Vavouri et al., 2009). This liability may drive the tightly regulated expression of proteins with IDRs in general and of “Q/N-rich” proteins in particular (Gsponer et al., 2008). Our computational and experimental analyses indicate that Qs, specifically, increase the propensity for toxic interactions by disordered proteins, which, in turn, may contribute to the pathology of Q-rich proteins in disease. Although more comparisons will be needed between proteins expressed in, and applied to, a variety of cell types and compartments, the toxicity of non-amyloid species seems to be related to multi-factorial effects on intracellular protein-protein interaction promiscuity and extracellular membrane permeabilizing activities. Both intracellularly and extracellularly, amyloid formation reduced toxicity, consistent with previous suggestions for protective roles for amyloids (Takahashi et al., 2008; Truant et al., 2008; Treusch et al., 2009). The genetic tractability of yeast prions provides a tool for investigating this very difficult problem.
The conformational transitions and protein::protein interactions of IDRs govern diverse biological processes, from regulatory networks to protein-misfolding diseases to protein-based inheritance. Further elucidating the conformational preferences of disordered proteins will be key to understanding their central roles in both normal biology and disease.
EXPERIMENTAL PROCEDURES
Cloning and gene synthesis
Cloning procedures were essentially performed as described previously (Alberti et al., 2009). Variant versions of PrDs and Huntingtin exon 1 were generated synthetically as described in the Supplemental Information.
Yeast techniques
Standard genetic manipulations, media conditions, fluorescence microscopy, and SDD-AGE were as described (Alberti et al., 2009). Details are in the supplement.
Protein purification
All proteins were expressed and purified from E. coli BL21-AI essentially as described (Alberti et al., 2009), using either pRH1 (for fusing a 7×His tag to the C-termini of Sup35 PrD-M, Ure2 PrD, and Lsm4 PrD variants) or pRH2 (for fusing a Sup35 M domain plus 7×His tag to the C-termini of Ure2 PrD variants). Details are in the supplement.
In vitro aggregation assays
Amyloid assembly reactions monitored by ThT fluorescence were performed in microplate format essentially as described (Alberti et al., 2009). Other aggregation experiments were performed in 1.5 ml Eppendorf tubes. Details are in the supplement.
Membrane disruption assay
Toxilight Bioassay kit measures leakage of adenylate kinase from the cells to the extracellular medium due to the loss of cell integrity (damage of plasma membrane). 2 × 105 SH-SY5Y cells were seeded in 24-well plates and grown overnight in a 1:1 mixture of DMEM and Ham’s F12 and 10% FBS. Fresh or pre-aggregated proteins of Sup35 PrD-M-His7 variants (2.5 μM) were prepared in serum-free medium and applied for 12–15 hrs. Cells were briefly spun at 800 rcf and 30 μl of the medium was carefully removed and used for the toxicity assay as recommended by the manufacturer.
Molecular simulations
Simulations were performed as described in the supplement.
Supplementary Material
Highlights.
N-rich proteins more efficiently form amyloids and prions than Q-rich proteins
N-richness reduces proteotoxicity relative to Q-richness
Molecular simulations suggest that N-richness decreases non-specific interactions
Acknowledgments
We thank members of the Lindquist lab for valuable discussions and comments on the manuscript, and Charles Glabe (U. C. Irvine) for providing the A11 polyclonal antibodies. We thank the G. Harold and Leila Y. Mathers Charitable Foundation and the NIH (grants GM025874 to SL and NS056114 to RVP) for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem. 2007;282:3004–3013. doi: 10.1074/jbc.M609597200. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dong J, Castro CE, Boyce MC, Lang MJ, Lindquist S. Optical trapping with high forces reveals unexpected behaviors of prion fibrils. Nat Struct Mol Biol. 2010;17:1422–1430. doi: 10.1038/nsmb.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueholm MS, Petersen SV, Sonderkaer M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, et al. Functional amyloid in Pseudomonas. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci U S A. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Finkelstein AV. Rate of beta-structure formation in polypeptides. Proteins. 1991;9:23–27. doi: 10.1002/prot.340090104. [DOI] [PubMed] [Google Scholar]
- Fiumara F, Fioriti L, Kandel ER, Hendrickson WA. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Grimminger V, Richter K, Imhof A, Buchner J, Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- Hammer ND, Wang X, McGuffie BA, Chapman MR. Amyloids: friend or foe? J Alzheimers Dis. 2008;13:407–419. doi: 10.3233/jad-2008-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- Jenkins J, Pickersgill R. The architecture of parallel beta-helices and related folds. Prog Biophys Mol Biol. 2001;77:111–175. doi: 10.1016/s0079-6107(01)00013-x. [DOI] [PubMed] [Google Scholar]
- Kawai-Noma S, Pack CG, Kojidani T, Asakawa H, Hiraoka Y, Kinjo M, Haraguchi T, Taguchi H, Hirata A. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J Cell Biol. 2010;190:223–231. doi: 10.1083/jcb.201002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci U S A. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, Alexandrov IM, Salnikova AB, Ter-Avanesian MD, Chernoff YO, Sherman MY. Endocytosis machinery is involved in aggregation of proteins with expanded polyglutamine domains. FASEB J. 2007;21:1915–1925. doi: 10.1096/fj.06-6878com. [DOI] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu RV, Wang X, Vitalis A, Crick SL. A polymer physics perspective on driving forces and mechanisms for protein aggregation. Arch Biochem Biophys. 2008;469:132–141. doi: 10.1016/j.abb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009 doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques. Proc Natl Acad Sci U S A. 2002;99:5596–5600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Windle AH. Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature. 2001;412:143–144. doi: 10.1038/35084141. [DOI] [PubMed] [Google Scholar]
- Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured? Biochemistry. 2005;44:321–328. doi: 10.1021/bi047964d. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci U S A. 2005a;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005b;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- Salazar AM, Silverman EJ, Menon KP, Zinn K. Regulation of synaptic Pumilio function by an aggregation-prone domain. J Neurosci. 2010;30:515–522. doi: 10.1523/JNEUROSCI.2523-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, Wickner RB. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Singh GP, Chandra BR, Bhattacharya A, Akhouri RR, Singh SK, Sharma A. Hyper-expansion of asparagines correlates with an abundance of proteins with prion-like domains in Plasmodium falciparum. Mol Biochem Parasitol. 2004;137:307–319. doi: 10.1016/j.molbiopara.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Vendruscolo M. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev. 2008;37:1395–1401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Thomas S, Rajagopala SV, Chiba T, Ito T, Uetz P. Transcriptional activators in yeast. Nucleic Acids Res. 2006;34:955–967. doi: 10.1093/nar/gkj493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Cyr DM, Lindquist S. Amyloid deposits: protection against toxic protein species? Cell Cycle. 2009;8:1668–1674. doi: 10.4161/cc.8.11.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington’s disease: revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS J. 2008;275:4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Turoverov KK, Kuznetsova IM, Uversky VN. The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog Biophys Mol Biol. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci U S A. 2010;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Amyloidogenesis of natively unfolded proteins. Curr Alzheimer Res. 2008;5:260–287. doi: 10.2174/156720508784533312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Vitalis A, Lyle N, Pappu RV. Thermodynamics of beta-sheet formation in polyglutamine. Biophys J. 2009;97:303–311. doi: 10.1016/j.bpj.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A, Wang X, Pappu RV. Quantitative characterization of intrinsic disorder in polyglutamine: insights from analysis based on polymer theories. Biophys J. 2007;93:1923–1937. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A, Wang X, Pappu RV. Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization. J Mol Biol. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RH, Murphy RM. Examining polyglutamine peptide length: a connection between collapsed conformations and increased aggregation. J Mol Biol. 2009;393:978–992. doi: 10.1016/j.jmb.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins. 2006;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- Weathers EA, Paulaitis ME, Woolf TB, Hoh JH. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 2004;576:348–352. doi: 10.1016/j.febslet.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Williamson TE, Vitalis A, Crick SL, Pappu RV. Modulation of polyglutamine conformations and dimer formation by the N-terminus of huntingtin. J Mol Biol. 2010;396:1295–1309. doi: 10.1016/j.jmb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jeang KT. Glutamine-rich domains activate transcription in yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:22873–22876. doi: 10.1074/jbc.273.36.22873. [DOI] [PubMed] [Google Scholar]
- Zhang J, Muthukumar M. Simulations of nucleation and elongation of amyloid fibrils. J Chem Phys. 2009;130:035102. doi: 10.1063/1.3050295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.