Abstract
The analysis of a human thyroid serial analysis of gene expression (SAGE) library shows the presence of an abundant SAGE tag corresponding to the mRNA of thyroglobulin (TG). Additional, less abundant tags are present that can not be linked to any other known gene, but show considerable homology to the wild-type TG tag. To determine whether these tags represent TG mRNA molecules with alternative cleavage, 3′-RACE clones were sequenced. The results show that the three putative TG SAGE tags can be attributed to TG transcripts and reflect the use of alternative polyadenylation cleavage sites downstream of a single polyadenylation signal in vivo. By screening more than 300 000 sequences corresponding to human, mouse and rat transcripts for this phenomenon we show that a considerable percentage of mRNA transcripts (44% human, 22% mouse and 22% rat) show cleavage site heterogeneity. When analyzing SAGE-generated expression data, this phenomenon should be considered, since, according to our calculations, 2.8% of human transcripts show two or more different SAGE tags corresponding to a single gene because of alternative cleavage site selection. Both experimental and in silico data show that the selection of the specific cleavage site for poly(A) addition using a given polyadenylation signal is more variable than was previously thought.
INTRODUCTION
Polyadenylation of eukaryotic mRNA is characterized by the cleavage of the precursor-RNA and the addition of a poly(A) tail. Polyadenylation of a primary RNA transcript is suggested to have a function in RNA metabolism such as export of the mature mRNA, stability and recognition by ribosomes. The signals that determine the site of polyadenylation have been studied extensively in in vitro experiments. The upstream element AAUAAA, together with a downstream U/GU-rich element, is present in nearly all eukaryotic pre-RNAs and is proposed to regulate the complex machinery of proteins necessary to complete 3′-processing (reviewed in 1–4). An extensive in silico analysis of 3′ processing control signals showed a general similarity to published experimental findings (5). The site of cleavage in the pre-RNA is determined by the position of the regulatory elements together with the nucleotide composition of the cleavage region. This region is located 11–24 nt downstream from a AAUAAA element and 10–30 nt upstream from a U/GU-rich element. The suggested preferred nucleotide at the cleavage site is ordered A>U>C>>G (6). Although not emphasized in the literature, in general it is assumed that in any given pre-RNA sequence, there is one preferred cleavage site used by the polyadenylation machinery, although mutational analysis has shown that in in vitro cleavage experiments, neighboring nucleotides can be used (7,8).
Many studies have investigated the use of alternative polyadenylation signals in 3′ RNA processing. Different polyadenylation signals in the 3′ untranslated region of pre-RNA molecules can be used to generate different mRNA transcripts (9). In sequence databases (e.g. GenBank, Unigene), mRNA reference sequences are represented by the most abundant transcript from every gene since this transcript will have been preferentially cloned. When looking at a large number of 3′ expressed sequence tag sequences (ESTs) corresponding to these genes differences can be often found which result from alternatively spliced or polyadenylated mRNA sequences. A genome-wide study showed that 29% of human mRNAs displayed two or more polyadenylation signals that were able to start polyadenylation (10). However, the selection of the actual nucleotide at which the pre-RNA is cleaved using a given polyadenylation signal has, thus far, not been addressed in the literature. In this study we have focussed exclusively on alternative polyadenylation as a result of heterogeneous cleavage downstream from a single polyadenylation hexanucleotide signal. We therefore term this event ‘alternative cleavage site selection’ to avoid confusion with alternative polyadenylation caused by the usage of different polyadenylation hexanucleotide signals.
When analyzing the data from a human thyroid expression profile generated using a serial analysis of gene expression (SAGE) library (11) we found different SAGE tags corresponding to alternatively polyadenylated thyroglobulin (TG) mRNA molecules. The mRNA sequences found here have different polyadenylation cleavage sites downstream from a single polyadenylation signal. In the analysis of growing amounts of SAGE-generated expression data this effect could mean that if a SAGE tag sequence is located in the cleavage region of a mRNA transcript the resulting tag corresponding to this transcript can be heterogeneous. We studied the incidence of this phenomenon in silico in three mammalian genomes (human, mouse and rat) by screening their corresponding Unigene databases. Unigene provides a unique data source for obtaining information about variation in cleavage sites. Unigene clusters consist of grouped GenBank sequences that belong to the same gene and which therefore also include the sequences of alternative cleavage sites. Consequently, analysis of the sequences in such cluster reveals possible cleavage site variation for that particular gene. In addition, we determined whether alternative cleavage sites could be accounted for in vivo by sequencing 3′-RACE clones. The implications for analyzing SAGE-generated expression data are discussed.
MATERIALS AND METHODS
SAGE library
The expression library was constructed from human normal thyroid tissue according to an original protocol (12) and as described previously (11).
3′-RACE–PCR
First-strand RACE cDNA was synthesized from the same thyroid mRNA as used in the SAGE library. Oligo-dT10–18 with a 3′ linker 5′-GCATGCCAGAATTCTGGATCC was used as a primer (13). The linker sequence was used as a reverse priming site in subsequent amplification of the TG 3′ region. TG-specific forward primer TG-2F 5′-GAGAAGATCTCCTAAGCCTC was designed according to the TG cDNA sequence (GenBank accession no. U93033) generating a PCR product of ∼200 bp. PCR amplification was performed using standard conditions, with 2 mM MgCl2 and 30 cycles of 95°C for 1 min, 55°C for 1 min and 72°C for 1 min.
Cloning of 3′-RACE TG fragments
TG 3′-RACE–PCR fragments were cloned using the pGEM/T-easy vector (Promega). A ligation reaction was transformed into Escherichia coli (DH5α) cells and plated. DNA from TG-positive clones was isolated using the Wizard Mini-Prep kit (Promega).
Amplification of TG exon 48
Genomic DNA was isolated from the same thyroid tissue as was used for the 3′-RACE experiments using Trizol (Gibco BRL) according to the manufacturer’s protocol. An aliquot of 100 ng DNA was used in a PCR amplification of exon 48 using intronlocated primers with M13 linkers attached. TG48-for, 5′-AGAGAAGTCCTAATCTGGCTTG; TG48-rev, 5′-CTGGTGCATAACAGATGCTCAT (GenBank accession no. AF080484).
Sequencing
TG 3′-RACE clones and the exon 48 PCR product were sequenced with the Dyenamic Direct cycle sequencing kit (Perkin Elmer) using M13 forward and reverse priming sites in the vector or PCR fragment. Samples were run on an ABI377XL Automatic Sequencer (Perkin Elmer) and analyzed using Sequence Analysis 3.0 software.
Sequence screening
For the in silico determination of alternative polyadenylation cleavage sites the Unigene databases for human, mouse and rat were used. Every sequence in a cluster was scanned for a poly(A) stretch and a poly(A) signal in the 5′–3′ (+) or 3′–5′ (–) orientation. Sequences that contained a poly(A) stretch with a minimum length of 5 nt and included the AATAAA or ATTAAA signal within 50 bp upstream were selected for further analysis. Unigene clusters containing less than four 3′ EST sequences were excluded because the following analysis needs at least four eligible sequences to be able to calculate a realistic result. For all selected sequences of a particular Unigene cluster the partial sequences between the poly(A) signal and start of the poly(A) stretch were aligned to identify alternative cleavage sites. Since the length of these partial sequences was only ∼18 nt on average, we allowed one mismatch during the pair-wise alignment of these sequences to account for sequencing errors. In the case of two or more mismatches, the sequence that was less frequent was removed from the set. Multiple mismatches in sequence alignments easily occur as a result of ESTs that were incorrectly classified in a Unigene cluster. Finally, from the remaining set of partial sequences, sequences that occurred only once were discarded for this set to avoid scoring of sequence errors. From the remaining sequences the number of alternative cleavage sites were counted.
RESULTS
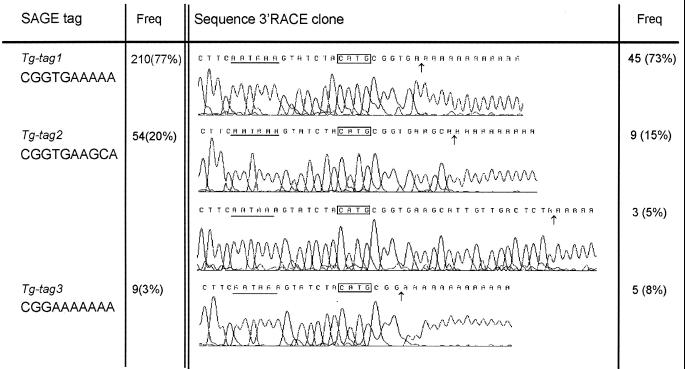
In the thyroid SAGE data (11) the sequence tag corresponding to the thyroid-specific mRNA TG presents itself as 5′-CGGTGAAAAA and is scored 210 times out of a total number of 10 994 sequenced tags. This tag partly spans the poly(A) tail. Two tags not corresponding to any known human mRNA, 5′-CGGTGAAGCA and 5′-CGGAAAAAAA, are present 54 and nine times, respectively. Because of the similarity of these tags to the most abundant (wild-type) SAGE tag we opted to clone TG 3′-RACE fragments to determine whether these tags correspond to TG mRNA. As shown in Figure 1, sequencing of 62 TG 3′-RACE clones resulted in four different polyadenylation sites. Forty-five sites corresponded to the wild-type TG mRNA, i.e. 5′-CGGTGAAAAA (TG-tag 1), nine clones had a polyadenylation site 4 bp downstream and corresponded to 5′-CGGTGAAGCA (TG-tag 2) and five clones showed a polyadenylation site 3 bp upstream, 5′-CGGAAAAAAA (TG-tag 3). Additionally, three TG cDNA clones with a polyadenylation site 16 bp downstream of the wild-type cleavage site were present, showing a fourth polyadenylation variant that is also represented by TG-tag 2. The extra 16 bp TG 3′-UTR in these three clones correspond to the sequence as published for the last TG coding exon (14). The 150 bp sequence upstream of the polyadenylation site was identical for all 62 clones and to the TG genomic sequence showing no additional polyadenylation signal sequences. To exclude any mutations in the pre-RNA sequence downstream from the poly(A) site putatively responsible for the variance in polyadenylation, the last coding exon of TG from the thyroid tissue used in these experiments was sequenced. The genomic sequence of TG in this particular thyroid gland is identical to the published TG exon 48 sequence (Fig. 2). Within this sequence the relevant regulating sequences have been indicated as well as the four different polyadenylation cleavage sites in human TG mRNA.
Figure 1.
Alternative cleavage sites in TG mRNA. Results are summarized from SAGE and 3′-RACE. Sequence for each variant is shown together with their relative abundance. SAGE-tag frequency is out of a total of 10 994 tags, 3′-RACE-clone frequency is out of a total of 62 clones.
Figure 2.
Genomic sequence (accession no. AF080484) of the last coding exon (48) of the human TG gene. Consensus regulating elements are indicated as well as the four described cleavage sites. Small font is coding sequence, large font represents pre-RNA containing regulating polyadenylation signals.
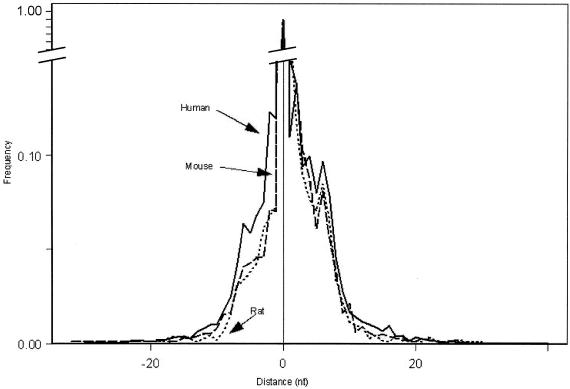
The in silico screening using Unigene databases of human, rat and mouse resulted in 302 975 sequences which passed the criteria out of a total of 3 037 962 sequences in the database. The selected sequences were represented in 9625 human Unigene clusters, 4424 mouse Unigene clusters and 5092 rat Unigene clusters. The number of alternative cleavage sites in these sets of genes are summarized in Table 1. The percentage of genes that encode mRNA molecules using more than one cleavage site in the pre-RNA sequence downstream from a single polyadenylation signal was 44% for human genes, 22% for mouse genes and 22% for rat genes. From these genes the majority used only one alternative site and the maximum amount of alternative cleavage sites used in a single gene for human, mouse and rat was nine, six and six, respectively. As a control we looked at the results of our in silico analysis in the TG Unigene cluster. The two most frequent variants out of the four that we scored by sequencing 3′-RACE clones are accounted for by this method. To get an impression of the region of the pre-RNA sequence in which this effect takes place the distance of the alternative cleavage sites relative to the most abundant and therefore arbitrarily called wild-type cleavage site was calculated. The distance was divided into upstream and downstream nucleotides. The maximal distance was 32 nt from the wild-type cleavage site, while the alternative sites were mostly in the immediate neighborhood (Fig. 3). In human, mouse and rat the majority of alternative cleavage sites were located between 8 nt upstream and 9 nt downstream of the wild-type cleavage site. The number of downstream cleavage sites was higher than that of upstream cleavage sites in all three organisms. For human, mouse and rat the percentages up/downstream were 43/57%, 34/66% and 33/67%, respectively. To investigate how often alternative cleavage sites can cause problems in the analysis of SAGE data, the number of SAGE tags located in an heterogeneous 3′ polyadenylation area were counted. In 267 out of a total of 9625 human genes, alternative SAGE tags resulting from alternative cleavage were distinguished. This result means that 2.8% of all human genes screened are represented by at least two SAGE tags. For mouse and rat these percentages were 1.3 and 1.2%, respectively. The lower percentages for mouse and rat are in concordance with the lower percentage of cleavage site heterogeneity in these organisms.
Table 1. Number of cleavage sites as scored in the screening of sequences in human, mouse and rat Unigene databases.
| Number of cleavage sites per gene | ||||||||||
| |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
Total |
| Human |
5430 |
2754 |
963 |
326 |
115 |
26 |
14 |
5 |
4 |
9625 |
| Mouse |
3438 |
824 |
127 |
23 |
8 |
4 |
– |
– |
– |
4424 |
| Rat | 3982 | 953 | 122 | 27 | 7 | 1 | – | – | – | 5092 |
The table shows the number of genes with their respective number of cleavage sites as used in the 3′-UTR sequence downstream from a single polyadenylation signal.
Figure 3.
Distance of alternative cleavage sites relative to wild-type cleavage site in Unigene clusters with alternative polyadenylation. The distance is divided into the number of upstream (–) and downstream (+) nucleotides (x-axis). The most abundant cleavage site is used as a reference and is depicted as 0. Frequency is given on the y-axis. Arrows indicate human, mouse and rat data.
DISCUSSION
The results from the cloning and sequencing of human TG 3′-RACE fragments (Fig. 1) prove the in vivo existence of mRNA transcripts with alternative cleavage sites. In cloning 3′-RACE fragments, a representation of the TG mRNA pool is found concerning the region of interest, namely the polyadenylation region. The abundance of the four different TG mRNA molecules is similar to the abundance of the respective tags in the SAGE expression library. After examination of the downstream pre-RNA sequence present in the genomic sequence of the last TG exon 48, a consensus pattern concerning the wild-type polyadenylation (6) could be found (Fig. 2). The AAUAAA signal was present and for the three most abundant polyadenylation variants the distance to the poly(A) site was within the defined consensus distance (11–24 bp). As for the infrequent fourth variant, its low occurrence may be explained by the 28 bp distance to the poly(A) site. Following consensus downstream of the cleavage site a putative GU-rich site was present 18–23 bp downstream of the TG wild-type poly(A) site, while another was present between 39–44 bp from the wild-type poly(A) site. It seems that the first putative GU-rich region (TGTGTG) would be the most likely candidate responsible for the three most abundant cleavage sites, since the distance to this GU-rich region is 13–20 bp. The fourth variant is only 6 bp from this region and probably too close. It may use the second putative GU-rich region located 27 bp from its cleavage site. This second GU-rich region may have a lower preference for the polyadenylation machinery since the expression of this last variant is lower compared to the three most abundant poly(A) sites probably using the most upstream GU-rich region.
The results from the Unigene sequence database screen for variation in cleavage site selection indicate that a considerable percentage of mammalian transcripts (human 44%, mouse 22% and rat 22%) are present in two or more variants reflecting alternative cleavage sites used for polyadenylation. The numbers in this study are probably underestimated because of the strict selection criteria imposed on eligible sequence and Unigene clusters. This very strict clone selection scheme avoids too many false positives in the analysis but simultaneously increases the number of false negatives. If we look at the TG Unigene cluster, two out of four variants were scored in our analysis, because not enough sequences from the less abundant variants were available. We therefore estimate the actual percentage of alternative cleavage site selection to be even higher. There is a distinct difference in the percentage of cleavage site heterogeneity between human mRNA on the one hand and rat/mouse mRNA on the other (44 %versus 22% and 22%). A possible explanation for this could be that there is a selection on the more abundant of the mRNAs. Because of the strict selection criteria in the genome screen, all alternative cleavage sites that were scored in only one sequence were discarded to avoid counting sequencing errors. Gene clusters with many 3′ EST sequences would therefore be scored for alternative cleavage sooner than gene clusters with a low number of 3′ sequences. Since the number of human 3′ EST sequences in GenBank is considerably larger than that of mouse or rat this could explain the difference.
The implications for analyzing SAGE generated data are obvious. SAGE tags that are generated near to the region of heterogeneous cleavage can show the same heterogeneity. In our analysis, 2.8% of human transcripts show two or more alternative SAGE tags. When analyzing a typical SAGE library containing 20 000 unique tags this means that about 550 tags will correspond to transcripts with alternatively cleaved mRNA molecules. Because this phenomenon is especially prone to show up in abundant tags, SAGE data will have to be screened for this.
The mechanism behind polyadenylation and its biological implications has been the subject of many studies. Most studied have focussed, however, on alternative polyadenylation using different polyadenylation signals. This form of alternative polyadenylation can even be associated with regulatory properties (9). The phenomenon of alternative cleavage site selection using a single polyadenylation signal is not emphasized in the literature. It is generally assumed that this is highly heterogeneous in yeast (N.J.Proudfoot, personal communication). In yeast, in contrast to mammalian polyadenylation, there is a greater lag-time between cleavage and poly(A) addition giving 3′–5′ exonucleases the potential to remove nucleotides from the 5′ cleavage product, thus producing different polyadenylated transcripts (3). In our study, we have shown that there is a higher percentage of alternative cleavage downstream than upstream of the wild-type cleavage site. This indicates that differences in mammalian mRNA polyadenylation cleavage sites can not be attributed to exonucleotic degeneration of the uncapped 5′ pre-RNA molecule. A recent study in yeast polyadenylation identified a novel factor (Nab4p/Hrp1p) that seems to control the cleavage site selection process (15). If the specific cleavage site used by the polyadenylation machinery to add the poly(A) tail is regulated by a specific factor in yeast, considering the homology between yeast and mammalian polyadenylation, this may be expected to happen in mammalian cells as well. From our results, however, it seems that a considerable part of mammalian mRNA transcripts are not subject to strict regulation of cleavage site selection by any protein.
In summary, it seems that there is considerably more variation in cleavage site selection in mammalian mRNA transcripts variable than until now was assumed. This has implications for the analysis of SAGE expression data.
Acknowledgments
ACKNOWLEDGEMENT
We thank Joost de Gast for writing part of the software used for the screening of the Unigene databases.
DDBJ/EMBL/GenBank accession nos U93033 and AF080484
References
- 1.Wahle E. (1995) 3′End cleavage and polyadenylation of mRNA precursors. Biochim. Biophys. Acta, 1261, 183–194. [DOI] [PubMed] [Google Scholar]
- 2.Colgan D.F. and Manley,J.L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 3.Wahle E. and Ruegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA, 96, 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F., MacDonald,C.C. and Wilusz,J. (1995) Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res., 23, 2614–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheets M.D., Ogg,S.C. and Wickens,M.P. (1990) Point mutation in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res., 18, 5799–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreira A., Takagaki,Y., Brackenridge,S., Wollerton,M., Manley,J.L. and Proudfoot,N.J. (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms Genes Dev., 12, 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaudoing E., Freier,S., Wyatt,J.R., Claverie,J.M. and Gautheret,D. (2000) Patterns of variant polyadenylation signal usage human genes. Genome Res., 10, 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauws E., Moreno,J.C., Tijssen,M., Baas,F., deVijlder,J.J.M. and Ris-Stalpers,C. (2000) Serial analysis of gene expression (SAGE) as a tool to assess the expression profile of the human thyroid and to identify novel thyroidal genes. J. Clin. Endocrinol. Metab., 85, 1923–1927. [DOI] [PubMed] [Google Scholar]
- 12.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 13.Frohman M.A., Dush,M.K. and Martin,G.R. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA, 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendive F.M., Rivolta,C.M., Vassart,G. and Targovnik,H.M. (1999) Genomic organization of the 39 region of the human thyroglobulin gene. Thyroid, 9, 903–912. [DOI] [PubMed] [Google Scholar]
- 15.Minvielle-Sebastia L., Beyer,K., Krecic,A.M., Hector,R.E., Swanson,M.S. and Keller,W. (1998) Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J., 17, 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]